Abstract
Induced pluripotent stem cells (iPSCs) are somatic cells that have been transcriptionally reprogrammed to an embryonic stem cell (ESC)-like state. iPSCs are a renewable source of diverse somatic cell types and tissues matching the original patient, including nephron-like kidney organoids. iPSCs have been derived representing several kidney disorders, such as ADPKD, ARPKD, Alport syndrome, and lupus nephritis, with the goals of generating replacement tissue and ‘disease in a dish’ laboratory models. Cellular defects in iPSCs and derived kidney organoids provide functional, personalized biomarkers, which can be correlated with genetic and clinical information. In proof of principle, disease-specific phenotypes have been described in iPSCs and ESCs with mutations linked to polycystic kidney disease or focal segmental glomerulosclerosis. In addition, these cells can be used to model nephrotoxic chemical injury. Recent advances in directed differentiation and CRISPR genome editing enable more specific iPSC models and present new possibilities for diagnostics, disease modeling, therapeutic screens, and tissue regeneration using human cells. This review outlines growth opportunities and design strategies for this rapidly expanding and evolving field.
Keywords: ADPKD, ARPKD, cilia, proximal tubule, KIM-1, regeneration, diabetes mellitus, diabetes insipidus, Wolfram syndrome, Alport syndrome, lupus nephritis, focal segmental glomerulosclerosis, podocyte, podocalyxin, in vitro clinical trials, macular degeneration, ESC, genome editing, CRISPR, transcriptome, organ replacement, cell therapy
I. Fundamental Concepts
1. Introduction
Induced pluripotent stem cells (iPSCs) are somatic cells that have been transcriptionally reprogrammed to an embryonic stem cell (ESC)-like state. Similar to ESCs, iPSCs have the potential to be used to bioengineer immunocompatible tissue or to model patient-specific disease in the laboratory. The kidney is the most commonly transplanted human organ. Many different disorders can lead to chronic kidney disease (CKD), each with specific pathophysiologies. For the vast majority of kidney diseases, there are no specific human disease models available, and no specific treatments or biomarkers. Human iPSCs from patients with kidney diseases represent a new model system in which to investigate pathophysiology and develop more effective therapeutics. Here, we review the potential of iPSCs for modeling kidney diseases based on the primary literature. Strategies for making effective comparisons between patient iPSCs are discussed. Although the focus is on the kidney, many of the principles are relevant to other organs.
2. Human pluripotent stem cells can differentiate into all somatic cell types
The term ‘human pluripotent stem cells (hPSCs)’ has been applied historically to a variety of different cell types, with distinct origins and properties. For the purposes of this review, hPSCs will be defined as the cultured equivalents of the specific cell population within the blastocyst-stage embryo that gives rise to the entire body. hPSCs are both pluripotent, meaning they can differentiate into any type of somatic cell in the body, and self-renewing, meaning they are capable of extensive replication without senescence or differentiation. This combination of pluripotency and self-renewal distinguishes hPSCs from other types of cultured cells, and makes them a powerful tool for regenerative medicine and human disease modeling.1 hPSCs include ESCs, which are primary cultures of human blastocyst-stage embryos, and iPSCs, which are somatic cells ‘reprogrammed’ to an ESC-like state.1,2 These two cell types are highly similar, to the point that genome-wide gene expression analysis cannot easily distinguish between them.3
The invention of iPSCs by Kazutoshi Takahashi and Shinya Yamanaka, who first described the technique in 2006, marks a significant advance for research involving hPSCs.4 To produce iPSCs, a combination of master transcription factors typically expressed in ESCs (such as OCT4, SOX2, KLF4, and c-MYC) are transiently expressed in somatic cells, inducing the expression of additional ESC-related genes. Over a period of weeks in ESC culture conditions, a small minority of somatic cells is transcriptionally reprogrammed into iPSCs, which are very similar to ESCs (Fig. 1).2,5–7 Although the efficiency of iPSC reprogramming is typically quite low (<1.0% of starting cells), the extensive self-renewal capability of iPSCs (like ESCs) enables production of essentially unlimited numbers of cells from a single iPSC colony. With respect to application, iPSCs are particularly significant for two reasons. First, they can be differentiated into transplantable cells and tissues, which are predicted to be fully immunocompatible with the original patient from whom they derive.8–12 Second, because iPSCs can be generated from any patient, they provide a means of producing in vitro models harboring naturally occurring genetic mutations.1,2,13–15 Such models, if indicative of a disease state, can be considered a type of patient-specific biomarker (see below Section 6, iPSCs Present Opportunities for Biomarker Discovery and Development). iPSC research represents a new paradigm for both human cellular therapy and disease modeling (Fig. 2). The invention of iPSCs in 2006 was so significant that Dr. Yamanaka was awarded the Nobel Prize in Medicine only six years later, in 2012.2,4
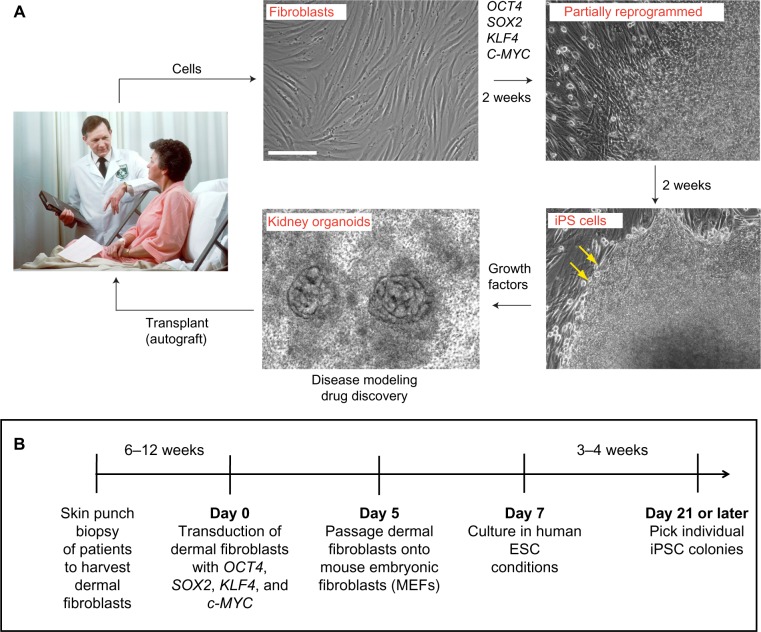
Figure 1.
Generation of iPSCs from kidney disease patients. (A) Representative images and (B) time line of iPSC generation from skin fibroblasts. At 4 weeks, mature iPSC colonies display characteristic rounded edges (yellow arrows). The figure is based on the protocol described in reference 2; alternative protocols, transcription factor combinations, and timelines are also possible. Scale bar, 200 μm.
Note: The image of doctor and patient in this figure was obtained from the National Cancer Institute and is the work of photographer Bill Branson. It is in the public domain.
