Abstract
Membrane-bound desaturases are physiologically and industrially important enzymes that are involved in the production of diverse fatty acids such as polyunsaturated fatty acids and their derivatives. Here, we identified amino acid residues that determine the substrate specificity of rat Δ6 desaturase (D6d) acting on linoleoyl-CoA by comparing its amino acid sequence with that of Δ5 desaturase (D5d), which converts dihomo-γ-linolenoyl-CoA. The N-terminal cytochrome b5-like domain was excluded as a determinant by domain swapping analysis. Substitution of eight amino acid residues (Ser209, Asn211, Arg216, Ser235, Leu236, Trp244, Gln245, and Val344) of D6d with the corresponding residues of D5d by site-directed mutagenesis switched the substrate specificity from linoleoyl-CoA to dihomo-γ-linolenoyl-CoA. In addition, replacement of Leu323 of D6d with Phe323 on the basis of the amino acid sequence of zebra fish Δ5/6 bifunctional desaturase was found to render D6d bifunctional. Homology modeling of D6d using recent crystal structure data of human stearoyl-CoA (Δ9) desaturase revealed that Arg216, Trp244, Gln245, and Leu323 are located near the substrate-binding pocket. To our knowledge, this is the first report on the structural basis of the substrate specificity of a mammalian front-end fatty acid desaturase, which will aid in efficient production of value-added fatty acids.
Keywords: site-directed mutagenesis, heterologous expression, mass spectrometry, homology modeling
Fatty acid desaturases are oxidases that introduce a double bond in the acyl chain of a fatty acid substrate by removing two hydrogens from adjacent carbon atoms using active oxygen. They comprise two types. Water-soluble desaturases are found in cyanobacteria and higher plants and act on the acyl chain bound to acyl carrier protein (ACP) (1), whereas membrane-bound desaturases from fungi, higher plants, and animals act on acyl-CoA or acyl-lipid substrates (2, 3). Some water-soluble enzymes such as castor Δ9 desaturase and ivy Δ4 desaturase are well characterized, and their crystal structures have revealed a molecular interaction between the ACP portion of the substrate and an amino acid located at the substrate-binding pocket of the enzyme, which could be the basis for change in the substrate specificity (4). The membrane-bound desaturases associate with endoplasmic reticulum membranes via two large hydrophobic domains that separate three hydrophilic clusters. The N-terminal hydrophilic region of some of these desaturases including mammalian Δ5 and Δ6 desaturases (D5d and D6d, respectively) and the C-terminal region of Saccharomyces cerevisiae Δ9 desaturase (OLE1p) contain a cytochrome b5-like heme-binding His-Pro-Gly-Gly (HPGG) motif. The histidine residue is indispensable for electron transfer from NADH-dependent cytochrome b5 reductase during the redox reaction (5, 6). Both this motif and that of diffused cytochrome b5 are necessary to fully express desaturase activity (7, 8). The other hydrophilic regions contain three histidine clusters (HX3-4H, HX2-3HH, and QX2-3HH) that form a catalytic center by coordinating nonheme diiron centers, and all of these histidine residues and the glutamine residue are essential for enzymatic activity (9, 10). D5d and D6d, as well as Δ4 desaturase, introduce a double bond at the respective Δ positions of fatty acid substrates between the carboxyl group and a preexisting double bond; therefore, these enzymes are called “front-end” desaturases (11). They are distinct from desaturases of ω-x and ν+x types that form double bonds at the methyl-terminal side.
The substrate specificity and regioselectivity (double bond positioning) of membrane-bound desaturases are defined by the structural fitness and interface affinity between the fatty acid substrate, including CoA and the lipid carrier, and the substrate-binding pocket with its surrounding residues. Protein engineering has been applied to understand the structure-function relationship. For instance, domain swapping has been used to identify the regioselective sites of nematode Δ12 and ω3 desaturases (12), a region determining the substrate specificity of Aspergillus nidulans Δ12 and ω3 desaturases (13), and a substrate recognition region of black currant Δ6 fatty acid desaturase and Δ8 sphingolipid desaturase (14). Site-directed mutagenesis based on amino acid sequence comparison has been used to identify amino acids participating in the substrate specificity of Mucor rouxii D6d (15), Siganus canaliculatus Δ4 and D5d/D6d (16), and marine copepod Δ9 desaturase (17). The regioselectivity of house cricket Δ12/Δ9 desaturase was investigated using chemical mutagenesis and yeast complementation assays (18). Moreover, fatty acid-modifying enzymes with protein structures similar to, but chemoselectivities different from, the fatty acid desaturases have been used to swap the function of Arabidopsis oleate 12-desaturase and hydroxylase (19) and to alter the product partitioning between Crepis alpina Δ12 desaturase and acetylenase (20) and Momordica conjugase itself (21).
In this study, we aimed to elucidate the structural basis of the substrate specificity of Rattus norvegicus D6d and D5d (22) by domain swapping and site-directed mutagenesis. The corresponding genes are positioned in a head-to-head configuration on the rat genome, suggesting a paralogous relationship (11). Although their primary structures are highly homologous, they are in charge of mutually exclusive substrates: D6d catalyzes the conversion of linoleic acid (LA; 18:2 Δ9,12) and α-linolenic acid (18:3 Δ9,12,15) into γ-linolenic acid (GLA; 18:3 Δ6,9,12) and stearidonic acid (18:4 Δ6,9,12,15), respectively, whereas D5d acts on dihomo-γ-linolenic acid (DGLA; 20:3 Δ8,11,14) and eicosatetraenoic acid (20:4 Δ8,11,14,17) to generate arachidonic acid (ARA; 20:4 Δ5,8,11,14) and eicosapentaenoic acid (20:5 Δ5,8,11,14,17), respectively. To identify and evaluate the amino acid residues important for substrate selection of D6d, we performed additional analyses on the basis of the primary sequence of zebra fish bifunctional Δ5/6 desaturase [zD5/6d (23)] and the recently reported crystal structure of human stearoyl-CoA (Δ9) desaturase (24, 25).
MATERIALS AND METHODS
Microorganisms, culture media, and reagents
Transformants of Escherichia coli DH5α were grown in LB medium (0.5% yeast extract, 1% NaCl, 1% Bacto tryptone, 2% agar for plates) or 2× YT medium (1.6% Bacto tryptone, 1% yeast extract, 0.5% NaCl) supplemented with ampicillin (50 μg/ml) at 37°C with rotary shaking at 160 rpm. Transformants of S. cerevisiae INVSc1 (Invitrogen, Carlsbad, CA) were selected on SD agar plates (0.67% yeast nitrogen base, 0.19% yeast synthetic dropout medium without uracil, 2% d-glucose, 2% agar) and cultivated in SCT medium (0.67% yeast nitrogen base, 0.19% yeast synthetic dropout medium without uracil, 4% raffinose, 0.1% Tergitol) or YPD medium (2% polypeptone, 1% yeast extract, 2% d-glucose) at 28°C and 160 rpm. Fatty acids were purchased from Sigma-Aldrich (St. Louis, MO) or Cayman Chemical (Ann Arbor, MI). Other guaranteed reagents were obtained from Nacalai Tesque (Kyoto, Japan), Sigma-Aldrich, Toyobo (Osaka, Japan), or Wako Chemicals (Osaka, Japan), unless otherwise indicated.
