Abstract
APOL1 gene renal-risk variants are associated with nephropathy and CVD in African Americans; however, little is known about the circulating APOL1 variant proteins which reportedly bind to HDL. We examined whether APOL1 G1 and G2 renal-risk variant serum concentrations or lipoprotein distributions differed from nonrisk G0 APOL1 in African Americans without nephropathy. Serum APOL1 protein concentrations were similar regardless of APOL1 genotype. In addition, serum APOL1 protein was bound to protein complexes in two nonoverlapping peaks, herein referred to as APOL1 complex A (12.2 nm diameter) and complex B (20.0 nm diameter). Neither of these protein complexes associated with HDL or LDL. Proteomic analysis revealed that complex A was composed of APOA1, haptoglobin-related protein (HPR), and complement C3, whereas complex B contained APOA1, HPR, IgM, and fibronectin. Serum HPR was less abundant on complex B in individuals with G1 and G2 renal-risk variant genotypes, relative to G0 (P = 0.0002–0.037). These circulating complexes may play roles in HDL metabolism and susceptibility to CVD.
Keywords: apolipoprotein L1, apolipoproteins, high density lipoprotein, kidney, proteomics, renal disease
A major breakthrough in human molecular genetics was identification of the powerful contribution of APOL1 gene G1 and G2 renal-risk variants to nondiabetic nephropathy susceptibility in African Americans (1). HIV-associated nephropathy, idiopathic focal segmental glomerulosclerosis, severe lupus nephritis, sickle cell nephropathy, and hypertension-attributed nephropathy strongly associate with APOL1 G1 and G2 renal-risk variants on chromosome 22q13.1 (1–3). Although APOL1 mRNA and APOL1 protein are present in human kidney (4, 5), the major APOL1 reservoir appears to be circulating protein (5, 6). APOL1 was initially discovered as a minor apolipoprotein of plasma HDLs (6); however, its distribution among HDL subfractions has not been well-defined. APOL1 nephropathy variants associate with HDL subfraction concentrations (7) and CVD risk, although controversial results have been reported with CVD (8–11). Trypanosome lytic factors (TLF1 and TLF2) contain APOL1 protein (12) and are minor HDL subfractions in humans that contribute to innate immunity via protection from infection, including from African trypanosomes (13).
Association was not observed between plasma APOL1 concentrations and APOL1 genotype in African Americans with treated HIV infection; plasma APOL1 levels also did not associate with the risk of HIV-associated nephropathy or chronic kidney disease (14). Whether serum APOL1 protein levels and their distribution among HDL particles are associated with APOL1 genotypes in healthy individuals is unknown. Because free APOL1 protein may be taken up by podocytes in vitro (5), it remains critical to determine whether serum APOL1 concentrations or the structure/composition of variant APOL1 proteins and their associated complexes are specific to the G1 and G2 renal-risk variants, relative to nonrisk G0.
To address these issues, serum APOL1 protein levels and size distribution were examined in age- and gender-matched African Americans without kidney disease based on APOL1 genotype using fast protein LC (FPLC) and immunoblot analysis. Proteomics analysis was performed to compare the composition of APOL1 protein-containing complexes. These results provide novel information on APOL1 genotype-specific circulating APOL1 protein and multiprotein complexes potentially involved in human CVD and HDL metabolism.
MATERIALS AND METHODS
Study subjects
Eighty-four unrelated healthy African Americans without nephropathy (estimated glomerular filtration rate >60 ml/min/1.73 m2 and urine albumin:creatinine ratio <30 mg/g) were selected from Natural History of APOL1-Associated Nephropathy (NHAAN) participants. These participants had first degree relatives with nondiabetic etiologies of end-stage kidney disease. Among them (42 female/42 male), mean ± SD age was 42.5 ± 13.8 years in females and 43.2 ± 13.4 years in males. Serum creatinine concentration (mean ± SD) was 0.79 ± 0.15 mg/dl in females and 1.05 ± 0.13 mg/dl in males. Seven men and seven women were included in each potential genotypic group, G0/G0, G1/G0, G2/G0, G1/G1, G2/G2, and G1/G2. Significant differences were not observed for age or kidney function with respect to APOL1 genotype. The Institutional Review Board at the Wake Forest School of Medicine approved the study and all participants provided written informed consent. Descriptive clinical data of the participants are provided in Table 1.
TABLE 1.
Serum APOL1 levels and other clinical data by APOL1 genotype in 84 African American subjects without kidney disease
| All | G0/G0 | G1/G0 | G2/G0 | G1/G2 | G1/G1 | G2/G2 | P | |
| N (M/F) | 84 (42/42) | 14 (7/7) | 14 (7/7) | 14 (7/7) | 14 (7/7) | 14 (7/7) | 14 (7/7) | 1.000 |
| Age (years) | 42.8 ± 13.5 | 42.5 ± 12.5 | 42.0 ± 13.6 | 43.5 ± 13.4 | 42.6 ± 14.0 | 42.3 ± 14.2 | 44.1 ± 15.7 | 0.999 |
| African ancestry (%) | 0.795 ± 0.089 | 0.773 ± 0.069 | 0.787 ± 0.119 | 0.795 ± 0.067 | 0.816 ± 0.096 | 0.796 ± 0.071 | 0.805 ± 0.108 | 0.864 |
| Creatinine (mg/dl) | 0.920 ± 0.192 | 0.934 ± 0.207 | 0.900 ± 0.254 | 0.861 ± 0.183 | 0.923 ± 0.133 | 0.988 ± 0.164 | 0.912 ± 0.200 | 0.256 |
| Cystatin C (mg/l) | 0.709 ± 0.124 | 0.666 ± 0.131 | 0.711 ± 0.110 | 0.689 ± 0.118 | 0.679 ± 0.129 | 0.803 ± 0.133 | 0.707 ± 0.090 | 0.060 |
| Urine Alb:Creat (mg/g) | 6.30 ± 5.71 | 4.66 ± 4.47 | 5.04 ± 5.27 | 6.11 ± 5.18 | 6.44 ± 4.01 | 8.59 ± 7.55 | 6.96 ± 7.04 | 0.344 |
| APOL1 (μg/ml) | 20.5 ± 30.8 | 16.2 ± 13.0 | 25.0 ± 24.9 | 15.6 ± 22.1 | 17.7 ± 11.9 | 32.6 ± 64.5 | 15.8 ± 15.5 | 0.723 |
| Triglycerides (mg/dl) | 99.4 ± 54.5 | 101.3 ± 77.0 | 93.8 ± 50.0 | 110.6 ± 66.2 | 86.0 ± 34.1 | 103.6 ± 54.9 | 100.9 ± 39.4 | 0.881 |
| TC (mg/dl) | 183.1 ± 36.5 | 179.5 ± 49.5 | 185.8 ± 35.0 | 186.8 ± 36.5 | 182.1 ± 29.9 | 175.6 ± 23.7 | 189.3 ± 43.5 | 0.945 |
| HDL (mg/dl) | 50.7 ± 11.1 | 46.9 ± 10.5 | 52.7 ± 13.1 | 53.0 ± 10.8 | 51.1 ± 9.19 | 52.5 ± 11.9 | 48.0 ± 11.2 | 0.532 |
| LDL (mg/dl) | 111.5 ± 32.1 | 111.4 ± 41.7 | 115.7 ± 34.8 | 109.2 ± 30.5 | 110.4 ± 22.6 | 102.3 ± 25.3 | 120.2 ± 37.0 | 0.864 |
| VLDL (mg/dl) | 19.9 ± 10.8 | 20.3 ± 15.4 | 18.7 ± 9.91 | 22.2 ± 13.2 | 17.3 ± 6.80 | 20.7 ± 10.9 | 20.1 ± 7.80 | 0.875 |
P values were determined by ANOVA, representing the difference across APOL1 genotypes. P values were calculated for log transformed serum creatinine, cystatin C, APOL1, triglycerides, TC, HDL, LDL, VLDL, and urine albumin/creatinine ratio (Urine Alb:Creat) due to the skewness of raw data from normal distribution. The P value for serum creatinine level by APOL1 genotype was additionally adjusted for gender. Data are presented as the mean ± SD.