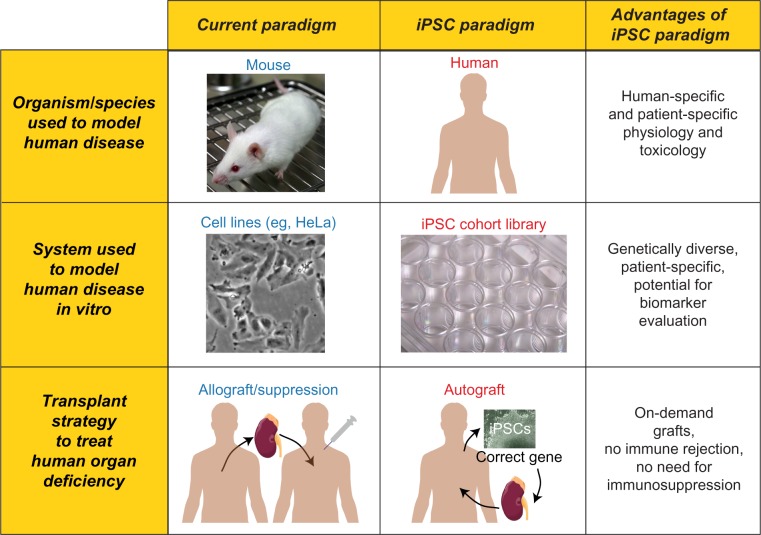
Figure 2.
Current paradigms and iPSC paradigms for research and therapeutics. Advantages of the iPSC paradigm are highlighted in the rightmost column. Top row: the current research paradigm relies primarily on nonhuman model species, such as mice, to model human disease. In contrast, the iPSC paradigm enables the study of human pathophysiology in human cells. Middle row: the current research paradigm relies on immortalized cell populations, such as HeLa cells, for disease modeling comparisons in vitro. In contrast, the iPSC paradigm promotes the development of genetically diverse iPSC cohorts/libraries. Bottom row: compared to the current therapeutic paradigm of kidney transplant, iPSC-derived autograft transplantation is envisioned, in combination with gene therapy.
Note: The image of the lab mouse in this figure, created by Rama and obtained from Wikimedia Commons, is reused under a CC-BY-SA license. The image of a multi-well cell culture plate, created by Lilli_M as part of Wikiproject LabSnap 2011, was obtained from Wikimedia Commons and reused under a CC-BY-SA license.
Pluripotent stem cells are believed to have the potential to generate any type of cell present in the embryonic or adult body. Using a technique called tetraploid complementation, entire mice have been generated from clonal populations of mouse ESCs and iPSCs.16,17 This experiment cannot currently be reproduced in humans, due to technical, ethical, and safety concerns. To demonstrate that human ESCs and iPSCs are pluripotent, they are implanted into immunodeficient animals and allowed to differentiate into large tumors called teratomas, which contain human tissues representing the three germ layers of the embryo.1,4 The complexity and diversity of these tissues demonstrates the potential of hPSCs as a next-generation cellular model for multiple organs (Fig. 3A). Using detailed regimens of specific growth factors, hPSCs can also been directed to differentiate into numerous types of cells and tissues, which in some cases can form functional xenografts in immunodeficient animals.10–12 The generation of kidney ‘organoids’ from hPSCs, containing multiple cell types in nephron-like architectures, is one recent example that illustrates the ability of differentiating hPSCs to self-assemble into complex structures (Fig. 3B; see below Section 7, Kidney Organoid Differentiation from hPSCs Enables Human Disease Models).18–21 hPSCs therefore provide a reproducible source of human tissues for investigation, both in vitro and in vivo.
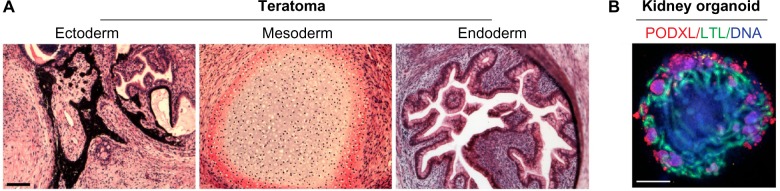
Figure 3.
Generation of complex tissues from hPSCs. (A) Teratoma sections stained with hematoxylin and eosin showing formation of pigmented epithelium (ectoderm), cartilage (mesoderm), and secretory crypts (endoderm) 10 weeks after injection of hPSCs beneath the kidney capsule of a NOD. SCID immunodeficient mouse. (B) Kidney organoid differentiated from a patient iPSC line, showing LTL+ tubules surrounded by peripheral clusters of PODXL+ podocyte-like cells. Image reproduced with permission from reference 20. Scale bars, 200 μm.
Note: Images are reused from Freedman BS, Brooks CR, Lam AQ, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nature Communications. Oct 23 2015;6:8715:1–13, under the terms of a CC-BY license.
3. hPSCs represent a possible source of new nephrons for research and transplant
Adult humans have a limited number of kidney nephrons and are incapable of generating new nephron units in a clinically useful capacity. Studies suggest that nephrogenesis in mammals ceases in utero or soon after birth, concomitant with the depletion of nephron progenitor cells (NPCs).22,23 In the adult, kidney tubular epithelial cells (KTECs) can proliferate and repair tubular segments after injury, but no adult cell population has been identified with the capacity to replace lost nephrons.24,25 KTECs are thus developmentally restricted to a more mature cell fate. In contrast to adult KTECs, hPSCs represent a very early developmental stage, well before the kidney has even formed. hPSCs self-renew extensively in vitro, without undergoing de-differentiation or senescence.1,2 This differs from fetal NPCs, which differentiate or apoptose when removed from their metanephric niche and serially passaged.26,27 hPSCs therefore represent the only long-term cultivatable cell type with the potential to generate new human nephrons.
For the kidney, iPSCs present two major opportunities (Figs. 1–2). The first is to produce new kidney tissue for transplantation, which could potentially be derived from patients and administered without immunosuppression as a renal replacement therapy.2,8,9 This goal may not be achievable in a short time frame, due to the complexities of kidney architecture, safety concerns, and the partially effective gold standards of dialysis and allograft transplant. The second application of iPSCs for the kidney is to develop ‘disease in a dish’ laboratory models of human kidney disease. As iPSCs self-renew extensively, cell lines from patients with kidney disease can be propagated indefinitely and distributed to any lab for reproducible experiments. For the vast majority of kidney diseases, iPSCs would be the only available human laboratory models, or perhaps the only laboratory models of any kind.
Whereas primary KTECs tend to dedifferentiate in culture, hPSCs differentiate into NPCs and subsequently KTECs along a developmental path. The epithelial structures that result from this differentiation process may more closely approximate morphogenesis and physiology than primary cells displaced from their tissue microenvironment in vivo. Another major advantage of hPSCs for disease modeling experiments is their extensive genetic diversity. iPSCs are now available representing hundreds, if not thousands, of patients.13,14 iPSCs harboring naturally occurring human mutations that cause renal disease can be generated directly from patient cells, without requiring targeted mutagenesis.15 Phenotypes identified in vitro can then be compared to clinical data from the original patient.