Construction of plasmids carrying desaturase genes
A FLAG DNA fragment was synthesized by PCR amplification with Takara Ex Taq (Takara, Kyoto, Japan) and the oligonucleotide primers FLAGf and FLAGr (Table 1), using 10 cycles of 95°C for 30 s, 50°C for 30 s, and 74°C for 30 s, without template. The fragment was subcloned in pGEM-T Easy vector (Promega, Madison, WI) and transformed into E. coli DH5α (pGEM-FLAG). The rat D6d gene (DDBJ accession number AB021980) was amplified from stock plasmid with KOD-Dash DNA polymerase (Toyobo) and the primers 24aF+ and 24R+ (Table 1), using 30 cycles of 95°C for 30 s, 68°C for 2 s, and 74°C for 30 s, and was digested with KpnI and XbaI. The product was ligated into KpnI/SpeI-digested pGEM-FLAG and the plasmid was transformed into E. coli DH5α (pGEM-FLAG-D6d). The rat D5d gene (DDBJ accession number AB052085) was amplified using KOD polymerase (Toyobo), the primers rD5df and rD5dr (Table 1), and a rat liver cDNA library (Clontech Laboratories, Palo Alto, CA) under the same thermal cycling conditions as for D6d, and was ligated into pGEM-FLAG (pGEM-FLAG-D5d). The nucleotide sequences of all plasmids were determined using the DYEnamic ET terminator cycle sequencing kit (GE Healthcare, Buckinghamshire, UK) or BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, Carlsbad, CA) with T7, SP6, and other appropriate primers (Table 1) on an ABI PRISM 310 or 3130 × 1 genetic analyzer (Life Technologies).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Nucleotide Sequence (5′ to 3′ Direction) | Purposea |
| Preparation of whole and partial regions of desaturases | ||
| FLAGf | GCAAAGCTTAAGATGGACTATAAGGATGATGATGAC | FLAG tag |
| FLAGr | CGTGGTACCCTTGTCATCATCATCCTTATAG | FLAG tag |
| 24aF+ | ACAGGTACCATGGGGAAGGGAGGTAACCAG | D6d |
| 24R+ | GTCTCTAGATTCATTTGTGGAGGTAGGCATCC | D6d |
| D5df | CCCGGTACCATGGCTCCCGACCCGGTGCAGACCC | D5d |
| D5dr | CCCCTGCAGCTATTGGTGAAGGTAAGCATCCAGCC | D5d |
| D6d-cytr | GGGCCGCGGAAGTACGAGAGGATGAACC | N-terminal region of D6d |
| D6d-cytf | CCCTTCCGCGGCAATGGCTGGATTCCC | Middle and C-terminal regions of D6d |
| D5d-cytr | GGGTTCCGCGGAAGATCCAAAGAGTGAGC | N-terminal region of D5d |
| D5d-cytf | CCCTTCCGCGGAACTTCCTTGGTGCCC | Middle and C-terminal regions of D5d |
| Amino acid substitution of D6d with D5d | ||
| d6d5-1 | ACCGTCATCACGGCCGTTCTGCTTGCTACCTCCC | F166V, V167L |
| d6d5-2 | ACGGCCTTTGTCCTTTCTACCGTCCAGGCCCAAGCTGGA | A169S, S171V |
| d6d5-3 | GGCTACAACATGATTTTGGCCACCTTTCTGT | Y182F |
| d6d5-4 | GCCACCTTTCTGTCTTTAGCACCTCCATATGGAAC | Y188F, K189S, K190T |
| d6d5-5 | TTTCTGTCTATAAGAAATCCACATGGAACCACATTGTC | I192T |
| d6d5-6 | TCCATATGGAACCACCTTGTCCACCATTTTGTCATTGGCCACTT | I196L, K199H |
| d6d5-7 | CACTTAAAGGGTGCCCCCGCCAGCTGGTGGAACCATCG | S209P, N211S |
| d6d5-8 | AACTGGTGGAACCATATGCATTTCCAGCACCAT | R216M |
| d6d5-9 | CATGCGAAGCCCAACTGCTTCCGCAAGGACCCCGACAT | I226C, H228R |
| d6d5-10 | GGACCCCGACATAAACATGCACGTGTTTGTCC | K234N, S235M, L236Δ |
| d6d5-11 | ATAAAGAGCCTGCACCCATTGGTGTTTGTCCTTGGA | Δ238P, Δ239L |
| d6d5-12 | ATAAAGAGCCTGCACTTCTTTGCCCTTGGAGAGTGGCA | V238F, V240A |
| d6d5-13 | GTGTTTGTCCTTGGAAAGGTGCTGCCCCTCGAGTATGG | E243K, W244V, Q245L |
| d6d5-14 | CTTGGAGAGTGGCAGTCCGTCGAGCTTGGCAAGAAGAAGCTG | P246S, L247V, Y249L |
| d6d5-15 | CTCGAGTATGGCAAGGAGAAGAAGAAATATCTGCCCTA | K252E, L254K |
| d6d5-16 | AAGAAGAAGCTGAAACATATGCCCTACAACCACC | Y256H, L257M |
| d6d5-17 | TACAACCACCAGCATAAATACTTCTTCCTGA | E264K |
| d6d5-18 | ATCTTGGGAGCCCTGTGTCTTTTCAACTTTATCAGGT | V321C, F322L, L323F |
| d6d5-19 | GCCCTGGTTTTCCTCTTCATTGTCAGGTTCCTGGAGA | N324F, F325I, I326V |
| d6d5-20 | AGGTTCCTGGAGAGCAACTGGTTTGTGTGGG | H332N |
| d6d5-21 | CAGATGAACCACATTCCCATGCACATTGATCTTGATCAC | V344P, E346H |
| d6d5-22 | TCATGGAGATTGATCATGATCGCTACCGGGACTGGTTCA | L349H, H351R |
| d6d5-23 | ATTGATCTTGATCACAACGTGGACTGGTTCAGCAGC | Y352N, R353V |
| d6d5-24 | CACTACCGGGACTGGGTCAGCACCCAGCTGGCAGCCAC | F356V, S358T |
| d6d5-25 | TTCAGCAGCCAGCTGCAAGCCACCTGCAATGT | A361Q |
| d6d5-26 | GCCACCTGCAATGTGCACCAGTCCTTCTTCA | E367H |
| d6d5-27 | AATGTGGAGCAGTCCGCCTTCAATAACTGGTTCAGCGGGC | F370A, D373N |
| d6d5-28 | TGCCAAGACACAACTACCACAAGGTTGCCCCACTGGTGA | L396Y, I399V |
| d6d5-29 | AAGATTGCCCCACTGGTGCAGTCTCTCTGCGCCA | K404Q |
| d6d5-30 | TCTCTCTGCGCCAAGTATGGCATTAAATACCAAGAGAAGC | H410Y, E413K |
| d6d5-31 | CATGGCATTGAATACGAATCGAAGCCGCTGCTGAG | Q415E, E416S |
| d6d5-32 | AGAAGCCGCTGCTGACGGCCTTCGCCGACATTGTGAGTTC | R421T, L423F, L424A |
| d6d5-33 | CTGCTCGACATTGTGTATTCACTGAAGAAGTC | S428Y |
| d6d5-34 | GTGAGTTCACTGAAGGAGTCTGGGCAGCTGTGGCTGGATG | K432E, E435Q |
| d6d5-35 | GATGCCTACCTCCACCAATGAATCTAGTGAA | K444Q |
| Amino acid substitution of D5d with D6d | ||
| d6d5-36 | CACTTAAAGGGTGCCTCCGCCAGCTGGTGGAAC | P209S |
| d6d5-37 | AGGGTGCCCCCGCCAACTGGTGGAACCAT | S211N |
| d6d5-38 | CTGGTGGAACCATCGACATTTCCAGCACCAT | M216R |
| d6d5-39 | GGACCCCGACATAAAGATGCACCCATTGGTGT | N234K |
| d6d5-40 | ACCCCGACATAAACAGCCACCCATTGGTGTTTG | M235S |
| d6d5-41 | CCGACATAAACATGCTGCACCCATTGGTGTTTG | Δ236L |
| d6d5-42 | ATAAACATGCACTTGGTGTTTGTCCTTGGA | P238Δ |
| d6d5-43 | ATAAACATGCACCCAGTGTTTGTCCTTGGA | L239Δ |
| d6d5-44 | GTGTTTGTCCTTGGAGAGGTGCTGCCCCTCGA | K243E |
| d6d5-45 | TTTGTCCTTGGAAAGTGGCTGCCCCTCGAGTA | V244W |
| d6d5-46 | GTCCTTGGAAAGGTGCAGCCCCTCGAGTATGG | L245Q |
| d6d5-47 | CAGATGAACCACATTGTCATGCACATTGATCTT | P344V |
| d6d5-48 | AACCACATTCCCATGGAGATTGATCTTGATCAC | H346E |
| Amino acid substitution of D6d with zD5/6d | ||
| d6zebd5-1 | TCGTACTTCGGCACTGGCTGGATTCCC | N156T |
| d6zebd5-2 | ACCGTCATCACGGCCGTTGTCCTTGCTACCTCC | F166V |
| d6zebd5-3 | TGGCTACAACATGATTTCGGCCACCTTTCTGTC | Y182F |
| d6zebd5-4 | CACCTTTCTGTCTTCAAGACCTCCATATGGAAC | Y188F, K190T |
| d6zebd5-5 | TCCATATGGAACCACCTCGTCCACAAGTTTGTC | I195L |
| d6zebd5-6 | GGACCCCGACATAAATATGCTGCACGTGTTTG | K234N, S235M |
| d6zebd5-7 | CTTGGAGAGGTCCAGCCCGTCGAGTATGGC | W245V, L248V |
| d6zebd5-8 | AAGAAGAAGCTGAAACACCTGCCCTACAACCAC | Y257H |
| d6zebd5-9 | TACAACCACCAGCATAAGTACTTCTTCCTGATC | E265K |
| d6zebd5-10 | TCCAGTACCAGATCTTCATGACCATGATCAG | I284F |
| d6zebd5-11 | GCCATCAGCTACTATGTTCGTTTCTTCTACACC | A305V |
| d6zebd5-12 | TTGGGAGCCCTGGTTCTCTTCAACTTTATCAGGTTC | F322L, L323F |
| d6zebd5-13 | GTTTTCCTCAACTTTGTCAGGTTCCTGGAGAGC | I326V |
| d6zebd5-14 | CAGATGAACCACATTCCCATGGAGATTGATCTTG | V344P |
| d6zebd5-15 | ATTGATCTTGATCACAACCGGGACTGGTTCAGCAG | Y352N |
| d6zebd5-16 | AATGTGGAGCAGTCCGCCTTCAATGACTGGTTC | F370A |
| d6zebd5-17 | TGCCAAGACACAACTATCACAAGATTGCCCC | L396Y |
| d6zebd5-18 | CTCTGCGCCAAGTACGGCATTAAGTACCAAGAGAAG | H410Y |
| d6zebd5-19 | GCCAAGTACGGCATTAAATACCAAGAGAAGCCG | E413K |
| d6zebd5-20 | CGCTGCTGAGGGCCTTCGCTGACATTGTGAGTTC | L423F, L424A |
Amino acids are indicated by single characters. Δ indicates a gap in the amino acid sequence alignment.