Measurement of serum APOL1 concentration
One microliter of sera from the 84 study participants was separated by 4–20% SDS-PAGE, transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA), and blocked for 1 h at room temperature with TBS containing 1% skim milk powder and 0.1% Tween 20 (TBST). Blots were incubated overnight at 4°C with a monoclonal anti-APOL1 antibody (1:1,000; Epitomics, 3245-1). Membranes were washed three times in TBST and incubated for 1 h in blocking buffer with HRP-conjugated anti-rabbit IgG (1:20,000; Jackson Immuno-Research, West Grove, PA). Bound antibodies were visualized using ECL (Super Signal West Pico; Thermo Pierce, Rockford, IL) and recorded on X-ray film. Bands were scanned and densities quantitated using ImageJ software (http://rsbweb.nih.gov/ij/). As a standard, an APOL1-maltose binding protein (MBP) fusion protein previously generated in our lab (15) was loaded at varying concentrations.
FPLC
One hundred fifty microliters of sera from four healthy individuals homozygous for APOL1 G0, four homozygous for G1, and four homozygous for G2 variants were injected into a Superose 6 analytical column (GE Healthcare Life Sciences, Pittsburg, PA) for FPLC fractionation. Fractions 17–48 were collected at a flow rate of 0.4 ml/min. A total cholesterol (TC) enzymatic assay was performed to examine TC distribution among samples. Fractions 22–46 (sizes ranging from ∼7 to 20 nm or ∼66 to 1,240 kDa, as determined by nondenaturing gradient gel electrophoresis) were analyzed by immunoblot and probed with APOL1, APOAI, and APOB antibodies, as described below.
Non-denaturing gradient gel electrophoresis
One microliter of sera from four healthy individuals homozygous for APOL1 G0, four homozygous for G1, and four homozygous for G2 variants was electrophoresed in 4–20% native gels (BioRad Ready Gel® Tris-HCl gel) for 900 V/h and transferred to polyvinylidene fluoride membranes at 35 V for 22 h at 4°C. Membranes were blocked in 5% nonfat dry milk/0.1% TBST at room temperature for at least 1 h. Anti-APOL1 (Epitomics, 1:1,000), anti-APOA1 (Meridian Life Science, 1:1,000), and anti-APOB (Academy Bio-Medical Co., 1:1,000) primary antibodies were incubated with membranes overnight at 4°C. Membranes were washed three times with 0.1% TBST, incubated with anti-rabbit (GE Healthcare, 1:15,000) or anti-goat (Santa Cruz Biotechnology, 1:6,000) HRP-conjugated secondary antibodies for 2 h, and washed three times with 0.1% TBST. SuperSignal® West Pico chemiluminescent substrate was added and membranes were visualized using a FUJIFILM LAS-3000 imager.
Density ultracentrifugation
Twenty microliters of plasma from four healthy individuals homozygous for APOL1 G0, four homozygous for G1, and four homozygous for G2 variants were brought to a final volume of 1 ml with addition of normal saline and adjusted to d = 1.225 g/ml with solid KBr. Samples were overlaid with an additional 1 ml d = 1.225 g/ml KBr solution and ultracentrifuged in a TLA 120.2 rotor at 100,000 rpm for 4 h at 15°C. Following ultracentrifugation, top (0.5 ml) and bottom (1.5 ml) fractions were collected. Top (20 μl) and bottom (60 μl) fractions were then precipitated with TCA and subjected to 12% SDS-PAGE followed by APOL1 and APOA1 immunoblot analysis, as described.
Agarose gel electrophoresis
One microliter of plasma was electrophoresed on 0.7% agarose gels at 100 mA for 2 h. Gels were transferred to a polyvinylidene fluoride membrane by pressure blotting for 2 h. Following transfer, membranes were blocked with 5% nonfat dry milk/0.1% TBST for 1 h at room temperature, and incubated with either rabbit anti-APOL1 (Sigma-Aldrich, 1:1,000) or goat anti-APOA1 (Meridian Life Science, 1:1,000) antibodies overnight at 4°C. Membranes were washed three times with 0.1% TBST, incubated with anti-rabbit (GE Healthcare, 1:15,000) or anti-goat (Santa Cruz Biotechnology, 1:6,000) HRP-conjugated secondary antibodies for 2 h, and washed three times with 0.1% TBST. SuperSignal® West Pico chemiluminescent substrate was added and membranes were visualized using a FUJIFILM LAS-3000 imager.
Isoelectric focusing electrophoresis
Plasma (1.5 ml) from four healthy individuals homozygous for APOL1 G0, four homozygous for G1, and four homozygous for G2 variants was injected into a Superose 6 preparative column (GE Healthcare) for FPLC fractionation. Fractions were collected at a flow rate of 1 ml/min. A TC assay was performed to examine distributions among samples. Combined peak FPLC fractions for complex B (fractions 43–47) and complex A (fractions 58–62) were concentrated 10-fold with Amicon Ultra-4 MWCO 10 kDa centrifugal filter units (Millipore, Billerica, MA). Ten microliters of glycerol were added to 50 μl of concentrated FPLC APOL1 complex A and complex B samples, 15 μl of which were loaded onto a Criterion pH 3–10 isoelectric focusing (IEF) gel and electrophoresed at 100 V for 1 h, 250 V for 1 h, and then 500 V for 30 min. The gel was transferred at 100 V for 1 h in 0.7% acetic acid. The primary antibody (Epitomics anti-APOL1) was diluted 1:3,000 and the secondary antibody (Jackson Immuno-Research goat anti-rabbit IgG) was diluted 1:20,000 for immunoblot analysis.
Co-immunoprecipitation
An N-terminal APOL1 (1-199 amino acids)-MBP fusion protein was obtained, as previously reported (5, 15). cDNA corresponding to APOL1 (amino acid residues 13-130 of the APOL1 reference sequence) was generated by PCR and cloned into the pMAL-C5E vector (New England Biolabs) to generate a MBP-APOL1 fusion protein in Escherichia coli. Primer sequences used for PCR were: forward, 5′-AAG GTA CCG GAG GAA GCT GGA GCG AGG-3′ reverse, 5′-ACC GTC GAC TCA CCT TCT TAT GTT ATC CTC-3′. A polyclonal antibody against this fusion protein was raised in a rabbit (Lampire Biological Labs, Pipersville, PA). Lampire anti-APOL1 IgG was purified with a Melon Gel IgG purification kit (Thermo) according to the manufacturer’s instructions. Twenty microliters of complex A and 200 μl of complex B APOL1 peak fractions, determined by FPLC, were diluted to 1 ml with PBS in a 1.5 ml centrifuge tube. Rabbit anti-APOL1 antibody (50 μl; Lampire) was then added. The antibody-immunoprecipitate mixture was incubated overnight at 4°C by gentle mixing on a shaker. Sequential incubation at 4°C was applied for 1 h after addition of 40 μl protein A agarose slurry (Invitrogen, Grand Island, NY). The tube was centrifuged again at 1,000–3,000 g for 2 min at 4°C, the supernatant removed, and the bead mixture washed three times using PBS. After washing, 20 μl 2× SDS-PAGE loading buffer was added to the bead-antibody mixture. The mixture was heated at 95°C for 5 min, the beads were spun down, and the supernatant kept for immunoblot analysis with mouse anti-APOL1 antibody (Novus), goat anti-APOA1, mouse anti-haptoglobin-related protein (HPR) (Abcam), and other antibodies against candidate partners of APOL1 complexes (antibody details in supplementary Table 1).