4. iPSCs complement mouse models and genetics
For the kidney, as for other tissues, iPSCs and mice are complementary model systems. Mice have several obvious advantages over iPSCs. iPSCs are unlikely, in the near term, to achieve the level of organ function and organization typical of an adult animal such as the mouse. Likewise, there exists no obvious way to integrate iPSC tissues from one organ system, such as the kidney, with other organ systems, such as the cardiovascular system. As kidney disease is multi-faceted and can both influence and be influenced by pathophysiologies in other organ systems, studies examining such relationships (eg, the role of hypertension in causing kidney failure) are likely to benefit more from animal models than iPSCs. From a genetics standpoint, mice can be crossed, whereas no equivalent technology exists for cultured human cell lines, including iPSCs.
iPSCs also have certain advantages over the mouse as a laboratory model. iPSCs can be readily generated from human patients, with naturally occurring genetic mutations that cause inherited disease. Such mutations may have species-specific effects which cannot be reproduced in mice.28 For the kidney, one example of such species specificity is autosomal dominant polycystic kidney disease (ADPKD). Human ADPKD is inherited as heterozygous loss-of-function mutations in either PKD1 or PKD2, which encode polycystin-1 (PC1) and polycystin-2 (PC2), respectively.29,30 In mice, however, germline Pkd1 and Pkd2 heterozygotes display only very mild cystic disease.31,32 Compared to rodents, iPSCs may therefore provide a more species-specific model for some aspects of human pathophysiology. iPSCs are also more accessible to microscopic inspection, experimental manipulation, and high-throughput analysis than living tissue in vivo. iPSCs are therefore well suited to drug discovery, being of human origin, capable of complex tissue differentiation, genetically diverse (derived from patients with assorted human diseases), and amenable to high-throughput screening approaches.33 One hope for the field is that iPSC research will lead more quickly to drugs that will work in humans, since many therapeutics that are effective in mice fail to confer benefit in human clinical trials.14
iPSCs are also a potentially valuable tool for human genetics research. Whole-exome and whole-genome sequencing technologies hold great promise for identifying disease-causative mutations. Ultimately, however, the human genome is extremely complex and every patient will have many ‘candidate’ mutations.34 Functional confirmation is therefore required to rigorously establish a causative relationship between genotype and phenotype. iPSCs derived from patients within the sequenced genetic cohort are well-suited for such experiments. The rate-limiting step is the development of assays which can reveal phenotypes specific to the disease in question. As these assays become more robust and accessible, iPSCs will be increasingly adopted by geneticists as a model system.
Conversely, while iPSCs can be derived from any patient, it becomes much more challenging to define a mutant phenotype when the disease mutations have not been sequenced. Many diseases and syndromes resemble each other phenotypically and many human disease genes are simply not known.34 For iPSCs, most phenotypic assays are still being developed and will require significant validation. Knowledge of the disease genes and the underlying mutations is particularly important for performing rescue experiments which validate the phenotype. In this regard, the best starting material for iPSC experiments are somatic cells from patients in which the disease-causative mutations have been identified with a strong likelihood.14 For some hereditary kidney diseases, such as ADPKD, the candidate genes are known and relatively few; for many others, there is a long list of candidate genes or they may not be known at all. In such cases, iPSCs may be derived from a cohort of patients in collaboration with geneticist.
II. Recent Advances
5. iPSC lines provide patient-specific models for studying kidney disease
iPSCs or hESCs have now been generated with genetic mutations relevant to several kidney diseases, including ADPKD, autosomal recessive PKD (ARPKD), renal cysts and diabetes syndrome (RCAD/MODY5), Alport syndrome, Wolfram syndrome (WFS), Wilms tumor, focal segmental glomerulosclerosis (FSGS), and systemic lupus erythematosus (Table 1).20,35–43 In an early study of ADPKD and ARPKD iPSCs, the first cell biological defect with potential relevance to kidney pathophysiology was identified. iPSCs from three unrelated patients with ADPKD expressed reduced levels of PC2 at the primary cilium, an antenna-like sensory organelle important to PKD etiology.35 In contrast, PC2 localized to cilia efficiently in iPSCs from healthy controls or patients with ARPKD. The global levels of PC2 were unchanged in ADPKD iPSCs, suggesting a trafficking defect.35 Interestingly, the mutations in these patients were in PKD1, suggesting that PC1 is required for efficient delivery of PC2 to the cilium.44,45 Consistent with this, overexpression of PC1 in iPSC-derived cells or mouse IMCD3 cells was sufficient to increase ciliary PC2 levels.35 This study marked the first iPSC phenotype established for a kidney disorder and suggested the possible utility of iPSCs for studying human PKD.35
Table 1.
hPSC lines for modeling kidney diseases.
| DISEASE | KIDNEY SYMPTOM | REFERENCE | CELL TYPE | GENE | PATIENTS | PHENOTYPE IN VITRO |
|---|---|---|---|---|---|---|
| Alport syndrome | Proteinuria, renal insufficiency | 43 | iPSC | COL4A5 | 3 | n.d. |
| Autosomal dominant polycystic kidney | Numerous cysts in adulthood | 35 | iPSC | PKD1 | 3 | Reduced ciliary PC2 |
| 36 | iPSC | PKD1 | 1 | n.d. | ||
| 37 | iPSC | n.d. | 1 | n.d. | ||
| 20 | ESC | PKD1 | (1) | Cysts from tubules | ||
| 20 | ESC | PKD2 | (1) | Cysts from tubules | ||
| Autosomal recessive polycystic kidney | Numerous cysts in childhood | 35 | iPSC | PKHD1 | 2 | n.d. |
| Fabry disease | Proteinuria, renal insufficiency | 39 | ESC | GLA | (1) | n.d. |
| Focal segmental glomerulosclerosis | Proteinuria, renal insufficiency | 20 | ESC | PODXL | (1) | Junctional defects in podocytes |
| Renal cysts and diabetes | Multiple cysts in childhood | 40 | iPSC | HNF1β | (3) | n.d. |
| Systemic lupus erythematosus | Nephritis | 36 | iPSC | n.d. | 1 | n.d. |
| 38 | iPSC | n.d. | (4) | n.d. | ||
| Wilms tumor | Nephroblastoma | 36 | iPSC | n.d. | 1 | n.d. |
| Wolfram syndrome | Diabetes insipidus | 41 | iPSC | WFS1 | 1,(3) | ER stress |
| 42 | iPSC | WFS1 | 3,(2) | ER stress |
Note: Parentheses () indicate unconfirmed clinical phenotype.
Abbreviation: n.d., not determined.