Construction of chimeric desaturase genes
DNA fragments corresponding to the N-terminal region (cyt) and the central and C-terminal regions (des) of D6d (D6cyt and D6des) and D5d (D5cyt and D5des) were amplified by PCR using KOD Dash, the template plasmids, and the following sets of oligo primers (Table 1): D6cyt (amino acids 1–154), 24aF+, and D6d-cytr; D6des (amino acids 155–444), D6d-cytrf, and 24R+; D5cyt (amino acids 1–156), D5df, and D5d-cytr; and D5des (amino acids 157–447), D5d-cytf, and D5dr. The products were digested with Sac II at the coupling site, incubated at 70°C for 15 min to inactivate the enzyme, and ligated with T4 DNA ligase in the following combinations: D6cyt-D6des, D6cyt-D5des, D5cyt-D6des, and D5cyt-D5des. Each of the resultant fragments was adenylated with Ex Taq (Takara) at 72°C for 10 min, subcloned into the pGEM-T Easy vector, digested with KpnI and SalI, and ligated into pGEM-FLAG.
Site-directed mutagenesis
The oligonucleotide primers d6d5-1–d6d5-48 and d6zebd5-1–d6zebd5-20 (Table 1) were designed to introduce nucleotide mutations for substitution of amino acids in D6d and D5d with each of their D5d, zD5/6d, or D6d counterparts (see Fig. 1). Each mutation site was flanked by at least 15 nucleotides in each primer. For multiple-site mutagenesis, 3 or 4 primers carrying mutation site(s) at least 15 amino acid residues apart from each other were mixed in an equivalent molar ratio. The primers were phosphorylated at the 5′ end with T4 polynucleotide kinase (Takara). Plasmids carrying single or multiple mutation(s) were synthesized using pGEM-FLAG-D6d or pGEM-FLAG-D5d as a template, the phosphorylated primers, and the QuikChange multi site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) or a combination of the AMAP multi site-directed mutagenesis kit (MBL International, Woburn, MA) and Pfu DNA polymerase (Thermo Scientific Fermentas, Carlsbad, CA), according to the manufacturer’s instructions. The reaction mix was used to transform E. coli DH5α or XL10-Gold ultracompetent (Agilent Technologies) cells, and transformants were randomly selected to check the nucleotide sequences of the cloned DNA fragments.
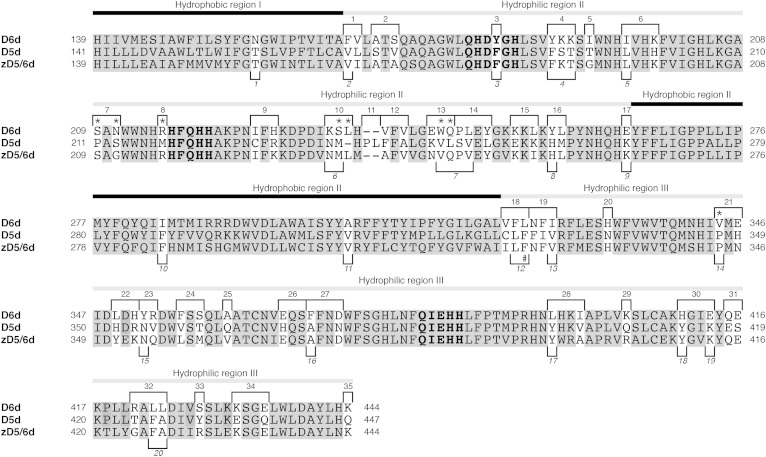
Fig. 1.
Alignment of the amino acid sequences of rat D6d and D5d and zebra fish zD5/6d. Site-directed mutagenesis was applied to create mutations at the sites shown with a white background using oligonucleotide primers indicated by respective numbers above (d6d5-1–d6d5-35) and below (d6zebd5-1–d6zebd5-20) the alignment. Conserved histidine clusters are indicated by bold letters. Asterisks indicate mutation sites that altered the substrate specificity from D6d-type to D5d-type; “#” indicates the mutation site that gave rise to Δ5/6 bifunctionality of D6d.
Expression of desaturase genes in yeast
The wild-type, chimera, and mutant desaturase genes were obtained by digestion of the pGEM-based plasmids with HindIII and EcoRI and were ligated into the yeast expression vector pYES2 (Invitrogen). The desaturase expression vectors were introduced into S. cerevisiae INVSc1 by using the lithium acetate method (26). Transformants were selected on uracil-deficient SD plates and cultivated at 28°C for 6 h with rotary shaking at 160 rpm in 15 ml of SCT medium supplemented with LA or DGLA at a concentration of 0.25 mM. After addition of galactose (2%, w/v) and further cultivation for another 16 h, yeast cells were recovered by centrifugation for fatty acid and protein analyses.
Fatty acid analysis
The yeast cells from ∼15 ml of broth were washed with distilled water and then vigorously vortexed in 2 ml of chloroform-methanol (2:1, v/v) plus 0.5 ml of distilled water. The chloroform phase was recovered by centrifugation, and methanolysis of total lipid was carried out by adding 1 ml of 10% methanolic hydrochloric acid (Tokyo Kasei, Tokyo, Japan) and heating at 60°C for 2 h. After evaporation of the solvents, fatty acid methyl esters (FAMEs) were extracted twice and dissolved in hexane. Fatty acid composition was determined using a gas chromatographic system (GC-17A and GC-2014; Shimadzu, Kyoto, Japan) equipped with a capillary column (TC-70, 0.25 mm × 30 m, GL Sciences, Tokyo, Japan; or Omegawax 250, 0.25 mm × 30 m, Sigma-Aldrich), a split injector (split ratio at 1:20–25; 270°C), and a flame ionization detector (270°C). The temperature of the column oven was maintained at 180°C (TC-70) or raised from 210°C to 225°C at 0.5°C/min (Omegawax 250). FAMEs were identified by comparing their retention time with those of the 37-Component FAME mix (Supelco, Bellefonte, PA) and by analyzing their molecular mass using MS. For GC/MS analysis, total lipid or FAME extracts were dissolved in 0.5 ml of 2-amino-2-methyl-1-propanol preheated at 75°C and were heated at 180°C for 24 h to form 4,4-dimethyloxazoline (DMOX) derivatives of fatty acids. After cooling to 75°C and adding 2 ml of distilled water preheated at 75°C, the DMOX derivatives were extracted several times with n-hexane/dichloromethane (2:3, v/v), dehydrated with anhydrous sodium sulfate, and analyzed on a GC/MS system consisting of a gas chromatograph (7890A, Agilent Technologies) equipped with a ZB-1HT Inferno capillary column (0.25 mm × 30 m; Phenomenex, Torrance, CA) and an electron ionization mass spectrometer (70 eV, JMS-T100GCV; JEOL, Tokyo, Japan). The enzymatic activity of the desaturase expressed in yeast was evaluated using the conversion ratio, which was determined as the ratio of the amount of product to the sum of the amounts of substrate and product and was expressed as a percentage.