Proteomics analysis
Concentrated complex A and B fractions (see the IEF electrophoresis section above) were analyzed by non-denaturing gradient gel electrophoresis (NDGGE) and IEF. Gels were stained with SYPRO® Ruby (Invitrogen) overnight. After destaining, bands were excised from the gel and subjected to proteomics analysis. In-gel digestion was performed with standard reduction (dithiothreitol) and alkylation (iodoacetamide) followed by proteolysis using MS grade trypsin (Pierce) in 50 mM NH4HCO3 overnight at 37°C. Peptides were extracted from the gel, concentrated, and analyzed on a Dionex UltiMate 3000 splitless nanoLC system coupled to a Thermo Orbitrap Velos Pro high resolution mass spectrometer. LC solvents were as follows: buffers A (98% water, 2% MeCN, 0.1% formic acid) and B (80% MeCN, 20% water, 0.1% formic acid). Peptides were loaded onto a nano trap C18 column (Acclaim PepMap 100, 100 μm × 2 cm, 5 μm) with a flow of 5 μl/min of 100% buffer A for 5 min, then separated using an analytical nano C18 column (Acclaim PepMap RSLC C18, 75 μm × 15 cm, 2 μm) with a gradient of 2–85% B over 160 min and a flow rate of 300 nl/min. The columns were held at 35°C. Following separation, peptides were introduced to the mass spectrometer via positive nanospray with the following settings: spray voltage 1.9 kV, capillary temperature 200°C. The mass spectrometer was operated in data-dependent top 10 mode using Xcalibur 2.1 (Thermo). MS spectra were acquired over the range of m/z 150–2,000 at a resolution of 60,000. Precursor ions were fragmented using collision-induced dissociation (normalized collision energy of 35%, activation time 10 ms) and fragment ions detected in the linear ion trap. Acquired raw data were processed and searched using Proteome Discoverer version 1.4 with the Mascot search engine and the SwissProt human proteomic database. Search parameters allowed for two missed trypsin cleavages, a precursor mass tolerance of 10 ppm, and fragment mass tolerance of 0.8 Da. N-terminal acetylation, cysteine carbamidomethylation, and methionine mono-oxidation were selected as variable modifications. Identities of proteins within the complex were filtered at a false discovery rate of 0.01.
RESULTS
Serum APOL1 concentrations in healthy African Americans
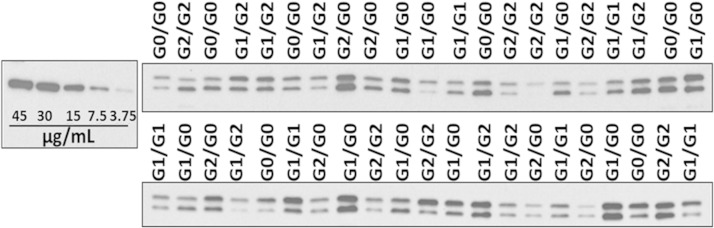
Serum samples from 84 age- and sex-matched NHAAN participants who had first-degree relatives with nondiabetic kidney disease were analyzed by immunoblot to determine whether circulating APOL1 concentrations were associated with APOL1 renal-risk variant genotypes in African Americans without kidney disease. Seven male and seven female NHAAN participants in each genotype group (G0/G0, G1/G0, G2/G0, G1/G2, G1/G1, and G2/G2) were analyzed. Consistent with previous reports (6), two bands were seen, reflecting the two APOL1 isoforms (39 kDa and 42 kDa) present in the circulation. Although APOL1 concentrations varied among individuals, effects were genotype-independent (Fig. 1). Regardless of APOL1 genotype, mean serum APOL1 concentrations were ∼15 μg/ml compared with an APOL1-MBP fusion protein standard of known concentration (Fig. 1). Mean serum APOL1 concentrations for each genotype are shown in Table 1.
Fig. 1.
APOL1 serum concentration is variable and genotype independent. Representative immunoblots of serum samples from homozygous reference (G0/G0), homozygous G1 (G1/G1), homozygous G2 (G2/G2), heterozygous G1 (G1/G0), heterozygous G2 (G2/G0), and compound heterozygous (G1/G2) African Americans (n = 14 per genotype) compared with an APOL1-MBP fusion protein standard of known concentrations (left).
Distribution of APOL1 on lipoprotein subfractions
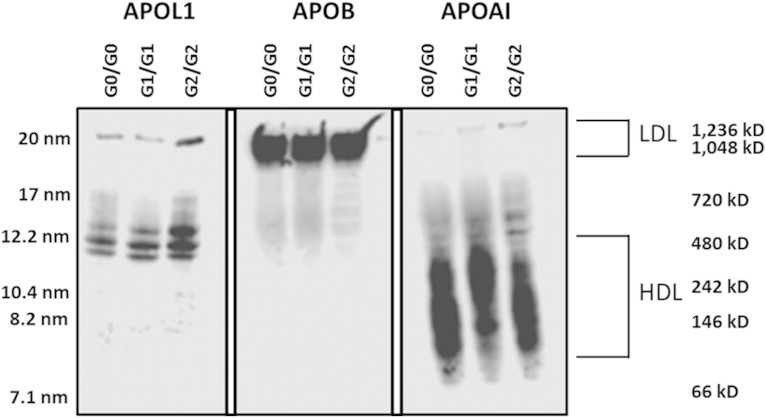
To examine potential differences in size distribution among APOL1-containing particles, NDGGE fractionation and immunoblot were performed on serum samples from African Americans lacking nephropathy with different APOL1 renal-risk variant genotypes. Based upon APOA1 and APOB immunodetection, the vast majority of APOL1-containing particles appeared to distribute between typical HDL- and LDL-sized particles, irrespective of genotype (Fig. 2). The predominant band was ∼12 nm in diameter (500 kDa molecular mass), with a smaller APOL1-containing particle band near the LDL migration position (20 nm diameter; 1,000 kDa molecular mass). Little, if any, serum APOL1 protein was in the unbound region of the gel (<7.1 nm) where lipid-poor and lipid-free apolipoproteins typically migrate (16) (Fig. 2). These results suggest that serum APOL1 is distributed on particles other than typically sized serum HDL and LDL.
Fig. 2.
Apolipoprotein distribution among serum lipoprotein subfractions. One microliter of serum from homozygous APOL1 (G0, G1, and G2) individuals was fractionated on 4–20% nondenaturing gradient gels and subjected to immunoblot analysis for APOL1, APOB, and APOA1. The average hydrodynamic diameter of standard proteins is shown on the left side of the blot and the molecular mass range is shown on the right side along with the approximate size range for LDL and HDL.
Serum samples were fractionated by FPLC, lipoprotein elution was monitored by measuring TC, and individual fractions were analyzed by immunoblot to better define APOL1-associated lipoprotein complexes. APOL1 primarily eluted in fractions between the LDL and HDL (fractions 31–38) and, to a lesser extent, coeluted with large LDL particles (fractions 26–28) (Fig. 3A, B). Herein, we refer to these fractions as APOL1 complex A and complex B, respectively, corresponding to the 12 nm- and 20 nm-diameter APOL1-containing particles identified using NDGGE (Fig. 2).
Fig. 3.
APOL1 mainly elutes with particles similar in size to large HDL. Human serum was size-fractionated by FPLC and eluted fractions were monitored for TC) (A) and apolipoprotein distribution (B). A: TC distribution for representative serum sample for homozygous APOL1 (G0, G1, and G2) individuals. B: Individual FPLC fractions (fxn) were immunoblotted for APOB, APOL1, and APOA1. Two column regions enriched in APOL1 were identified; one containing most of the serum APOL1 (complex A; fractions 31–38) and a minor fraction (complex B; fractions 26–28).