Notably, at the time this study was performed, efficient protocols for differentiation of hPSCs into KTECs had not yet been developed. As surrogates for KTECs, other types of somatic epithelial cells were investigated, as well as undifferentiated hPSCs, which were shown to be ciliated and to express the PKD disease genes.35,46 Reduced ciliary PC2 levels were previously observed in cyst-lining epithelia from PKD1-mutant patients and mice, although in such cases it was not clear whether the phenotype was a primary defect or a secondary consequence of damage to the kidney.44,45 In contrast, the iPSCs were derived from skin fibroblasts, a cell type not known to be affected by PKD. Recently, strong interdependence of PC1 and PC2 ciliary trafficking was also shown in kidney epithelia and embryonic fibroblasts from Pkd1−/− and Pkd2−/− mice.47,48 These findings, together with the iPSC data, strongly support the hypothesis that ciliary trafficking of PC2 is directly regulated by PC1 in diverse cell types, in contrast to a previous study.49 Further mechanistic studies are required addressing the pathophysiological consequences of PC1 and PC2 ciliary trafficking and the possibility of modulating this process as a therapeutic strategy. Notably, the primary cilium has been previously identified as a biomarker in the adaptation of mesenchymal stem cells to low oxygen tensions.50 Loss of PC2 from ADPKD iPSC cilia may similarly represent a highly-sought biomarker for ADPKD, for which genetic diagnostics are complex and have limited predictivity.35,51
Kidney disease is complex and may involve other organ systems. In addition to the disorders listed in Table 1, several studies have described iPSCs from patients with diabetes mellitus (DM).15,40,52–55 Diabetic nephropathy resulting from DM is a major cause of chronic kidney disease. Studies of DM iPSCs have focused on modeling the primary defect in these patients in iPSC-derived pancreatic β cells and have not yet examined downstream effects on the kidney. Another disorder, Wolfram syndrome (WFS), is associated with both DM and diabetes insipidus (DI), the latter characterized by excessive thirst and dilute urine. WFS-associated DI appears to result from the inability of the hypothalamus to efficiently secrete arginine vasopressin.56 In two studies with potential relevance to the kidney, fibroblasts from WFS patients, including several with clinically documented DI, were reprogrammed into iPSCs.41,42 WFS iPSCs differentiated into the pancreatic lineage exhibited a phenotypic defect in insulin production, which was linked to ER stress. This phenotype was absent in healthy control iPSCs, as well as WFS iPSCs expressing a ‘rescue’ allele of WFS1, which regulates protein processing in the endoplasmic reticulum (ER).41 In a second study, WFS iPSCs differentiated into neuronal progenitor cells were shown to have increased sensitivity to thapsigargin-induced cell death, compared to iPSCs from control patients. The FDA-approved small molecule dantrolene was found to be protective in these iPSC-derived neuronal cells, suggesting a possible therapeutic.42 These findings provide new insight into the causes of both β cell and neuronal dysfunction in WFS patients, with potential relevance to both DM and DI. Studies modeling dysfunction in other organ systems using iPSCs may therefore have implications for understanding and treating kidney disease.
Clinically, the hallmark of CKD is not any specific pathophysiology but rather a decline in kidney function. Traditional methods such as pathway mapping, metabolic studies and molecular pharmacology have revealed that there is no single, generalizable ‘kidney disease.’ Rather, kidney disease encompasses a wide spectrum of different conditions, each with its own unique pathophysiology, which in many cases remains poorly understood. This in turn affects the suitability of iPSCs as a model system. The examples provided above indicate that iPSCs may be useful in modeling individual kidney pathophysiologies at the cellular level. Experimental design, and data interpretation, are likely to be easier for conditions in which at least some progress has been made in understanding the molecular pathway involved, such as ADPKD. As iPSCs are a cellular system, they are a promising tool for delving deeper into these pathways, to better understand pathophysiology at the cellular and molecular level. In contrast, more systemic features of kidney disease, such as its cause-and-effect relationship with hypertension, may be too complex to be modeled with iPSCs in their current state, and are better suited to animal models or clinical studies.
6. iPSCs present opportunities for biomarker discovery and development
A biomarker has been defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.”57 Disease modeling with iPSCs presents new opportunities for further development of existing biomarkers and the discovery of novel ones (Fig. 4). In contrast to mouse models, iPSCs are potentially high-throughput, and can recapitulate human species-specific biology. iPSC-derived somatic cells are now positioned to compete effectively with immortalized and primary cell lines currently in use in industry. A recent study revealed that immortalized cell lines representing the cardiac, renal, and hepatic lineages were unable to predict organ-specific toxicologies in vitro.58 hPSC-derived cells and organoids may provide more accurate models for toxicology, using human biomarkers as a readout of organ-specific injury. This opens up a new frontier for biomarker research.
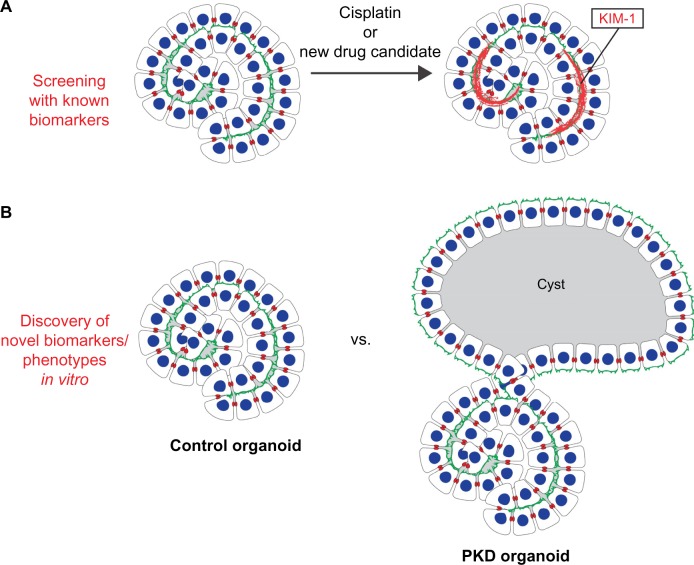
Figure 4.
Biomarker applications for iPSCs. (A) Injury of kidney oganoids results in upregulation of KIM-1, a known proximal tubule injury biomarker (references 20, 21). Such assays may provide a basis for screens testing nephrotoxicity of candidate therapeutics. (B) Organoids with PKD1™/™ and PKD2™/™ mutations form cysts from kidney tubules, which are not observed in organoids without PKD mutations (reference 20). This assay may provide a novel, patient-specific biomarker for PKD in vitro. A similar approach could be taken for other kidney diseases.