SDS-PAGE and Western blotting
Yeast cells recovered from 1 ml of broth were washed with distilled water and suspended in 0.1 ml of 50 mM Tris-HCl (pH 7.5) containing 4 μl EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). An equivalent volume of glass beads (0.5 mm in diameter) was added to disrupt the cells by eight rounds of vortexing for 30 s and chilling on ice for 30 s. The homogenate was centrifuged at 5,000 g for 10 min and the supernatant was subjected to SDS-PAGE (27). The proteins separated in the gel were transferred to an Immobilon membrane (Merck Millipore, Darmstadt, Germany) using a semidry blotter. The membrane was blocked by immersing in 5% skim milk in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, 0.05% (w/v) Tween-20), and then moved to the same buffer containing mouse anti-FLAG antibody (Sigma-Aldrich; 1:5,000). After shaking for 1 h and washing with PBST, the membrane was probed with rabbit anti-mouse IgG (1:20000) for 1 h. The FLAG-tagged proteins were detected by using ECL plus (GE Healthcare) and exposure to X-ray film.
Statistical analysis
All experiments were performed at least twice. Student’s t-test was used to compare experimental values between groups where applicable. P < 0.05 was considered significant.
RESULTS
The N-terminal region of desaturase is not involved in substrate specificity
To examine the involvement of the N-terminal hydrophilic regions of D6d and D5d, including the cytochrome b5-like domain, in the substrate specificity of both desaturases, chimeras D5cyt-D6des and D6cyt-D5des were constructed and expressed in yeast in the presence of LA or DGLA. The results indicated that D5cyt-D6des and D6cyt-D6des converted LA into GLA, but did not act on DGLA, whereas D6cyt-D5des and D5cyt-D5des generated ARA from DGLA, but did not use LA as a substrate (Table 2). No other fatty acids, except spontaneous ones, were detected in all cases (data not shown). These results indicated that the N-terminal domains of both enzymes do not determine the specificity toward the corresponding substrates. However, the rate of conversion by D5cyt-D6des (8%) was substantially lower than that by D6cyt-D6des (40%), suggesting that a specific interaction between the N-terminal region and the central and C-terminal regions may contribute to maximum activity of D6d through conformational stabilization of the enzyme.
TABLE 2.
Substrate specificity of chimeric desaturases
| Desaturase | Rate of Substrate Conversion (%) | |
| LA to GLA | DGLA to ARA | |
| D6cyt-D6des | 40 | 0 |
| D5cyt-D6des | 8.0 | 0 |
| D6cyt-D5des | 0 | 45 |
| D5cyt-D5des | 0 | 45 |
Identification of amino acids responsible for D6d activity
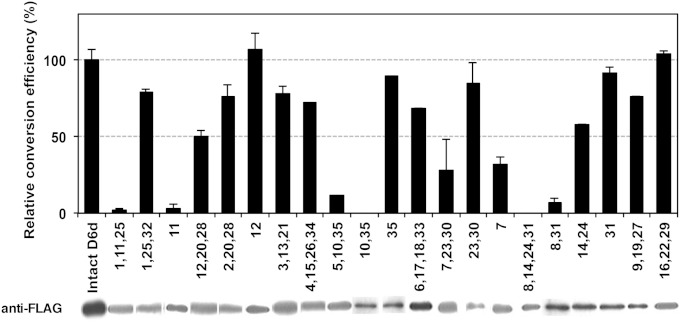
The amino acid sequence homology between D6d and D5d was 66% (67/101 amino acids) in the central hydrophilic region (hydrophilic region II) and 73% (91/124) in the C-terminal region (hydrophilic region III). To identify the amino acids involved in substrate specificity, site-directed mutagenesis was applied to the 67 nonidentical amino acids, which had been organized into 35 groups of 1–3 amino acid substitutions as depicted in Fig. 1. Multi site-directed mutagenesis using mixtures of three or four groups of oligonucleotide primers (d6d5-1–d6d5-35; Table 1) resulted in the generation of an array of mutant D6d genes encoding enzymes in which various (numbers of) amino acids were substituted with the corresponding D5d residues (Fig. 2). The mutant genes were individually expressed in S. cerevisiae in the presence of DGLA. Despite the successful expression of mutant proteins, none of the mutants generated ARA at a detectable level (data not shown). A series of expression experiments was then performed in the presence of LA to see whether the D6d activity of the mutants had been changed. As shown in Fig. 2, the D6d activity of four mutants constructed using the primer sets d6d5-1/11/25 (introducing the mutations F166V+V167L, Δ238P+Δ239L, A361Q), d6d5-5/10/35 (I192T, K234N+S235M+L236Δ, K444Q), d6d5-7/23/30 (S209P+N211S, Y352N+R353V, H410Y+E413K), and d6d5-8/14/24/31 (R216M, P246S+L247V+Y249L, F356V+S358T, Q415E+E416S) was significantly lower than that of intact D6d. Further analysis of the mutants generated by using fewer or single primer(s) revealed that the amino acid change(s) introduced by each of the primers d6d5-7 (S209P+N211S), d6d5-8/31 (R216M, Q415E+E416S), d6d5-10/35 (K234N+S235M+L236Δ, K444Q), and d6d5-11 (Δ238P+Δ239L) yielded decreased or null D6d activity. Because the single primer mutations d6d5-31 and d6d5-35 did not affect the D6d activity, the decreases in the activity by the mutations d6d5-8/31 and d6d5-10/35 could be due to the mutations by d6d5-8 and d6d5-10, respectively.
Fig. 2.
The D6d activity of D6d mutants. The D6d mutants were generated by site-directed mutagenesis using one or more oligonucleotide primers as indicated by the numbers, which correspond with the primer numbers (d6d5-1–d6d5-35; Table 1). The mutants carrying D5d amino acid(s) were expressed in yeast S. cerevisiae in the presence of LA, and the fatty acid composition was measured as described in the Materials and Methods. D6d activity was determined as the conversion rate from LA to GLA and is shown as the average value relative to that of intact D6d, together with the standard deviation (n = 3). FLAG-tagged D6d proteins were detected by Western blotting of total yeast protein using anti-FLAG antibody.