Fractionation of serum APOL1
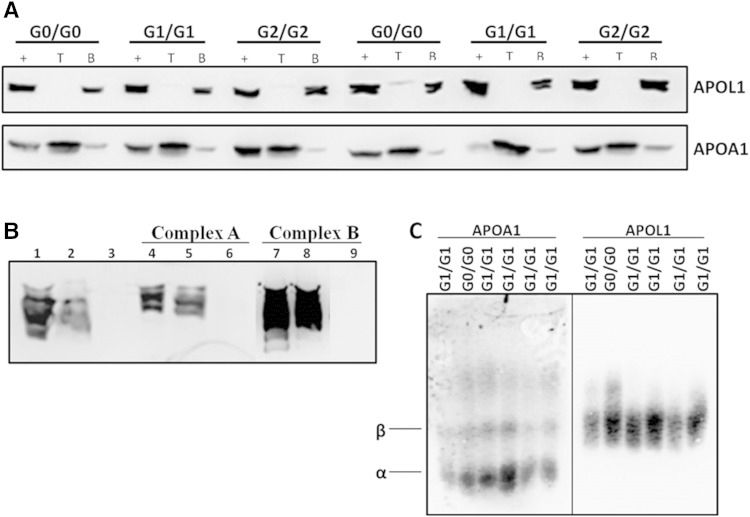
To determine whether APOL1-containing complexes were stably bound to HDL or LDL, density ultracentrifugation at d = 1.225 g/ml KBr was performed using sera from healthy African Americans with different APOL1 genotypes. Based on immunoblot analysis, >90% of APOL1 protein was found in the d > 1.225 gm/ml (bottom) fraction, whereas >90% of APOA1 was in the d < 1.225 gm/ml (top) fraction, suggesting that APOL1 was not as strongly associated with HDL as APOA1 (Fig. 4A). This finding was independent of APOL1 genotype. Moreover, ultracentrifugation or presence of high salt (i.e., 1.225 gm/ml KBr) did not impact the FPLC elution position of APOL1, suggesting that APOL1 was in a large complex that did not contain sufficient lipid to allow floatation at d < 1.225 gm/ml (Fig. 4B). Another method (agarose gel electrophoresis) was used that separates lipoproteins primarily by charge, rather than size, to determine whether APOL1 comigrated with HDL. Serum APOL1 migrated in the β/pre-β position characteristic of lipid-free APOA1, LDL, and VLDL, not in the α position where HDL particles migrate (Fig. 4C). This also suggests that serum APOL1 is not bound to HDL particles. Immunoblot analysis of d > 1.225 gm/ml (bottom) serum fractions subsequently separated by NDGGE demonstrated that APOL1 in complexes A and B exhibited similar size distributions as in serum that was not subjected to ultracentrifugation. Combined results using three different lipoprotein separation procedures that rely on size, density, and charge support the conclusion that APOL1 is likely a component of multiprotein complexes similar in size to large HDL and large LDL particles.
Fig. 4.
APOL1 does not associate with HDL in human serum. A: African American serum was subjected to density ultracentrifugation at d = 1.225 g/ml. Top (T) and bottom (B) fractions were then analyzed by immunoblot to visualize APOL1 and APOA1. Immunoblots of bottom fractions indicate that most of the APOL1 in circulation does not associate with HDL, whereas APOA1 floats in the top fractions. The + represents serum that did not undergo density ultracentrifugation. Results are representative of homozygous APOL1 (G0, G1, and G2) individuals. B: Following ultracentrifugation, bottom fractions were size-fractionated by FPLC. Complex A, complex B, and the lipid-free fraction (fractions 45–48, Fig. 3) were pooled and separated by nondenaturing gradient gel electrophoresis to determine the elution position of APOL1. Lanes 1–3: serum was adjusted to d = 1.225 g/ml prior to FPLC fractionation (lane 1, starting serum; lanes 2 and 3, complex A and lipid-free fraction, respectively, from FPLC fractionation). Results demonstrate that complex A was not disrupted by 1.225 g/ml KBr. Lanes 4–6: bottom fraction (d > 1.225 g/ml) from ultracentrifugation of serum was size-fractionated by FPLC (lane 4, unfractionated bottom fraction; lanes 5 and 6, complex A and lipid-free fraction, respectively, from FPLC fractionation of bottom fraction). Lanes 7–9: bottom fraction (d > 1.225 g/ml) from ultracentrifugation of serum was size-fractionated by FPLC (lane 7, unfractionated bottom fraction; lanes 8 and 9, complex B and lipid-free fraction, respectively, from FPLC fractionation of bottom fraction). C: One microliter of human serum from African Americans homozygous for APOL1 (G0 and G1) was electrophoresed in 0.7% agarose and immunoblotted for APOA1 and APOL1 to examine apolipoprotein migration in agarose gels. APOL1 does not comigrate with α particles, which correspond to HDL.
Composition of APOL1-containing complexes
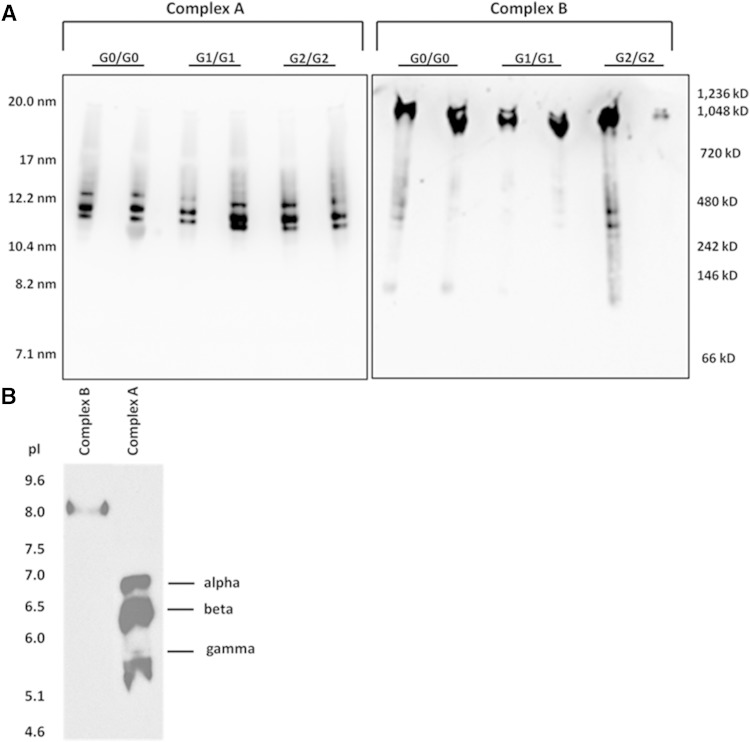
To determine whether there was a difference in the size of these complexes in African Americans with different APOL1 genotypes, a two-step fractionation of sera was used involving FPLC followed by NDGGE for serum samples (1.5 ml) from six individuals with different APOL1 genotypes (G0/G0, G1/G1, and G2/G2; N = 2 per genotype). APOL1 immunoblot of NDGGE gels demonstrated that complex A (∼12 nm diameter) was composed of three distinct size particles, defined as complexes A-α, A-β, and A-γ, whereas only a single band was apparent for complex B (∼20 nm diameter) (Fig. 5A).
Fig. 5.
Determining the composition of APOL1-containing complexes. Five FPLC fractions surrounding APOL1 complex A and complex B were pooled and concentrated. Samples were loaded onto 4–20% nondenaturing gradient gels (A) or IEF gels (B), followed by immunoblot analysis.
Concentrated complex A and complex B were separated by IEF based on isoelectric point (pI). Complex A separated into three bands with distinct pI values (assigned as A-1, A-2, and A-3 from top to bottom) and complex B remained as one band (Fig. 5B). Bands were carefully excised from gels for protein identification via MS. The distinct size components of complex A (α, β, and γ) detected by NDGGE were consistent with A-1, A-2, and A-3 on the IEF gel, respectively. Hereafter, these complexes are referred to as A-α, A-β, and A-γ, respectively.