Recent work indicates that hPSC-derived cell types are capable of expressing injury- or disease-specific biomarkers, with implications for drug discovery. hESC-derived cardiomyocytes exposed to doxorubicin, a chemotherapeutic associated with cardiotoxicity, were found to release two clinical indicators of cardiac injury, cardiac troponin T and fatty acid binding protein 3.59 In a proposed model of cardiac hypertrophy, iPSC-derived CMs treated with endothelin-1 responded with increased cell size, reactivation of fetal cardiac genes, and secretion of B-type natriuretic peptide, a biomarker of myocardial dysfunction.60 This assay could be miniaturized to a 384-well format amenable to high-throughput screens, which showed reproducible characteristics between different lots of the same iPSC line.60 In the neuronal lineage, iPSC-derived neurons purchased from Cellular Dynamics International were shown to be highly sensitive to Clostridium botulinum neurotoxins, using cleavage of SNARE proteins as a bioindicator and rat spinal cord neurons as a positive control.61 In another example, oligomers of amyloid-β peptide, a biomarker of Alzheimer’s disease that forms insoluble plaques in patient brains, were shown to accumulate intracellularly in patient iPSC-derived neurons, leading to ER stress.62
In principle, the same approach may also be useful for kidney biomarkers, such as those used to detect acute kidney injury (AKI). Because assessment of individualized kidney tissue derived from iPSCs would take months, the application of these techniques is unlikely to be of clinical utility in treating AKI in patients, which is an acute syndrome with a narrow therapeutic window. Rather, the availability of a human model system with characteristics of AKI in vitro, for instance the specific upregulation of AKI biomarkers under stress conditions, might provide a useful pre-clinical diagnostic with which to screen candidate therapeutics for chemical nephrotoxicity, a common side effect that can be a major problem in clinical trials. For instance, kidney injury molecule-1 (KIM-1) is a biomarker of proximal tubular injury, which can be used to diagnose AKI in the clinical setting.63,64 Primary KTECs, however, lose the ability to specifically upregulate KIM-1 after injury, which has restricted the use of KIM-1 to the diagnosis of individual patients on a limited basis.63,64 Recent studies suggest that hPSC-derived human kidney tubules are capable of KIM-1 expression under nephrotoxic conditions, a finding that extends the use of KIM-1 to drug toxicity screening in vitro (Fig. 4A).20,21 Treatment with either cisplatin and gentamicin, two agents that commonly cause drug-induced nephrotoxicity, caused dose-dependent toxicity to hPSC-derived proximal tubules, identified based on their high affinity for Lotus tetragonolobus lectin (LTL) and their arrangements relative to other nephron cell populations (see below Section 7, Kidney Organoid Differentiation from hPSCs Enables Human Disease Models).19–21 Treatment with these agents induced KIM-1 upregulation in proximal tubules, but not in similarly treated distal tubule-like structures or in undifferentiated hPSCs, suggesting this is a tissue-specific response.20,21 A similar approach might be applied in a disease- or segment-specific way to analyze additional renal biomarkers, such as neutrophil gelatinase-associated lipocalin (NGAL) and other markers.65–67 The application of biomarkers such as KIM-1 and N-GAL to drug toxicity screening in vitro represents a new area of research, which expands the utility of these biomarkers beyond clinical practice. Notably, in one of these studies, kidney organoids were produced in 96-well plates, a format amenable to high-throughput screens to identify nephrotoxic or alternatively renoprotective agents.20 Testing of many additional nephrotoxic and non-nephrotoxic agents in this assay is required to determine the degree to which the system is specific and predictive of biomarker responses in human patients.
In a more general sense, disease-specific phenotypes in iPSCs may also be considered a type of biomarker since they can be assessed on a personalized medicine level using patient-derived iPSCs. If the phenotype is sufficiently robust and clinically predictive, it may be used to diagnose a patient. This is particularly important in cases where the genetic defects underlying a hereditary disease cannot be easily determined. In such cases, iPSC phenotypes could conceivably predict inheritance of a disease, decades before the symptoms first manifest in patients. For example, kidney tubules derived from iPSCs with PKD mutations might be expected to exhibit a cystogenesis phenotype in vitro, compared to tubules from iPSCs without PKD mutations, as suggested by recent experiments using gene editing in hESCs (Fig. 4B; see below Section 7, Kidney Organoid Differentiation from hPSCs Enables Human Disease Models).20 In such a case, cystogenesis in iPSC-derived kidney tissue could be considered a biomarker for PKD, since it is a quantifiable, tangible characteristic that can be used as an indicator of a current or future disease state. Such a biomarker could be utilized to predict PKD and possibly gauge its severity in patients, well before clinical symptoms become apparent. iPSC phenotypes are a particularly powerful kind of biomarker, because they can also be used to test therapeutics capable of rescuing the disease. Using such an approach, it may one day be possible to model a patient’s response to specific drugs in vitro, before initiating clinical treatment. A limitation of this approach, however, is that it requires iPSC generation, differentiation, and phenotyping for each individual patient. Given the cost involved, significant advances in each of these areas will be required for personalized iPSC disease modeling to become a reality.
7. Kidney organoid differentiation from hPSCs enables human disease models
Protocols for directed differentiation of hPSCs into NPCs and KTECs have been described by several groups.18–21,68–71 In these studies, hPSCs were treated with growth factors and chemicals, such as the GSK3β inhibitor CHIR99021, to promote their differentiation into mesendodermal progenitor cells. Subsequent treatment with factors such as FGF2 induced expression of NPC markers, including WT1 and PAX2, in a mesenchyme-like population. Spontaneous epithelialization was observed in a subset of cells during longer incubations (~2 weeks). These studies relied primarily on descriptive methods such as immunofluorescence, histology, and gene expression data to categorize the derived cells as kidney.66–70
Whereas earlier studies did not clearly demonstrate nephron-like segmentation in these cell cultures, several recent studies, the earliest being the paper by Taguchi et al. in 2014, have shown that differentiating hPSCs can self-organize into epithelial structures containing continuous segments of distal tubules or ducts (ECAD+), proximal tubules (LTL+CUBN+), and podocytes (PODXL+WT1+).18–21 These structures bear a striking resemblance to the architecture of the nephron, and are therefore referred to as kidney ‘organoids’ (see Fig. 3B). The term ‘organoid’ is used in different ways by different groups, but generally refers to an organized collection of cells in vitro that structurally and functionally resembles a body organ, tissue, or multicellular subunit thereof.72 In one recent study, RNA-Seq analysis was performed on hPSC-derived kidney organoids and thirteen human somatic fetal tissues. Clustering of gene expression profiles based on 85 classifier genes suggested that the organoids were most similar to first-trimester kidneys, although the concordance was not perfect, and similarities with the embryonic gonad were also noted.19 Further functional experiments are therefore required to conclusively identify these cells and their precise embryological correlates.