Switching the substrate specificity of D6d
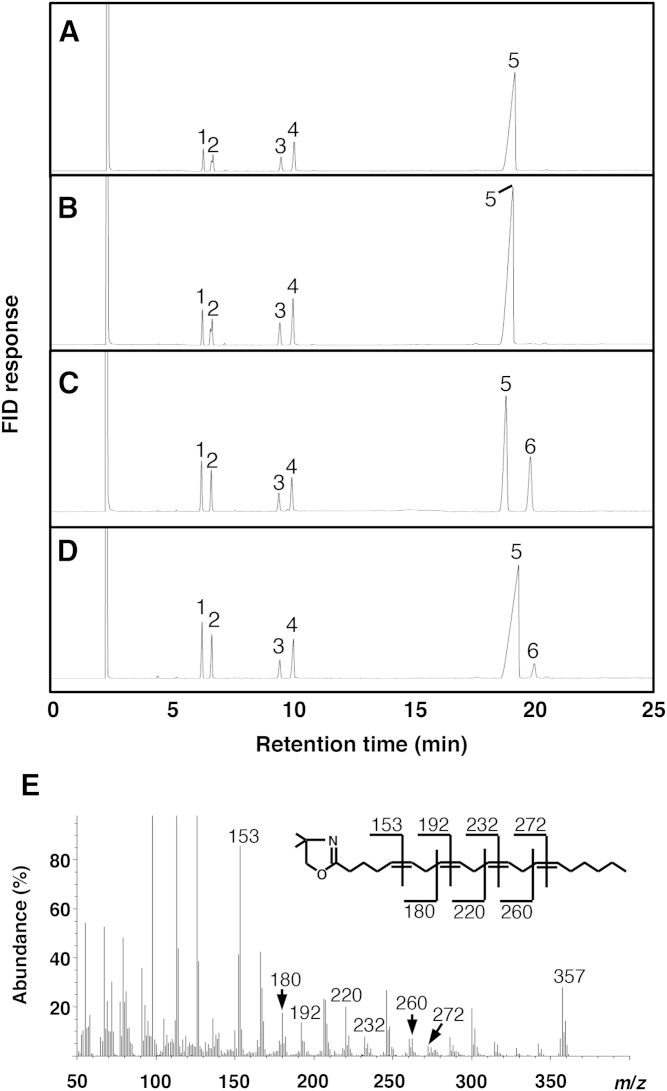
As expected, a D6d mutant made by using a mixture of the four primers d6d5-7, 8, 10, and 11 showed neither D6d nor D5d activity. Additional mutations were introduced into this mutant using several sets of primers randomly selected from d6d5-1 to d6d5-35, and D5d activity of the resultant mutants was investigated using DGLA substrate. Fig. 3D shows that one D6d mutant, namely, D6d-Z2, made using the primers d6d5-13 (E243K+W244V+Q245L) and d6d5-21 (V344P+E346H) in addition to d6d5-7/8/10/11 (S209P+N211S, R216M, K234N+S235M+L236Δ, Δ238P+Δ239L), gave a peak with a retention time similar to that of ARA (peak 6, Fig. 3D) on the chromatogram. By GC/MS analysis of its DMOX derivative, the generated fatty acid was identified as ARA on the basis of its total mass (m/z 357), the MS pattern of fragment ions, and a mass peak at m/z 153, characteristic of a Δ5 double bond (Fig. 3E). The conversion efficiency from DGLA to ARA of D6d-Z2 was 12% of that of intact D5d, whereas the efficiencies of the mutants made using the primers d6d5-13 (E243K+ W244V+Q245L) and d6d5-21 (V344P+E346H) were 8% and 2%, respectively. Other mutants made by using the primer sets d6d5-2/30 (A169S+S171V, H410Y+E413K), d6d5-5/16/24/33 (I192T, Y256H+L257M, F356V+S358T, S428Y), and d6d5-19/26/28 (N324F+F325I+I326V, E367H, L396Y+I399V) also generated ARA from DGLA; however, the efficiency was <2% of that of D5d in all cases. Because the enzymes carrying the mutations introduced by d6d5-13 (E243K+W244V+Q245L) showed the highest D5d activity and only the mutations introduced by d6d5-21 (V344P+E346H) were located at the C-terminal region of D6d, the mutant D6d-Z2, possessing both of those mutated regions, was used for further analysis.
Fig. 3.
Conversion of ARA from DGLA by D6d desaturase mutant D6d-Z2 carrying 12 D5d amino acids. Intact D6d (B), intact D5d (C), and D6d-Z2 (D) were expressed in yeast in presence of DGLA (20:3 Δ8,11,14; peak 5), and the generation of ARA (20:4 Δ5,8,11,14; peak 6) was detected by GC. Yeast harboring the pYES2 vector was used as negative control (A). Other peaks in A–D are 16:0 (peak 1), 16:1 Δ9 (peak 2), 18:0 (peak 3), and 18:1 Δ9 (peak 4). DMOX-derivative of ARA generated by the mutant D6d-Z2 (peak 6 in D) was identified by GC/MS analysis (E) as described in the Materials and Methods. Predicted molecular mass numbers of daughter ions of ARA are shown on the structural formula.
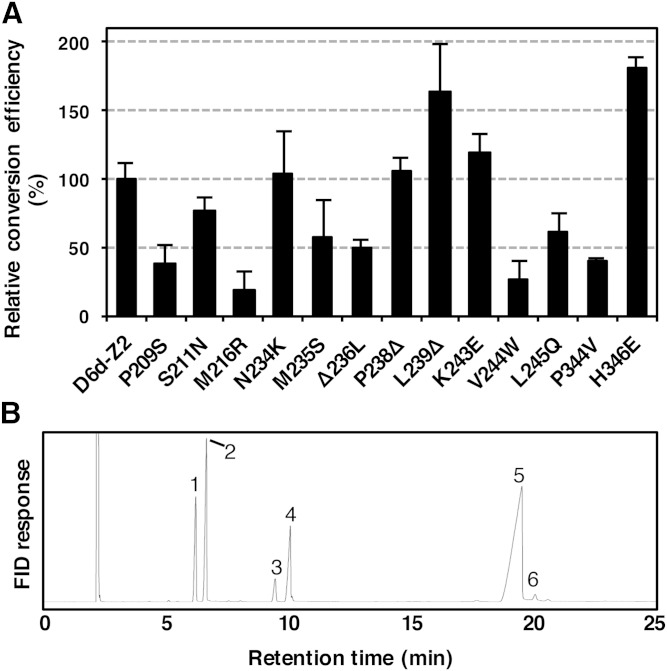
To exclude the mutations that do not contribute to the D5d activity of D6d-Z2, each of its 12 D5d amino acids was restored to its D6d counterpart by using the primers d6d5-36–d6d5-48. Expression analysis of the restored mutants in yeast revealed that mutations P209S, S211N, M216R, M235S, Δ236L, V244W, L245Q, and P344V decreased the D5d activity compared with that of D6d-Z2, whereas the mutations L239Δ and H346E boosted the activity (Fig. 4A). Thus, a new D6d mutant, namely, D6d-mut8, carrying the eight mutations S209P, N211S, R216M, S235M, L236Δ, W244V, Q245L, and V344P, was made and expressed in yeast in the presence of DGLA (Fig. 4B). The D5d activity of D6d-mut8 was 1.4%, equivalent to 30.1% and 4.7% of that of D6d-Z2 and intact D5d, respectively (Fig. 3 and supplementary Figure 1). Another D6d mutant carrying the substitutions R216M and W244V that yielded the lowest activity in the restoration experiment (Fig. 4A) was made, and its activity was examined. Although a fatty acid with a retention time similar to that of ARA was detected, GC/MS analysis did not allow structural identification due to an insufficient amount of fatty acid (data not shown).
Fig. 4.
The D5d activity of point mutants obtained from D6d-Z2. A: Each of D5d amino acid changes or the gap in D6d-Z2 was restored to its D6d counterpart by site-directed mutagenesis using oligonucleotide primers (d6d5-36–d6d5-48; Table 1). The mutants were expressed in yeast in the presence of DGLA, and the generation of ARA was detected by GC. D5d activity was determined as the conversion rate of DGLA to ARA and shown as the average value relative to that of D6d-Z2, together with the standard deviation (n = 3). B: D6d-mut8 carrying eight D5d-type amino acids was expressed in yeast in the presence of DGLA (peak 5), and the generation of ARA (peak 6) was detected by GC. Other peaks in B are 16:0 (peak 1), 16:1 Δ9 (peak 2), 18:0 (peak 3), and 18:1 Δ9 (peak 4).
Mutations conferring bifunctionality to D6d
The swapping of different amino acids within D6d with their D5d counterparts provided D6d with D5d activity as mentioned above; however, D6d activity was lost (data not shown). Thus, the rat desaturases were compared with the zebra fish [zD5/6d, which acts on both LA and DGLA (23)]. Twenty-five amino acids shared by D5d and zD5/6d, but not by D6d (Fig. 1), were selected as target sites for mutation. A D6d mutant carrying the 25 corresponding D5d and zD5/6d amino acids, namely, D6d-25m, was made by multi site-directed mutagenesis using the oligonucleotide primers d6zebd5-1–d6zebd5-20 (Table 1). We examined its substrate specificity using the yeast expression system and GC/MS analysis. D6d-25m could act on both LA and DGLA to generate GLA and ARA at conversion rates of 26.3% and 6.8%, respectively (Table 3; supplementary Fig. 2), proving the acquisition of D5d activity without losing D6d activity.
TABLE 3.