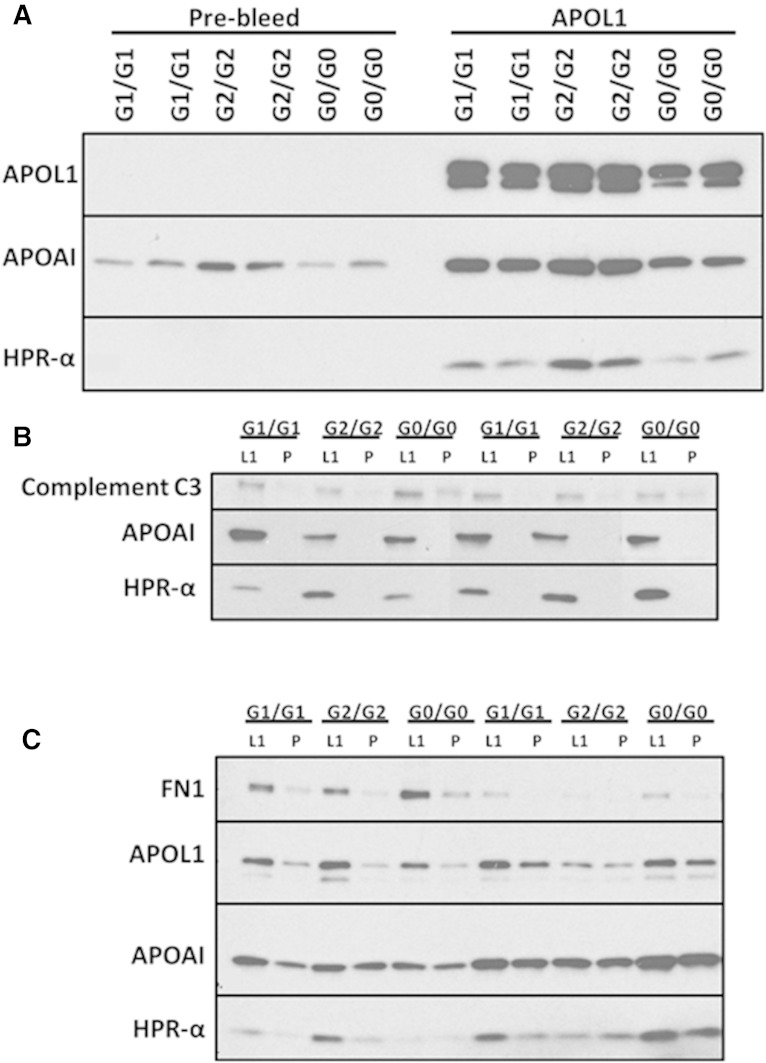
Table 2 lists the candidate proteins that were identified by MS from complex A and complex B (on both NDGGE and IEF gels) in six healthy African Americans. Immunoprecipitation of APOL1 complex A and complex B using a rabbit anti-APOL1 antibody directed at the N terminus of APOL1 was performed using 12 additional serum samples from African Americans with different APOL1 genotypes (G0/G0, G1/G1, and G2/G2; N = 4 per genotype). The anti-APOL1 immunoprecipitates were next examined by immunoblot analysis using antibodies targeting proteins that were identified by MS. APOL1, HPR, and APOA1 appeared on both complex A and complex B; however, complement C3 was unique to APOL1 complex A (Fig. 6B). Fibronectin appeared as a unique polypeptide on APOL1 complex B (Fig. 6C), in addition to IgM (17). The other candidate polypeptides in Table 2 were not verified because their antibodies failed to distinguish the intensity of corresponding polypeptide signals via immunoblot between co-immunoprecipitation of APOL1 peak FPLC fractions with Lampire rabbit anti-APOL1 (N terminus) IgG and that with the same amount of pre-bleed rabbit IgG; antibodies to candidate polypeptides on APOL1 complexes have been verified in human sera by immunoblot with clean bands of expected sizes.
TABLE 2.
Identification of candidate polypeptides on APOL1 complexes A and B by proteomics analysis
| Candidate Protein | Accession Number | Molecular Mass (kDa) | |||
| Complex A | TLF1-α | TLF1-β | TLF1-γ | ||
| Complement C3a | sp\P01024\CO3_HUMAN | 187 | ✓ | ✓ | ✓ |
| HPRa | sp\P00739\HPRT_HUMAN | 39 | ✓ | ✓ | ✓ |
| HP | sp\P00738\HPT_HUMAN | 45 | ✓ | ✓ | ✓ |
| Alpha-2-macroglobulin | sp\P01023\A2MG_HUMAN | 163 | ✓ | ✓ | ✓ |
| APOA1a | sp\P02647\APOA1_HUMAN | 31 | ✓ | ✓ | ✓ |
| APOL1a | sp\O14791\APOL1_HUMAN | 44 | ✓ | ✓ | ✓ |
| Prothrombin | sp\P00734\THRB_HUMAN | 70 | ✓ | ✓ | |
| Complement C5 | sp\P01031\CO5_HUMAN | 188 | ✓ | ✓ | |
| Complement C1r subcomponent | sp\P00736\C1R_HUMAN | 80 | ✓ | ✓ | |
| APOE | sp\P02649\APOE_HUMAN | 36 | ✓ | ✓ | |
| Clusterin | sp\P10909\CLUS_HUMAN | 52 | ✓ | ||
| Complement factor H | sp\P08603\CFAH_HUMAN | 139 | ✓ | ||
| Complex B | |||||
| Ig mu chain C regiona | sp\P01871\IGHM_HUMAN | 49 | ✓ | ||
| Fibronectina | sp\P02751\FINC_HUMAN | 263 | ✓ | ||
| Alpha-2-macroglobulin | sp\P01023\A2MG_HUMAN | 163 | ✓ | ||
| HP | sp\P00738\HPT_HUMAN | 45 | ✓ | ||
| Thrombospondin-1 | sp\P07996\TSP1_HUMAN | 129 | ✓ | ||
| APOA1a | sp\P02647\APOA1_HUMAN | 31 | ✓ | ||
| HPRa | sp\P00739\HPTR_HUMAN | 39 | ✓ | ||
| Vitamin K-dependent protein S | sp\P07225\PROS_HUMAN | 75 | ✓ | ||
| APOE | sp\P02649\APOE_HUMAN | 36 | ✓ | ||
| APOL1a | sp\O14791\APOL1_HUMAN | 44 | ✓ |
Check marked proteins are present in both IEF and native gel APOL1-positive bands for pooled complexes A and B from FPLC peak fractions.
Indicates those candidate proteins that were confirmed via subsequent co-immunoprecipitation analysis.
Fig. 6.
Confirmation of APOL1-associated protein components. A: Co-immunoprecipitation of APOL1 with protein components in whole serum using the N-terminal rabbit anti-APOL1 antibody (APOL1) and pre-bleed normal IgG (Pre-bleed). APOL1, APOA1, and HPR are confirmed to be present in co-immunoprecipitated APOL1 complexes. B: Co-immunoprecipitation of APOL1 in complex A FPLC peak fractions. APOA1, HPR, and complement C3 are confirmed to be present in co-immunoprecipitated APOL1 complex A. C: Co-immunoprecipitation of APOL1 in complex B FPLC peak fractions. Fibronectin, APOA1, and HPR are confirmed to be present in co-immunoprecipitated APOL1 complex. L1, anti-APOL1 antibody (Lampire); P, pre-bleed normal IgG from the same rabbit.
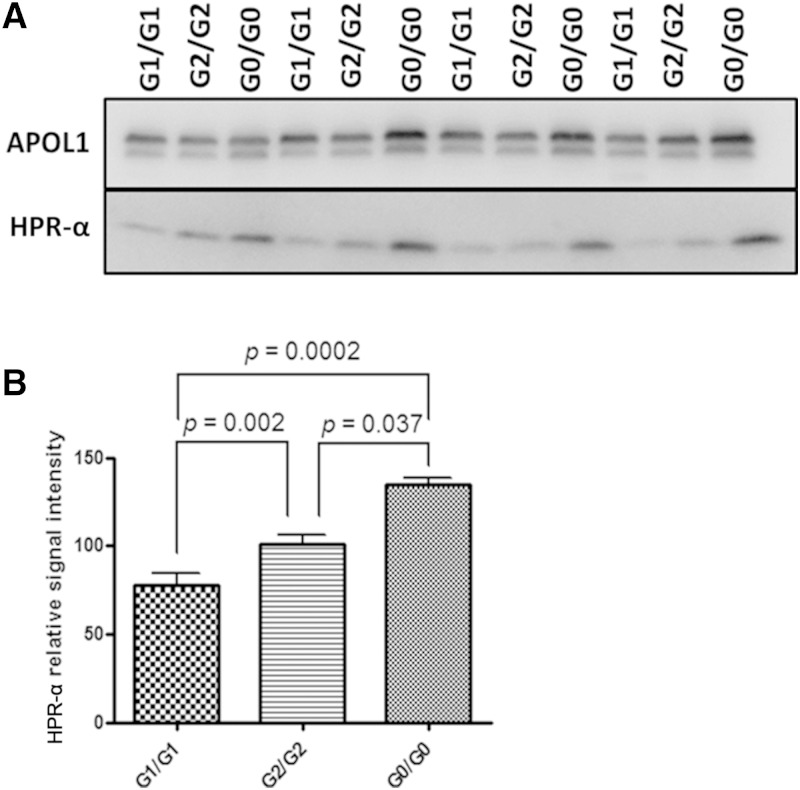
APOL1 protein concentrations in complex A and complex B peak fractions were calculated in 12 unrelated African Americans with APOL1 G0/G0, G1/G1, and G2/G2 genotypes (N = 4 per genotype) via immunoblot against a standard curve of known concentrations of MBP-APOL1 (N terminal) fusion protein using methods previously described (5). When FPLC complex B peak fractions containing 10 ng of APOL1 were probed with HPR antibody for immunoblot analysis, HPR-α, a proteolytic fragment of HPR (see below), was less abundant on complex B in individuals with APOL1 G1/G1 and G2/G2 genotypes, relative to those with G0/G0 genotypes (Fig. 7A, B). However, no significant pattern for HPR-α was observed on complex A in individuals with different APOL1 genotypes.