Among these recent publications describing kidney organoid differentiation, one study further utilized these organoids to address the possibility of modeling kidney physiology and disease in the kidney lineage, using hPSCs.20 To model disease in KTECs, gene editing of hESCs was performed using the Cas/CRISPR (clustered regularly interspersed short palindromic repeats) system to introduce biallelic, truncating mutations into PKD1 or PKD2, the autosomal dominant PKD disease genes. Kidney organoids derived from PKD1™/™ or PKD2™/™ hESCs both produced cyst-like structures, which were not observed in control ESCs of identical genetic background that lacked these targeted mutations (Fig. 4B).20 The cyst-like structures appeared approximately two weeks after differentiation of the kidney organoids, and were shown to be hollow by confocal microscopy. Notably, the cyst-like structures were derived from KTECs in proximal tubules, as demonstrated both by their high affinity for LTL as well as time-lapse imaging showing cystic expansion from tubular organoid structures.20 Demonstration of PKD-specific cystogenesis from kidney tubules represents a significant advance for modeling PKD in cell cultures, which were previously limited to cell lines or primary cells that readily form cysts even in the absence of any mutations in the PKD disease genes.73–75 With regard to PKD, one unanswered question is whether KTECs derived from ESCs and iPSCs with PKD1 mutations will exhibit reduced ciliary PC2 levels, similar to undifferentiated iPSCs and hepatic descendant cells with such mutations.35 The cellular and molecular details of cyst formation in this system remain to be determined, and it is anticipated that KTECs with PKD mutations may also exhibit additional PKD phenotypes, such as increased proliferation or apoptosis, which are believed to contribute to aberrant cystogenesis.30–32,73–75 Such defects may be difficult to detect in lineages other than the kidney, which is the most severely affected organ in PKD. Notably, despite the presence of severe, biallelic loss-of-function mutations in the PKD1™/™ or PKD2™/™ ESC lines, a cystogenesis phenotype was observed only in a minority of kidney organoids in each dish, suggesting that the phenotype is less than 100 % penetrant.20 As PKD is typically thought to be a loss-of-function disease that requires a somatic loss of heterozygosity (the two-hit hypothesis),26–28 it seems unlikely that iPSCs from patients with PKD, in whom the mutations are typically less severe than biallelic truncations, would also form cysts from tubular cells under these conditions. Notably, cyst-like structures can also arise in cultures of unmodified (non-PKD) hPSCs during the process of directed differentiation.68 Careful controls, ideally of the same genetic background, are therefore essential in iPSC studies of PKD cystogenesis to avoid false positives (see below Section 8, Making Meaningful Comparisons Between iPSCs from Different Patients).
Disease modeling with iPSCs may be particularly valuable for kidney cell types which have been difficult to study or derive using other systems. This includes podocytes, whose morphology is not well preserved in primary cultures from kidney biopsy tissue.76 It remains unclear how reproducibly iPSCs can be differentiated into mature podocytes with bona fide foot processes, much less a complete renal corpuscle. Several papers, however, have now demonstrated that hPSC-derived kidney organoids contain pockets of podocyte-like (WT1+PODXL+) cells, which form tightly clustered aggregates continuous with S-shaped tubular structures.18–21 One of these studies has further utilized this system to model glomerular disease in these podocytes. In this study, the CRISPR/Cas was applied to hESCs to introduce truncating, biallelic mutations into PODXL,20 a candidate gene for FSGS.77 PODXL encodes podocalyxin, a negatively-charged plasma membrane sialomucin expressed strongly in podocytes and frequently downregulated in glomerular disease states.78 In podocytes derived from wild-type hESCs, podocalyxin localized to the exterior of the aggregates, whereas synaptopodin and zonula occludens-1 localized in linear tracks inside the aggregate, in a reciprocal pattern to podocalyxin. Podocytes derived from CRISPR-mutant PODXL™/™ hESCs exhibited a change in the localization of these junctional components, which correlated with a decrease in the spacing between the nuclei of adjacent podocytes, as detected by confocal microscopy.20 Notably, junctional defects were not observed in undifferentiated PODXL™/™ hESCs, suggesting this phenotype may be specific to kidney podocytes.20 The findings are consistent with podocalyxin’s proposed role as an anti-adhesive and a regulator of junctional organization in podocytes, which has been previously demonstrated in Podxl™/™ mice.79 Further mechanistic and ultrastructural studies are required to better understand the precise function of podocalyxin in human podocyte junctional organization. This phenotype provides proof of principle for modeling podocyte disorders with hPSCs. Notably, a genetic basis has yet to be identified for the majority of glomerular diseases. As podocalyxin downregulation and cytoskeletal defects are typical of glomerular disease states, it will be interesting to see whether junctional phenotypes such as those described in this study might establish a more generally diagnostic biomarker for podocyte injury and disease states.
As iPSCs represent a relatively early stage of development, they may be particularly useful for studying early developmental cell types, such as NPCs, which are difficult to sustain in primary cultures from mouse embryos.26 Progenitor cell populations expressing SIX2, a marker of the metanephric mesenchyme, have been reported in several of the published protocols for hPSC differentiation.18,68–70 In the mouse metanephros, SIX2 is required to sustain the nephrogenic mesenchyme in a self-renewing state and prevent its premature differentiation into ectopic tubules.26,80,81 The SIX2+ population therefore represents a potential target for functional experiments in the NPCs derived from hPSCs. Here, too, it is important to further define the extent to which the observed SIX2+ population specifically represents NPCs, as opposed to SIX2+ cells found in various other tissues.82
It is not yet clear whether the cells/organoids generated from hPSCs using these recent protocols represent pronephric, mesonephric, or metanephric cells, or possibly some other type of ductal lineage with gene expression patterns similar to the kidney. It is very difficult to conclusively answer this question using marker expression. In contrast to primary cells, which are isolated from specific body parts, there are no spatial cues available to identify hPSC-derived tissues a priori. This means that marker expression must be compared to the entire soma, for which a comprehensive and specific list of markers (adult or developmental) remains very far away. The ideal proof for kidney function would be therapeutic transplantation in a small animal model of kidney insufficiency. Accomplishing such a task will require significant advances in directed differentiation, bioengineering, and transplantation. A complementary approach is to reproduce specific disease phenotypes in kidney cells derived from iPSCs. Many kidney diseases specifically affect the kidney; if kidney-like cells display a phenotype consistent with these syndromes, it would provide strong evidence for lineage specificity. The recent study demonstrating PKD and FSGS-related phenotypes in organoid cells derived from CRISPR-mutant hPSCs, but not in the corresponding undifferentiated hPSCs, therefore supports the conclusion that these organoids represent the kidney lineage rather than another ductal organ.20
III. Technical Considerations
8. Making meaningful comparisons between iPSCs from different patients
iPSC models have been published for many different human diseases.13,14 In most cases, these consist of a side-by-side comparison of iPSCs derived from a few disease patients to control iPSCs from healthy patients. It is critical to note, therefore, that ESCs or iPSCs from different individuals differ significantly in their ability to differentiate along certain lineages.3,83,84 To gauge line-to-line variability, a set of 20 hESC and 12 hiPSC lines were subjected to whole-genome transcriptional and epigenetic profiling in the pluripotent state.3,84 Hierarchical clustering of hESCs and hiPSCs revealed no clear-cut break, indicating the two cell types were essentially the same. However, variabilities in both gene expression and epigenetic marks were observed between different individual hPSC lines, both hESCs and hiPSCs. Each line was then subjected to stochastic and directed differentiation protocols, and differentiation efficiencies estimated either based on RNA sequencing or marker expression.3,84 These studies demonstrate that individual iPSCs and ESCs, while essentially the same cell type, do exhibit significant line-to-line variabilities, which can affect their potential for differentiation into various lineages. The authors used the data to create a prototype ‘lineage scorecard,’ proposed to predict the differentiation tendencies of hPSC lines based on the expression levels of 500 genes in the undifferentiated state.3,84 One caveat to this scorecard, which has limited its wider-scale adoption and validation, is that it requires significant resources and specialized instrumentation to perform and analyze.