D5d and D6d activities of D6d mutants carrying zD5/6d-type amino acids
| Substitution | 1C-1 | 1C-2 | 1C-3 | 2C-1 | 2C-2 | 2C-3 | 2C-4 | 3C-1 | 3C-2 | 3C-3 | 4C-1 | 4C-2 | 4C-3 | 4C-4 | 5C-1 | D6d-25m |
| N156T | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| A305V | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Y182F | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| K234N | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| S235M | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| E365K | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| F166V | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| L423F | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| L424A | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| L248V | + | + | + | + | + | + | + | + | + | + | + | |||||
| F322L | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| L323F | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| I195L | + | + | + | + | + | + | + | + | + | + | ||||||
| W245V | + | + | + | + | + | + | + | + | + | + | ||||||
| L396Y | + | + | + | + | + | + | + | + | + | |||||||
| Y257H | + | + | + | + | + | + | + | + | ||||||||
| V344P | + | + | + | + | + | + | + | + | ||||||||
| I284F | + | + | + | + | + | + | + | |||||||||
| F370A | + | + | + | + | + | |||||||||||
| Y352N | + | + | + | + | + | |||||||||||
| I326V | + | + | + | + | + | |||||||||||
| Y188F | + | + | + | |||||||||||||
| K190T | + | + | + | |||||||||||||
| H410Y | + | + | + | |||||||||||||
| E413K | + | |||||||||||||||
| D5d activity (%) | 0 | 0 | 0 | 0 | 2.5 | 2.6 | 1.3 | 2.4 | 2.8 | 2.6 | 3.4 | 6.2 | 4.4 | 4.8 | 5.3 | 6.8 |
| D6d activity (%) | 16.3 | 17.4 | 13.7 | 5.3 | 11.6 | 10.0 | 13.5 | 13.0 | 13.0 | 16.0 | 13.8 | 11.0 | 21.3 | 21.3 | 26.8 | 26.3 |
D5d and D6d activities were represented as conversion rate from substrates (18:2 Δ9,12 for D6d and 20:3 Δ8,11,14 for D5d) to products (18:3 Δ6,9,12 for D6d and 20:4 Δ5,8,11,14 for D5d, respectively). The plus sign (+) indicates mutation points of each D6d mutant carrying zD5/6d-type amino acids.
To determine the mutations responsible for the bifunctional nature of D6d-25m, 15 mutants harboring fewer mutations, obtained during D6d-25m construction, were examined for their D5d activity. A remarkable increase in D5d activity was observed by introducing the mutations F322L/L323F (mutant 2C-2), I326V (4C-2), and E413K (D6d-25m) to the respective backgrounds (Table 3). Further analysis of single mutants for each of these four amino acids introduced into wild-type D6d demonstrated that only L323F led to the generation of ARA from DGLA at a conversion rate of 2.3% (Fig. 5 and supplementary Figure 3). Because addition of the mutations I326V and I326V/E413K to L323F did not significantly increase D5d activity, amino acids other than those might contribute to the full activity of D6d-25m.
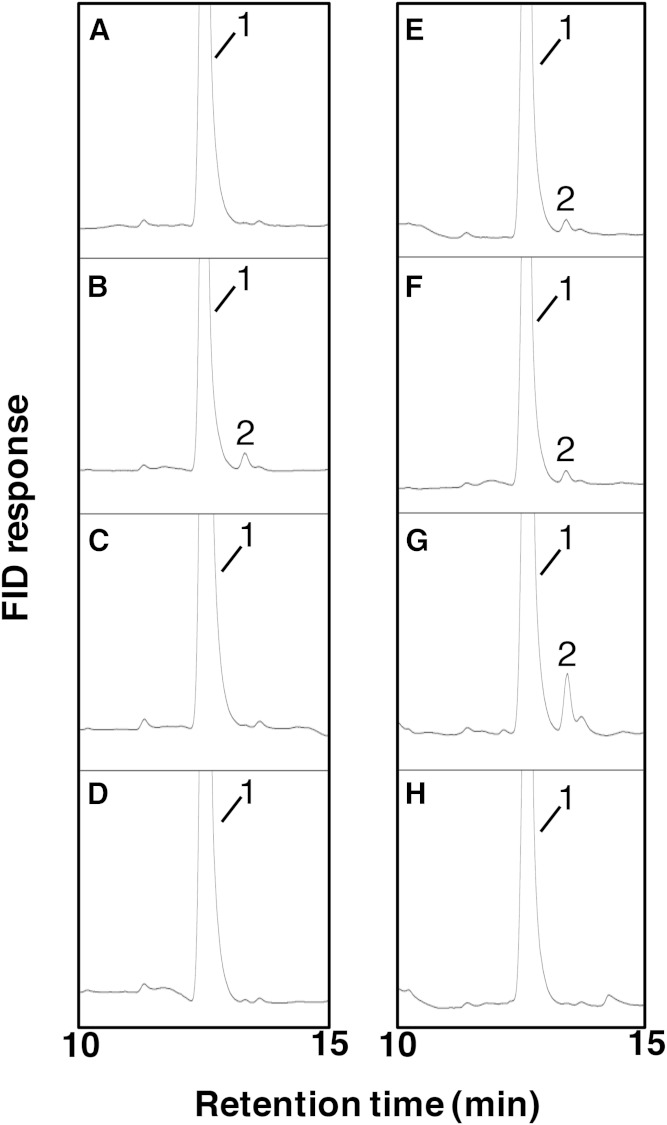
Fig. 5.
Conversion of ARA from DGLA by desaturase mutants with point mutation(s). A: D6d-F322L. B: D6d-L323F. C: D6d-I326V. D: D6d-E413K. E: D6d-L323F-I326V. F: D6d-L323F-I326V-E413K. G: D6d-25m. H: Intact D6d. Each mutant was expressed in yeast in the presence of DGLA (peak 1), and the generation of ARA (peak 2) was detected by GC.
Structure-function relationship
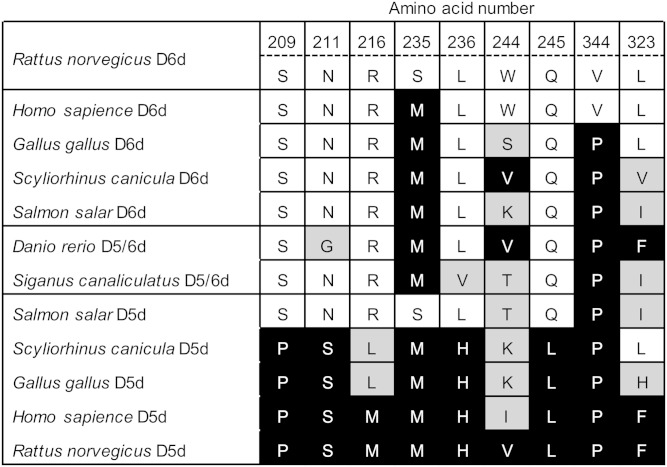
To explore the molecular evolution and functional divergence of front-end fatty acid desaturases, the mutations that conferred D5d activity to D6d in this study were compared with the corresponding residues in desaturases from various vertebrates (23, 28–34) as shown in Fig. 6. Most of the amino acids at positions 209, 211, 216, 236, and 245 of rat D6d are conserved among each group of D6d and D5d, and amino acids at those sites are D6d-type in teleost bifunctional desaturases. Therefore, these specificity-determining residues might be targets for functional modification of these types of desaturases. Further, the amino acids at positions 235 and 344 are highly conserved among D6d and D5d from most vertebrates, whereas those at positions 244 and 323 are variable. It is possible that these conserved amino acids cooperatively contribute to substrate recognition.
Fig. 6.
Comparison of the amino acid residues involved in the substrate specificity of rat D6d with corresponding residues in desaturases from various vertebrates. Amino acid residues identical to those in rat D6d (top row) and D5d (bottom row) are indicated with white and black backgrounds, respectively, and other amino acid residues are shown with a gray background.