Fig. 7.
HPR-α is less abundant on complex B in APOL1 G1/G1 and G2/G2 individuals relative to G0/G0. A: APOL1 complex B peak fractions from AA individuals with different APOL1 genotypes (G1/G1, G2/G2, and G0/G0) were first quantified for APOL1 concentration by denatured immunoblot using Epitomics anti-APOL1 antibody and comparison with a series of APOL1 fusion proteins with known concentrations (see Fig. 1). Each well contains 10 ng APOL1. Samples were subjected to 4–20% SDS-PAGE and immunoblotting using specific antibody to APOL1 (Lampire anti-APOL1 polyclonal antibody) and HPR. The signal intensity of APOL1 was similar among complex B samples of different APOL1 genotypes. However, the relative amount of HPR-α residing on APOL1 complex B appeared to be less abundant for homozygous G1 carriers. B: Comparison of the relative signal intensity of HPR-α on serum APOL1 complex B containing the same amount of APOL1 (10 µg) of different genotypes (G1/G1, G2/G2, G0/G0), n = 3 each. P values were determined by t-test (2-sided).
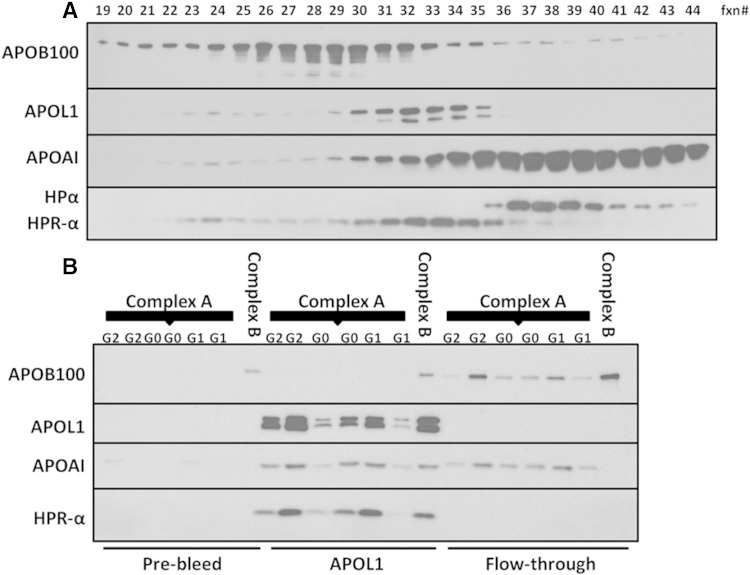
The HPR-α distribution pattern was consistent with that of APOL1 following FPLC fractionation (Fig. 8A). HPR-α was present on APOL1-containing protein complexes, whereas HPR-α was absent in the supernatant following APOL1 co-immunoprecipitation (Fig. 8B). After normalization for APOL1 content, no other differences were observed in APOL1 genotype-dependent distributions with other components of APOL1 complexes.
Fig. 8.
HPR-α distribution is consistent with that of APOL1 following FPLC. A: FPLC fractions (fxn#) were subjected to 4–20% SDS-PAGE and probed with APOB, APOA1, APOL1, and HPR antibodies via immunoblot analysis. APOA1 and APOB100 peak fractions represent the enrichment of HDL and LDL, respectively. The peak fractions for APOL1 and HPR-α are consistent, indicating the positions of complexes A and B. B: APOL1 complex A peak fractions from G1/G1, G2/G2, and G0/G0 individuals and one sample of peak APOL1 complex B from a G0/G0 subject were co-immunoprecipitated with rabbit anti-N-terminal APOL1 antibody (APOL1) or pre-bleed IgG from the same rabbit (Pre-bleed). APOL1 and HPR-α only appeared in co-immunoprecipitated APOL1, but not in the supernatant (Flow-through), indicating the coexistence of APOL1 and HPR in serum complexes A and B. The absence of APOL1 and HPR in the APOL1 supernatant suggests that anti-APOL1 IgG was sufficient to precipitate all APOL1 in the sample. The absence of APOL1, APOA1, and HPR in the pre-bleed rabbit IgG co-immunoprecipitation indicates the specificity of the antibodies. Because APOB is enriched in complex B fractions and APOB also appeared in the pre-bleed rabbit co-immunoprecipitation, it is undetermined whether APOB is specifically present on complex B.
Potential surface epitope conformational changes of APOL1 protein related to G1 and G2 variants
To detect potential surface epitope conformational differences between reference APOL1 (G0) and renal-risk (G1, G2) variants, the avidity of monoclonal APOL1 antibody (Epitomics) to APOL1 was tested on FPLC-isolated complex B from healthy African Americans with different APOL1 genotypes. Peak fractions from the aforementioned 12 individuals containing 10 ng APOL1 were separated by NDGGE. Immunoblot analysis was performed using an Epitomics APOL1 antibody targeting amino acids 300-330. Signal intensity was significantly higher in complex B peak fractions from those with APOL1 G1/G1 or G2/G2 variants relative to G0/G0 (Fig. 9). Potential differences among APOL1 genotypes were also examined in complex A; however, no significant differences were observed (data not shown).
Fig. 9.
Estimate of potential surface epitope conformational change of APOL1 protein caused by G1 and G2 variants. APOL1 complex B peak fractions containing 10 ng of APOL1 (quantified by Epitomics anti-APOL1 antibody shown in Fig. 7) were run on a 4–20% nondenaturing gradient gel at 150 V for 1 h at room temperature, transferred to a cellulose membrane, and probed with the same Epitomics anti-APOL1 antibody (immunogen within amino acids 300-330) via immunoblot. APOL1 signal intensity was significantly higher for complex B in G1/G1 and G2/G2 individuals compared with G0/G0, n = 3/genotype.
DISCUSSION
Differences between circulating APOL1 G0 and mutant G1 and G2 renal-risk variant proteins were analyzed in an attempt to better understand potential roles in human disease. APOL1 is predominantly present in the circulation and to a lesser extent in the liver and other solid organs, including the kidneys. Kidney APOL1 protein is enriched in glomerular podocytes, relative to proximal tubule and endothelial cells (5). Due to the abundance of APOL1 in human serum and uptake of free APOL1 into podocytes in vitro (5), it was initially hypothesized that circulating APOL1 protein might contribute to APOL1-associated nephropathy. Subsequent kidney transplantation studies failed to support this hypothesis (18–20); however, potential roles for circulating APOL1 protein in CVD and HDL-cholesterol metabolism remain plausible (7).
No association was observed between APOL1 G1 and G2 risk variant genotypes and serum APOL1 concentration by immunoblot in 84 African Americans lacking kidney disease. Individuals comprised all possible renal-risk variant genotype combinations (G0/G0, G1/G0, G2/G0, G1/G1, G2/G2, and G1/G2) with 14 healthy individuals of similar age and equal sex distributions in each group. We estimated the presence of ∼0.75 power to detect a 1-fold difference in serum APOL1 protein in genotypic groups (versus G0/G0), assuming α = 0.05 (two-sided). Comparing subjects with two APOL1 renal-risk alleles versus zero or one, estimated power reached ∼0.80 for detecting 50% differences in serum APOL1. These results confirm the observations of Bruggeman et al. (14) in African Americans with HIV infection. As serum APOL1 protein concentrations are genotype independent, it appears more likely that potential risk for disease may relate to protein function rather than quantity (14).