It is not yet clear to what extent the variability in differentiation tendency observed among different hPSC lines is an intrinsic property of the cell lines and their genetic backgrounds, or an indicator of insufficiently-robust protocols for directed differentiation. Either way, such variability is a concern for many disease modeling studies using iPSCs, which rely heavily on comparing iPSCs from disease patients to iPSCs from healthy control patients. Given the small sample size and the genetic heterogeneity between these cell lines, how reliable are such comparisons? Work in this field over several years has suggested several strategies for making meaningful comparisons between iPSCs (Table 2). It is critical that iPSCs used for side-by-side comparisons should be as similar as possible, both in their derivation history and their characteristics in culture.3,84 Although the precise reasons for iPSC variability are poorly understood, there are certain measures researchers can take in order to reduce variability and improve the chances for correct interpretation of the data. Early iPSCs were generated using lentiviral transgenes which integrated into the genome.2,5,85 Although these viral genomic DNA sequences were efficiently silenced in the resulting undifferentiated iPSCs, in some cases they could become re-activated during subsequent differentiation.84 Newer approaches use non-integrating episomes or Sendai virus that are less likely to persist after reprogramming.85–87
Table 2.
Technical challenges and strategies for iPSC comparisons.
| TECHNICAL CHALLENGE | POSSIBLE CAUSE | EXPERIMENTAL STRATEGY |
|---|---|---|
| Variability between iPSCs | Genetic background | Increase sample size; use genome editing or RNAi on isogenic cells |
| Gender/age differences | Include gender/aged matched controls | |
| Clone-to-clone variability | Study 2 or more clones per patient | |
| Lentiviral integration | Use non-integrating episomal vectors or Sendai virus to reprogram | |
| Experimental conditions | Plate and assay lines side-by-side with controls | |
| Inconsistent differentiation | iPSC culture conditions | Equilibrate all lines to one growth condition for all lines prior to starting the experiment |
| Partially reprogrammed lines | Validate morphology, markers, self-renewal, and teratoma formation for each line | |
| Protocol insufficiently robust | Optimize differentiation protocol | |
| Disease gene affects differentiation | Increase sample size and perform rescue experiments | |
| Memory of somatic cell lineage | Establish controls from same somatic cell type | |
| Inconsistent mutant phenotype | Outlier iPSC lines | Present individual patient data alongside pooled data |
| Allelic differences | Sequence genes and assess genotype-phenotype relationship | |
| Secondary defects in source tissue | Reprogram a cell type unaffected by the disease |
In addition, the choice of starting cell for reprogramming may be important for subsequent experiments. Aside from fibroblasts, many other types of somatic cells have been successfully reprogrammed, including blood cells, hair follicle cells, kidney mesangial cells, and urinary cells.14,28,38,43,88–91 It may not be acceptable to compare iPSCs derived from fibroblasts to iPSCs derived from blood, since they might retain transcriptional or epigenetic ‘memory’ of the parental lineage, which could affect subsequent differentiation.92,93 The key consideration here is that iPSCs from disease patients and iPSCs from control patients should be derived similarly, from the same starting cell type, and ideally side-by-side.
Even iPSC lines reprogrammed exactly the same way should be carefully inspected to make sure that they resemble one another with respect to morphology, marker expression, and pluripotency. Otherwise, there is the risk that some lines may be only partially reprogrammed – with some but not all the features of bona fide iPSCs (see Fig. 1A).94,95 To reduce intrinsic variability, multiple patients and clones should be examined side-by-side, and phenotypic data should be presented for each individual patient or line alongside the averaged data pooled from the entire cohort of iPSCs. For major conclusions, differences should be subjected to appropriate statistical tests to demonstrate significance or lack thereof.
9. Technical strategies for efficient disease modeling with iPSCs
A major practical challenge for iPSC disease modeling is the time required to establish and characterize the disease model. Because iPSC derivation involves patient recruitment, approval of an institutional review board is typically required, which can take several months. Up to twelve weeks may be required to grow the primary cultured cells from the patient into somatic cell lines amenable to reprogramming. An additional twelve weeks is required to establish and expand the iPSC lines, and to perform minimal quality control assessments such as karyotyping and stochastic differentiation assays. Thus, merely generating the cells can easily take half a year. Directed differentiation protocols can take anywhere from 2 to 10 weeks from the time of initially plating the cells. Furthermore, differentiation may be less than 100% reproducible from one experiment to the next, presumably due to minor differences in technique and the inherent stochasticity of the differentiation process. Failure to efficiently differentiate can result in weeks of lost labor for the investigator. This problem becomes significantly compounded when dealing with multiple lines (each with its own tendencies) and when the objective of the experiment is not merely to reach a specific stage of differentiation but also to rigorously analyze a cellular phenotype. In general, cells in these differentiation protocols do not freeze well or behave the same after dissociation, so stopping mid-way or growing up a large batch of cells may not be an option.
There are several strategies that the experimentalist can adopt to minimize the risk of failure during lengthy differentiation protocols (Table 2). First, as many iPSC lines as can be reasonably cultured at one time (typically, six) should be differentiated and analyzed side-by-side. This saves effort in the long run and also minimizes experiment-to-experiment variabilities, so that the comparisons made are more meaningful. Second, experiments should be staggered so that new differentiated cells are available on a regular basis (every week or second week). This way, the lag time of differentiation is limited to the very first round of experiments. Third, if parameters affecting the differentiation are known, it is frequently helpful to try a few different variations of the differentiation protocol within each round of differentiation. For instance, this may involve modulation of the concentration of a specific growth factor, or plating density. Including multiple variations reduces the risk that the differentiation protocol will fail completely. Of course, comparisons between different iPSC lines must always be made using a single differentiation condition, at least within any given experiment.
It is critical to perform a rescue experiment to more rigorously establish the specificity of any discovered phenotype. For instance, stable expression of wild-type WFS1 in WFS iPSCs significantly improved insulin secretion from derived pancreatic cells.41 Such rescue experiments can pose a significant technical challenge in iPSCs, for which exogenous gene expression techniques are still being optimized. In the case of PKD iPSCs, ADPKD iPSC-derived hepatoblasts were transfected with full-length human PKD1 to rescue PC2 localization at the primary cilium.35 Transfection was relatively inefficient, due to the difficulty of transfecting these confluent differentiated cell layers and the huge size of the PKD1 construct. The experiment was therefore repeated in the more traditional model of IMCD3 cells, with similar results. A hypothesis generated from experiments in iPSCs may therefore be further tested and validated in other, more developed model systems.