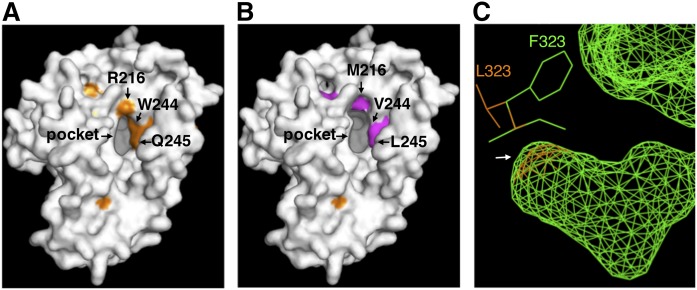
It is reasonable to assume that the substrate specificity and positioning are determined by the electric charge and polarity of the particular desaturase amino acids, which affect their affinity to the acyl chain and carrier portion of the substrate, and by the depth and angle of substrate insertion into the binding pocket (21). To evaluate the above-mentioned results of the protein engineering analysis, homology modeling of D6d was carried out on the basis of the recently reported crystal structure of human SCD1 (Protein Data Bank ID 4YMK) (24), using the structure prediction program Phyre2 (35). Amino acid residues of R216, W244, and Q245, located near the substrate-binding pocket (Fig. 7A), are considered to form hydrogen bonds with the pantothenic acid portion and the carbonyl group of acyl-CoA substrate, according to the findings for SCD1. Substitution of these amino acids with the corresponding D5d residues (M, V, and L, respectively) that do not form hydrogen bonds might alter the substrate-binding strength (Fig. 7B). Simultaneously, the substitutions R216M and W244V seem to cancel the steric hindrance just around the threshold of the pocket, allowing the substrate acyl chain to be inserted much deeper, resulting in the introduction of a carbon-carbon bond at the Δ5 position close to the catalytic site. L323 was predicted to be situated at the bottom of the substrate-binding pocket. The mutation L323F, which conferred Δ5/6 bifunctionality to D6d but might not alter the position of catalytic site nor deform the threshold of the pocket, would strengthen the hydrophobic affinity with the methyl terminus of the substrate acyl chain and/or create more space for insertion of the acyl chain (Fig. 7C).
Fig. 7.
Homology modeling of three-dimensional structures of rat D6d (A) and the mutants D6d-mut8 (B) and D6d-L323F (C). The models were generated by using the protein structure prediction program Phyre2, with the crystal structure of human stearoyl-CoA desaturase as a template. Amino acid residues that were identified as determinants for substrate specificity and that are located around the entrance of the substrate-binding pocket are indicated by arrows. C: Compares the predicted structures of the bottom of the internal substrate-binding cavity (an arrow) and the vicinal amino acid residues of intact D6d (orange) and D6d-L323F (green).
DISCUSSION
On the basis of the structural similarity of the enzymes, the desaturase family is also considered to include hydroxylase that produces hydroxyl fatty acids such as plant surface coating wax (36), conjugase that produces conjugated fatty acids with anticarcinogenesis activity (37), and acetylenase and epoxidase that produce fatty acids with a triple bond (38) and an epoxy group (39), respectively. Elucidation of the molecular basis of substrate recognition and regio- and chemoselectivity of the enzymes enables us to design new bioactive lipids and to produce them efficiently. In this study, rat D6d and D5d with highly homologous primary structures were used as a model to identify the sites critical for their mutually exclusive substrate specificity.
Heterologous expression analysis of chimeric enzymes of D6d and D5d, in which the cytochrome b5-like domains were swapped, demonstrated that these domains do not contribute to substrate recognition (Table 2). However, the D6d activity of the chimera D5cyt-D6des was significantly lower than that of intact D6d, suggesting that the cytochrome b5-like domain might be necessary for full activity of D6d, but not D5d. Therefore, a D6d mutant with D5d activity (equivalent to D6cyt-D5des) would be preferable over the reverse to detect declined desaturase activity. Indeed, the D6d-based mutant D6d-mut8 barely showed D5d activity (Fig. 4B), whereas the introduction of mutations at the corresponding sites in D5d did not result in the generation of detectable D6d product (data not shown). Moreover, given that the distance from the carboxyl group of the fatty acid substrate to the position to be desaturated is considered to be larger in D6d than in D5d, the substrate range of D5d is expected to be wider than that of D6d (40). Thus, site-directed mutagenesis was applied to the D6d gene to readily observe successful conversion of substrate specificity.
We assumed that structural differences between D6d and D5d due to some of the 67 nonconserved amino acids in their hydrophilic regions (Fig. 1) would confer altered substrate specificity. To our knowledge, this study is the first to identify specific positions that are involved in alteration of the substrate selectivity of a mammalian front-end fatty acid desaturase. On the basis of heterologous expression analyses of a series of D6d mutants, we identified eight mutations (S209P, N211S, R216M, S235M, L236Δ, W244V, Q245L, and V344P) that abolished D6d activity from D6d but conferred D5d activity (Fig. 4B). It is obvious that several amino acid residues are necessary to determine the substrate specificity as well as to support maximum enzymatic activity. The K218 residue of M. rouxii D6d has been reported to be involved in binding of the substrate (15); however, mutation of the corresponding amino acid in rat, R216, did not yield D5d activity (data not shown). Compared with the D6d-Z2 mutant carrying 12 mutations and with D6d activity 4.6% that of the wild-type D6d (Fig. 3), the D6d-mut8 mutant carrying eight mutations showed much lower activity (1.4%; Fig. 4B), and the D6d product was not detected in the double mutant R216M/W244V (data not shown). The fact that all of the restored mutants shown in Fig. 4A retained D5d activity suggested that more than two critical amino acid residues exist in each group of mutations introduced with the primers d6d5-7/8/10/11 and d6d5-13/21.
The mammalian D6d and D5d and zebra fish zD5/6d might have evolved from a common ancestor enzyme (33). Site-directed mutagenesis targeting the residues identical between D5d and zD5/6d but not D6d resulted in the generation of a mutant D6d-25m possessing bifunctional activity (Table 3) and pinpointed L323F as responsible for providing D5d activity to D6d (Fig. 5). However, the mutation L323F was overlooked in the first mutagenesis experiment based on the sequence comparison of only D5d and D6d and the use of multimutagenic primers. This might be because the D5d activity of the D6d-L323F mutant was below the detection limit and/or the other amino acid mutations introduced by the primer (d6d5-18; V321C+F322L+L323F) counteracted its effect. By using two different approaches in the mutagenesis experiments, a broad-horizon search was achieved, resulting in the determination of the amino acid residues responsible for both switching and adding the substrate specificity of D6d.
In addition, the predicted D6d tertiary structure supported our findings at least in part on the molecular basis of substrate specificity of the fatty acid desaturases. This knowledge will largely contribute to furthering our understanding of the structure-function relationship and the molecular evolution of the desaturase family and to generating structurally and functionally novel fatty acyl compounds for industrial applications.
Supplementary Material
Acknowledgments
The authors thank Dr. Amimoto for her kind support with MS analyses.