The size distribution of APOL1-containing particles in human sera, relative to HDL and LDL, was assessed using NDGGE. This fractionation procedure is less disruptive to HDL particle distribution than ultracentrifugation (21, 22). APOL1-containing particles were present in two major protein complexes: APOL1 complex A (12 nm diameter, 500 kDa) and APOL1 complex B (20 nm diameter, 1,000 kDa); patterns were consistent with those seen on fractionation using FPLC. APOL1 complex A is larger than typical HDL particles (8–12 nm), with similar distributions noted across APOL1 G0/G0, G1/G1, and G2/G2 genotypes. Following density ultracentrifugation of plasma adjusted to d = 1.225 g/ml KBr (standard for plasma lipoprotein isolation) (23), more than 90% of APOL1 (G0, G1, and G2) failed to “float” with other apolipoproteins such as APOA1, which was surprising because it had been assumed that APOL1-containing particles were a stable component of HDL (6). Lipids were not directly measured in complex A or complex B; however, both are likely to be lipid poor because they reside in the bottom fraction after density ultracentrifugation. TLF1 floated following density ultracentrifugation at a slightly higher density (d = 1.25) in a report by Thomson et al. (17). That report did not examine APOL1; as such, the ratios of APOL1 in top and bottom fractions remain unknown. Additionally, agarose gel electrophoresis revealed that APOL1 does not migrate to the position of serum HDL particles, evidenced by its migration at the β/pre-β position, compared with HDL that migrates at the α position. This is consistent with NDGGE data showing that APOL1 has a larger size distribution than APOA1 and suggesting that APOL1 is not associated with normal size HDL particles (Fig. 2). Based on the results from three fractionation procedures, APOL1-containing complexes do not appear to reside in “typical” serum HDL particles, as defined by density, charge, and size. Previous studies have shown that a small percentage (∼5%) of plasma HDL particles containing APOA1 are lipid poor, smaller in size (<7 nm diameter) than most plasma HDL, and migrate in the pre-β position on agarose gels. These so called “pre-β HDLs” are likely newly assembled nascent HDL particles generated by ATP binding cassette transporter A1 or products of plasma HDL remodeling by lipases or lipid transfer proteins (24–26). Another study used immunoaffinity chromatography to isolate lipid-poor (∼3% lipid) phospholipid-transfer protein-APOA1 complexes containing proteins involved in innate immunity (27), suggesting that HDL should be more generically defined as APOA1-containing complexes/particles, particularly those lipid-poor APOA1 complexes involved in innate immunity.
Proteomic analyses were performed to detect potential differences in the composition of APOL1 complex A and complex B in healthy African Americans with APOL1 G0/G0, G1/G1, and G2/G2 genotypes. Following FPLC fractionation of sera, complexes A and B were applied to IEF and NDGGE to further separate complexes by pI and particle size, respectively. This was an attempt to narrow the number of candidate proteins by prioritizing common polypeptide identities through different separation methods for subsequent proteomic analyses. Subsequent immunoblot and immunoprecipitation verification of results support the conclusion that circulating APOL1 complexes are likely to be the previously identified TLF1 and TLF2, based on complex size (13) and matching APOL1 and HPR-α FPLC elution profiles (Fig. 8A). Co-immunoprecipitation of APOL1 captured nearly all HPR-α (Fig. 8B), providing additional evidence that APOL1 and HPR-α coexist in one serum protein complex. HPR is a core component of TLFs based on its ability to induce trypanosome lysis. HPR is synthesized as a 45 kDa protein that is proteolytically cleaved to form an α-chain and a β-chain, which remain connected via a disulfide bond (28). The HPR-α chain is a 13 kDa N-terminal HPR fragment that retains its signal peptide (29, 30). Because haptoglobin (HP) and HPR are highly homologous, HPR/HP antibodies recognize both HPR and HP. The size differences between HPR-α (13 kDa) and HP1-α (16 kDa) or HP2-α (18 kDa) is the only way to distinguish the probed polypeptide by the HPR antibody (28), which recognizes the N terminus of HP/HPR. Therefore, the HPR-α here actually represents HPR. The trypanolytic activity profile (17) is consistent with our APOL1 distribution pattern following FPLC fractionation. The presence of APOA1 and HPR in complex A is consistent with that of TLF1 (29) and the presence of APOA1, HPR, and IgM is consistent with that of TLF2 (29). APOL1 complexes A and B both contain APOL1, APOA1, and HPR. Novel components for both APOL1-containing complexes were identified (complement C3 in complex A or TLF1; fibronectin in complex B or TLF2).
An APOL1 epitope conformational change is expected for the SRA interaction domain that surrounds the G1 and G2 renal-risk variants (31). However, the surface epitope exposure also appeared to be altered in the region between the membrane-addressing and SRA-interacting domains. Increased avidity to APOL1 antibody (Epitomics) for the G1/G2 variants was observed (Fig. 9). We speculate that the increased APOL1 signal for complex B in individuals with the G1 or G2 variant following NDGGE (Fig. 9) could represent increased exposure of APOL1 amino acids 300-330 and/or lower binding of other protein(s) in that region on the surface of the protein complex; the lower signal of HPR-α on complex B in individuals with G1 or G2 variants (Fig. 7) may partially explain its increased avidity to Epitomics anti-APOL1 antibody, although the G1 and G2 variants are located beyond amino acid 340 (1).
Based on APOL1 co-immunoprecipitation experiments and serum APOL1/HPR distribution patterns following FPLC, serum APOL1 coexists with HPR. We could not determine whether serum APOL1 is exclusively complexed with HPR because existing HPR antibodies do not allow complete and specific immunoprecipitation of HPR. HPR and APOA1 are secreted from the liver and their mRNAs are absent in kidney. However, HPR and APOA1 proteins are present in kidney tubule cells. It appears likely that APOL1 protein in podocytes is predominantly locally synthesized (4, 5). A considerable amount of HDL APOA1 is catabolized in the kidney in human APOA1 transgenic mice and nonhuman primates (32, 33). APOL1 (5), APOA1, and HPR proteins are present in renal tubule cells, but not glomeruli, whereas HPR and APOA1 mRNAs are absent in glomeruli (data not shown). It is possible that circulating APOL1 complexes/TLFs are catabolized by renal tubule cells (34), instead of being directly involved in glomerulosclerosis, as we originally hypothesized. Although our results do not support a role for the APOL1 complexes in the pathogenesis of glomerulosclerosis due to the absence of APOA1 and HPR (mRNAs and proteins), the key components of APOL1 complexes (TLF1 and TLF2), in glomeruli, they clarify the perspective that locally synthesized APOL1 in the kidney, especially in glomerular podocytes, is the major player of focal glomerulosclerosis.
We conclude that the majority of circulating APOL1 resides in lipid-poor multiprotein complexes, the contents of which largely represent TLFs, not typical lipid-enriched HDL particles. APOL1 G1 and G2 renal-risk variant genotypes appear to alter the composition, and potentially the conformation, of these multiprotein complexes. Potential roles for genotype-specific alterations in APOL1-protein complexes require further study in human disease, particularly vascular disease and susceptibility to the development of calcified atherosclerotic plaque.
Supplementary Material
Footnotes
Abbreviations:
- FPLC
- fast protein LC
- HP
- haptoglobin
- HPR
- haptoglobin-related protein
- IEF
- isoelectric focusing
- MBP
- maltose binding protein
- NDGGE
- non-denaturing gradient gel electrophoresis
- NHAAN
- Natural History of APOL1-Associated Nephropathy
- pI
- isoelectric point
- TBST
- TBS containing 1% skim milk powder and 0.1% Tween 20
- TC
- total cholesterol
- TLF
- trypanosome lytic factor
This project was supported by National Institutes of Health Grants R01 DK070941 (B.I.F.) and R01 DK084149 (B.I.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All authors declare no competing interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Genovese G., Friedman D. J., Ross M. D., Lecordier L., Uzureau P., Freedman B. I., Bowden D. W., Langefeld C. D., Oleksyk T. K., Uscinski Knob A. L., et al. . 2010. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 329: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S., Rosset S., Shemer R., Yudkovsky G., Selig S., Tarekegn A., Bekele E., Bradman N., Wasser W. G., Behar D. M., et al. . 2010. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 128: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman B. I., Kopp J. B., Langefeld C. D., Genovese G., Friedman D. J., Nelson G. W., Winkler C. A., Bowden D. W., and Pollak M. R.. 2010. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J. Am. Soc. Nephrol. 21: 1422–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madhavan S. M., O’Toole J. F., Konieczkowski M., Ganesan S., Bruggeman L. A., and Sedor J. R.. 2011. APOL1 localization in normal kidney and nondiabetic kidney disease. J. Am. Soc. Nephrol. 22: 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L., Shelness G. S., Snipes J. A., Murea M., Antinozzi P. A., Cheng D., Saleem M. A., Satchell S. C., Banas B., Mathieson P. W., et al. . 2015. Localization of APOL1 protein and mRNA in the human kidney: nondiseased tissue, primary cells, and immortalized cell lines. J. Am. Soc. Nephrol. 26: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duchateau P. N., Pullinger C. R., Orellana R. E., Kunitake S. T., Naya-Vigne J., O’Connor P. M., Malloy M. J., and Kane J. P.. 1997. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 272: 25576–25582. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez O. M., Judd S. E., Irvin M. R., Zhi D., Limdi N., Palmer N. D., Rich S. S., Sale M. M., and Freedman B. I.. APOL1 nephropathy risk variants are associated with altered high-density lipoprotein profiles in African Americans. Nephrol. Dial. Transplant. Epub ahead of print. July 6, 2015; doi:10.1093/ndt/gfv229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K., Bick A. G., Flannick J., Friedman D. J., Genovese G., Parfenov M. G., Depalma S. R., Gupta N., Gabriel S. B., Taylor H. A. Jr., et al. . 2014. Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ. Res. 114: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detrano R., Guerci A. D., Carr J. J., Bild D. E., Burke G., Folsom A. R., Liu K., Shea S., Szklo M., Bluemke D. A., et al. . 2008. Coronary calcium as a predictor of coronary events in four racial or ethnic groups. N. Engl. J. Med. 358: 1336–1345. [DOI] [PubMed] [Google Scholar]
- 10.Langefeld C. D., Divers J., Pajewski N. M., Hawfield A. T., Reboussin D. M., Bild D. E., Kaysen G. A., Kimmel P. L., Raj D. S., Ricardo A. C., et al. . 2015. Apolipoprotein L1 gene variants associate with prevalent kidney but not prevalent cardiovascular disease in the Systolic Blood Pressure Intervention Trial. Kidney Int. 87: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman B. I., Langefeld C. D., Lu L., Palmer N. D., Smith S. C., Bagwell B. M., Hicks P. J., Xu J., Wagenknecht L. E., Raffield L. M., et al. . 2015. APOL1 associations with nephropathy, atherosclerosis, and all-cause mortality in African Americans with type 2 diabetes. Kidney Int. 87: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzureau P., Uzureau S., Lecordier L., Fontaine F., Tebabi P., Homble F., Grelard A., Zhendre V., Nolan D. P., Lins L., et al. . 2013. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 501: 430–434. [DOI] [PubMed] [Google Scholar]
- 13.Samanovic M., Molina-Portela M. P., Chessler A. D., Burleigh B. A., and Raper J.. 2009. Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog. 5: e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruggeman L. A., O’Toole J. F., Ross M. D., Madhavan S. M., Smurzynski M., Wu K., Bosch R. J., Gupta S., Pollak M. R., Sedor J. R., et al. . 2014. Plasma apolipoprotein L1 levels do not correlate with CKD. J. Am. Soc. Nephrol. 25: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D., Weckerle A., Yu Y., Ma L., Zhu X., Murea M., Freedman B. I., Parks J. S., and Shelness G. S.. 2015. Biogenesis and cytotoxicity of APOL1 renal-risk variant proteins in hepatocytes and hepatoma cells. J. Lipid Res. 56: 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulya A., Lee J. Y., Gebre A. K., Thomas M. J., Colvin P. L., and Parks J. S.. 2007. Minimal lipidation of pre-beta HDL by ABCA1 results in reduced ability to interact with ABCA1. Arterioscler. Thromb. Vasc. Biol. 27: 1828–1836. [DOI] [PubMed] [Google Scholar]
- 17.Thomson R., Genovese G., Canon C., Kovacsics D., Higgins M. K., Carrington M., Winkler C. A., Kopp J., Rotimi C., Adeyemo A., et al. . 2014. Evolution of the primate trypanolytic factor APOL1. Proc. Natl. Acad. Sci. USA. 111: E2130–E2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves-Daniel A. M., DePalma J. A., Bleyer A. J., Rocco M. V., Murea M., Adams P. L., Langefeld C. D., Bowden D. W., Hicks P. J., Stratta R. J., et al. . 2011. The APOL1 gene and allograft survival after kidney transplantation. Am. J. Transplant. 11: 1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee B. T., Kumar V., Williams T. A., Abdi R., Bernhardy A., Dyer C., Conte S., Genovese G., Ross M. D., Friedman D. J., et al. . 2012. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am. J. Transplant. 12: 1924–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedman B. I., Julian B. A., Pastan S. O., Israni A. K., Schladt D., Gautreaux M. D., Hauptfeld V., Bray R. A., Gebel H. M., Kirk A. D., et al. . 2015. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am. J. Transplant. 15: 1615–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung M. C., and Wolf A. C.. 1988. Differential effect of ultracentrifugation on apolipoprotein A-I-containing lipoprotein subpopulations. J. Lipid Res. 29: 15–25. [PubMed] [Google Scholar]
- 22.Kunitake S. T., and Kane J. P.. 1982. Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. J. Lipid Res. 23: 936–940. [PubMed] [Google Scholar]
- 23.Havel R. J., Eder H. A., and Bragdon J. H.. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane J. P., and Malloy M. J.. 2012. Prebeta-1 HDL and coronary heart disease. Curr. Opin. Lipidol. 23: 367–371. [DOI] [PubMed] [Google Scholar]
- 25.Lee J. Y., and Parks J. S.. 2005. ATP-binding cassette transporter AI and its role in HDL formation. Curr. Opin. Lipidol. 16: 19–25. [DOI] [PubMed] [Google Scholar]
- 26.Rosenson R. S., Brewer H. B. Jr., Davidson W. S., Fayad Z. A., Fuster V., Goldstein J., Hellerstein M., Jiang X. C., Phillips M. C., Rader D. J., et al. . 2012. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 125: 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung M. C., Vaisar T., Han X., Heinecke J. W., and Albers J. J.. 2010. Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation. Biochemistry. 49: 7314–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen M. J., Petersen S. V., Jacobsen C., Oxvig C., Rees D., Moller H. J., and Moestrup S. K.. 2006. Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood. 108: 2846–2849. [DOI] [PubMed] [Google Scholar]
- 29.Raper J., Fung R., Ghiso J., Nussenzweig V., and Tomlinson S.. 1999. Characterization of a novel trypanosome lytic factor from human serum. Infect. Immun. 67: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lugli E. B., Pouliot M., Portela Mdel P., Loomis M. R., and Raper J.. 2004. Characterization of primate trypanosome lytic factors. Mol. Biochem. Parasitol. 138: 9–20. [DOI] [PubMed] [Google Scholar]
- 31.Pays E., Vanhollebeke B., Vanhamme L., Paturiaux-Hanocq F., Nolan D. P., and Perez-Morga D.. 2006. The trypanolytic factor of human serum. Nat. Rev. Microbiol. 4: 477–486. [DOI] [PubMed] [Google Scholar]
- 32.Huggins K. W., Burleson E. R., Sawyer J. K., Kelly K., Rudel L. L., and Parks J. S.. 2000. Determination of the tissue sites responsible for the catabolism of large high density lipoprotein in the African green monkey. J. Lipid Res. 41: 384–394. [PubMed] [Google Scholar]
- 33.Lee J. Y., Timmins J. M., Mulya A., Smith T. L., Zhu Y., Rubin E. M., Chisholm J. W., Colvin P. L., and Parks J. S.. 2005. HDLs in apoA-I transgenic Abca1 knockout mice are remodeled normally in plasma but are hypercatabolized by the kidney. J. Lipid Res. 46: 2233–2245. [DOI] [PubMed] [Google Scholar]
- 34.Gregorini G., Izzi C., Ravani P., Obici L., Dallera N., Del Barba A., Negrinelli A., Tardanico R., Nardi M., Biasi L., et al. . 2015. Tubulointerstitial nephritis is a dominant feature of hereditary apolipoprotein A-I amyloidosis. Kidney Int. 87: 1223–1229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.