Footnotes
Abbreviations:
- ACP
- acyl carrier protein
- ARA
- arachidonic acid
- D5d
- Δ5 desaturase
- D6d
- Δ6 desaturase
- DGLA
- dihomo-γ-linolenic acid
- DMOX
- 4,4-dimethyloxazoline
- FAME
- fatty acid methyl ester
- GLA
- γ-linolenic acid
- LA
- linoleic acid
- zD5/6d
- zebra fish bifunctional Δ5/6 desaturase
The authors appreciate the financial support of all our group members of Core Research for Evolutional Science and Technology (CREST) and Japan Science and Technology Agency (JST). The authors have no conflicts of interest to declare.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.McKeon T. A., and Stumpf P. K.. 1982. Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-acyl carrier protein thioesterase from maturing seeds of safflower. J. Biol. Chem. 257: 12141–12147. [PubMed] [Google Scholar]
- 2.Watts J. L., and Browse J.. 1999. Isolation and characterization of a Δ5-fatty acid desaturase from Caenorhabditis elegans. Arch. Biochem. Biophys. 362: 175–182. [DOI] [PubMed] [Google Scholar]
- 3.Knutzon D. S., Thurmond J. M., Huang Y. S., Chaudhary S., Bobik E. G., Chan G. M., Kirchner S. J., and Mukerji P.. 1998. Identification of Δ5-desaturase from Mortierella alpina by heterologous expression in bakers’ yeast and canola. J. Biol. Chem. 273: 29360–29366. [DOI] [PubMed] [Google Scholar]
- 4.Guy J. E., Whittle E., Moche M., Lengqvist J., Lindqvist Y., and Shanklin J.. 2011. Remote control of regioselectivity in acyl-acyl carrier protein-desaturases. Proc. Natl. Acad. Sci. USA. 108: 16594–16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayanova O., Shewry P. R., and Napier J. A.. 1999. Histidine-41 of the cytochrome b5 domain of the borage Δ6 fatty acid desaturase is essential for enzyme activity. Plant Physiol. 121: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gostinčar C., Turk M., and Gunde-Cimerman N.. 2010. The evolution of fatty acid desaturases and cytochrome b5 in eukaryotes. J. Membr. Biol. 233: 63–72. [DOI] [PubMed] [Google Scholar]
- 7.Michinaka Y., Aki T., Inagaki K., Higashimoto H., Shimada Y., Nakajima T., Shimauchi T., Ono K., and Suzuki O.. 2001. Production of polyunsaturated fatty acids by genetic engineering of yeast. J. Oleo Sci. 50: 359–365. [Google Scholar]
- 8.Guillou H., Rioux V., Catheline D., Thibault J-N., Bouriel M., Jan S., D’Andrea S., and Legrand P.. 2003. Conversion of hexadecanoic acid to hexadecenoic acid by rat Δ 6-desaturase. J. Lipid Res. 44: 450–454. [DOI] [PubMed] [Google Scholar]
- 9.Shanklin J., Whittle E., and Fox B. G.. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 33: 12787–12794. [DOI] [PubMed] [Google Scholar]
- 10.Sayanova O., Beaudoin F., Libisch B., Castel A., Shewry P. R., and Napier J. A.. 2001. Mutagenesis and heterologous expression in yeast of a plant Δ6-fatty acid desaturase. J. Exp. Bot. 52: 1581–1585. [DOI] [PubMed] [Google Scholar]
- 11.Meesapyodsuk D., and Qiu X.. 2012. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 47: 227–237. [DOI] [PubMed] [Google Scholar]
- 12.Sasata R. J., Reed D. W., Loewen M. C., and Covello P. S.. 2004. Domain swapping localizes the structural determinants of regioselectivity in membrane-bound fatty acid desaturases of Caenorhabditis elegans. J. Biol. Chem. 279: 39296–39302. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M., Hornung E., Busch S., Kassner N., Ternes P., Braus G. H., and Feussner I.. 2007. A small membrane-peripheral region close to the active center determines regioselectivity of membrane-bound fatty acid desaturases from Aspergillus nidulans. J. Biol. Chem. 282: 26666–26674. [DOI] [PubMed] [Google Scholar]
- 14.Song L. Y., Zhang Y., Li S. F., Hu J., Yin W. B., Chen Y. H., Hao S. T., Wang B. L., Wang R. R. C., and Hu Z. M.. 2014. Identification of the substrate recognition region in the Δ6-fatty acid and Δ8-sphingolipid desaturase by fusion mutagenesis. Planta. 239: 753–763. [DOI] [PubMed] [Google Scholar]
- 15.Na-Ranong S., Laoteng K., Kittakoop P., Tanticharoen M., and Cheevadhanarak S.. 2006. Targeted mutagenesis of a fatty acid Δ6-desaturase from Mucor rouxii: role of amino acid residues adjacent to histidine-rich motif II. Biochem. Biophys. Res. Commun. 339: 1029–1034. [DOI] [PubMed] [Google Scholar]
- 16.Lim Z. L., Senger T., and Vrinten P.. 2014. Four amino acid residues influence the substrate chain-length and regioselectivity of Siganus canaliculatus Δ4 and Δ5/6 desaturases. Lipids. 49: 357–367. [DOI] [PubMed] [Google Scholar]
- 17.Meesapyodsuk D., and Qiu X.. 2014. Structure determinants for the substrate specificity of acyl-CoA Δ9 desaturases from a marine copepod. ACS Chem. Biol. 9: 922–934. [DOI] [PubMed] [Google Scholar]
- 18.Vanhercke T., Shrestha P., Green A. G., and Singh S. P.. 2011. Mechanistic and structural insights into the regioselectivity of an acyl-CoA fatty acid desaturase via directed molecular evolution. J. Biol. Chem. 286: 12860–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broun P., Boddupalli S., and Somerville C.. 1998. A bifunctional oleate 12-hydroxylase: desaturase from Lesquerella fendleri. Plant J. 13: 201–210. [DOI] [PubMed] [Google Scholar]
- 20.Gagné S. J., Reed D. W., Gray G. R., and Covello P. S.. 2009. Structural control of chemoselectivity, stereoselectivity, and substrate specificity in membrane-bound fatty acid acetylenases and desaturases. Biochemistry. 48: 12298–12304. [DOI] [PubMed] [Google Scholar]
- 21.Rawat R., Yu X. H., Sweet M., and Shanklin J.. 2012. Conjugated fatty acid synthesis: residues 111 and 115 influence product partitioning of Momordica charantia conjugase. J. Biol. Chem. 287: 16230–16237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aki T., Shimada Y., Inagaki K., Higashimoto H., Kawamoto S., Shigeta S., Ono K., and Suzuki O.. 1999. Molecular cloning and functional characterization of rat Δ6 fatty acid desaturase. Biochem. Biophys. Res. Commun. 255: 575–579. [DOI] [PubMed] [Google Scholar]
- 23.Hastings N., Agaba M., Tocher D. R., Leaver M. J., Dick J. R., Sargent J. R., and Teale A. J.. 2001. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc. Natl. Acad. Sci. USA. 98: 14304–14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Klein M. G., Zou H., Lane W., Snell G., Levin I., Li K., and Sang B-C.. 2015. Crystal structure of human stearoyl–coenzyme A desaturase in complex with substrate. Nat. Struct. Mol. Biol. 22: 581–585. [DOI] [PubMed] [Google Scholar]
- 25.Bai Y., McCoy J. G., Levin E. J., Sobrado P., Rajashankar K. R., Fox B. G., and Zhou M.. 2015. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 524: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito H., Fukuda Y., and Murata K.. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 28.Cho H. P., Nakamura M. T., and Clarke S. D.. 1999. Cloning, expression, and nutritional regulation of the mammalian Δ6 desaturase. J. Biol. Chem. 274: 471–477. [DOI] [PubMed] [Google Scholar]
- 29.Cho H. P., Nakamura M., and Clarke S. D.. 1999. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274: 37335–37339. [DOI] [PubMed] [Google Scholar]
- 30.International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 432: 695–716. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X., Tocher D. R., Dickson C. A., Bell J. G., and Teale A. J.. 2005. Highly unsaturated fatty acid synthesis in vertebrates: new insights with the cloning and characterization of a Δ6 desaturase of Atlantic salmon. Lipids. 40: 13–24. [DOI] [PubMed] [Google Scholar]
- 32.Hastings N., Agaba M. K., Tocher D. R., Zheng X., Dickson C. A., Dick J. R., and Teale A. J.. 2004. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar). Mar. Biotechnol. (NY). 6: 463–474. [DOI] [PubMed] [Google Scholar]
- 33.Castro L. F. C., Monroig Ó., Leaver M. J., Wilson J., Cunha I., and Tocher D. R.. 2012. Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS One. 7: e31950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y. Y., Hu C. B., Zheng Y. J., Xia X. A., Xu W. J., Wang S. Q., Chen W. Z., Sun Z. W., and Huang J. H.. 2008. The effects of dietary fatty acids on liver fatty acid composition and Δ6-desaturase expression differ with ambient salinities in Siganus canaliculatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151: 183–190. [DOI] [PubMed] [Google Scholar]
- 35.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckhardt M., Yaghootfam A., Fewou S. N., Zöller I., and Gieselmann V.. 2005. A mammalian fatty acid hydroxylase responsible for the formation of alpha-hydroxylated galactosylceramide in myelin. Biochem. J. 388: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyer J. M., Chapital D. C., Kuan J. W., Mullen R. T., Turner C., Mckeon T. A., and Pepperman A. B.. 2002. Molecular analysis of a bifunctional fatty acid conjugase/desaturase from tung. Implications for the evolution of plant fatty acid diversity. Plant Physiol. 130: 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperling P., Lee M., Girke T., Za U., Stymne S., and Heinz E.. 2000. A bifunctional Δ6-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus. Eur. J. Biochem. 267: 3801–3811. [DOI] [PubMed] [Google Scholar]
- 39.Hamberg M., and Fahlstadius P.. 1992. On the specificity of a fatty acid epoxygenase in broad bean (Vicia faba L.). Plant Physiol. 99: 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong H., Datla N., MacKenzie S. L., and Qiu X.. 2002. Isolation and characterization of a Δ5 FA desaturase from Pythium irregulare by heterologous expression in Saccharomyces cerevisiae and oilseed crops. Lipids. 37: 863–868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.