IV. Future Directions
10. Advanced genetic and genomic approaches comple ment disease modeling with iPSCs
Recent years have seen a significant increase in genome modification approaches for human cells. There are now several different technologies available for modifying the human genome, which have recently been reviewed for hPSC applications.96,97 These include traditional site-directed homologous recombination, zinc-finger nucleases (ZFNs), TALENs (transcription activator- like effector nucleases), piggyBac transposons, adeno-associated viruses (AAVs), and Cas/CRISPR. The principle behind these approaches is to direct nuclease activity to a specific sequence within the genome, and thereby introduce a double stranded break (DSB). The ensuing DSB repair process may introduce stochastic insertions or deletions (indels) which disrupt the gene, or alternatively allow for sequence-specific mutagenesis in the presence of an appropriate template for homology directed repair (HDR). Using any of these approaches, hPSCs can be derived harboring targeted mutations, which are otherwise isogenic with the parental hPSCs.96,97 For instance, the Cas9 nuclease has biallelic mutagenesis efficiencies of 4%–25% in hPSCs.98,99
Genome editing approaches have been applied to hPSCs to study several organs and diseases. In one example, introduction of a specific long-QT syndrome (LQTS) mutation via genome editing of hESCs resulted in a prolonged action potential in derived cardiomyocytes, similar to the phenotype previously described in LQTS iPSCs.100 Genome editing techniques have also been successfully applied to produce homologous recombinants which correct the genetic lesions in patient-derived iPSCs.97,101,102 This is a powerful way to demonstrate the specificity of the gene mutations to the observed phenotypes, which models gene therapy in the original patient. In one example, genetic correction of LMNA mutations significantly rescued senescence and nuclear abnormality phenotypes in progeria iPSCs.101 Similarly, correction of A1AT mutations in iPSCs from α1-antitrypsin deficiency patients led to a dramatic cell biological rescue of A1AT structure and enzyme function.102 Genome editing also enables selection strategies that have been used to correct trisomy of chromosome 21 in Down syndrome iPSCs.103 Genome-editing experiments represent a new genetic paradigm for human cells and are likely to be required for rigorous validation of phenotypes in iPSCs.
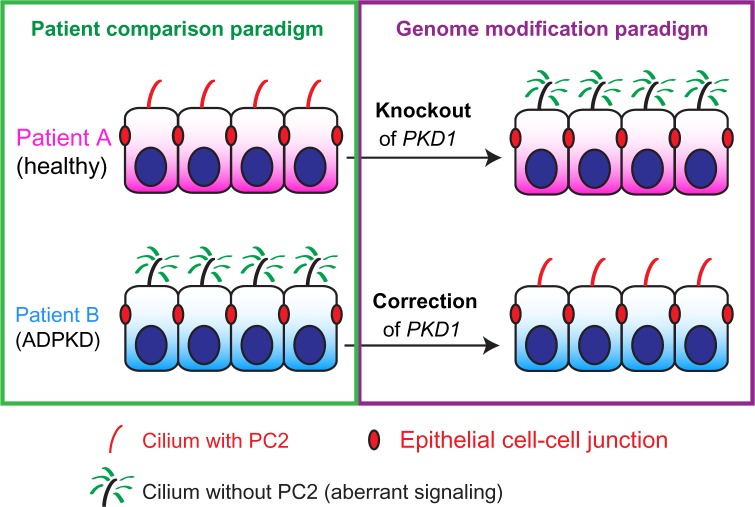
Recent work demonstrates that these same approaches are likely to be very informative for studies of kidney disease iPSCs. As described above (see section 7, Kidney Organoid Differentiation from hPSCs Enables Human Disease Models), a recent study applied the CRISPR/Cas system to generate hESCs with mutations in disease genes linked to PKD and FSGS.20 Disease-relevant phenotypes were observed in the derived CRISPR-mutant kidney organoids, in the appropriate cell types (KTECs for PKD, podocytes for FSGS). Critically, these phenotypes were not observed in unmodified, control hPSCs of identical genetic background that lacked these targeted mutations.20 This paper establishes a paradigm of gene editing that can be applied to validate kidney disease phenotypes observed in patient-derived iPSCs. For instance, CRISPR-mutant PKD1™/™ iPSCs could be compared to unmodified, healthy iPSC controls of otherwise identical genetic background to test the hypothesis that PC1 is required for ciliary trafficking of PC2 (Fig. 5).35 In this hypothetical experiment, the only difference between the wild-type and PKD cell lines is the mutation in PKD1, reducing the issue of variability between patient iPSCs (Table 2). Alternatively, correction of PC1 mutations in ADPKD iPSCs would be predicted to enhance PC2 trafficking to cilia (Fig. 5). The futuristic application of this would be to isolate iPSCs from the patient, correct the genetic defect, differentiate the cells, and validate that the disease phenotype has been rescued in vitro. Having validated the corrective approach, gene therapy could be performed directly in the patient’s tissues in situ, or alternatively, one could administer the gene-corrected, immunocompatible transplant to the patient to replace lost tissues (Figs. 1–2).
Figure 5.
Patient and genome-modification paradigms for iPSCs. In the patient comparison paradigm (left, green box), genetically heterogenous iPSCs are compared in cohorts, revealing specific phenotypes (loss of PC2 and ciliary signaling defects in ADPKD patient). In the genome-modification paradigm (right, purple box), the original iPSCs are genetically modified (arrows) to introduce or correct genetic mutations and the corresponding phenotypes in isogenic lines.
RNA interference (RNAi) is a useful alternative and complement to genome editing in hPSCs. RNAi can be performed transiently to replicate plates of starting cells. In this way, phenotypes produced with RNAi are free of any clonal aberrations which might affect genome-modified lines. RNAi is also less labor intensive than genome modification and has been performed efficiently in hPSCs for many years.104 For hPSCs, RNAi can be performed at any stage of the differentiation protocol, simplifying the interpretation of experiments involving genes important at multiple developmental stages. Interestingly, recent experiments suggest that outcomes of RNAi experiments may in some cases differ from those of genome editing.105,106 Rigorous, side-by-side comparisons at both the genome scale and that of individual genes are required to understand the specific differences between gene knockdown and knockout. Such studies are likely to shed light on our understanding of how gene expression levels affect cellular functions.
hPSCs also provide new opportunities to perform studies at the scale of the genome, epigenome, transcriptome, and proteome. Such studies can be challenging to interpret in animal tissues which typically include a variety of cell types expressing different genes, and may be further complicated by inflammation and injury in disease states. In contrast, hPSCs can be differentiated into cultures with defined cell types, under relatively simple and controllable culture conditions, enabling analysis of cell-type specific global gene expression patterns. In proof of principle for this approach, one recent study utilized the whole-genome bisulfite sequencing, chromatin immunoprecipitation sequencing, and RNA sequencing to characterize changes in gene expression during differentiation of hPSCs into each of the three embryonic germ layers (endoderm, mesoderm, and ectoderm).107 Flow cytometry was necessary to purify these sub-populations, underscoring the point that differentiated hPSC cultures are typically mixed populations and that purity remains a significant challenge. The results of this study supported an important role for chromatin modification and DNA methylation in regulating germ layer-specific transcriptional patterns.107 The logical next step for whole-genome studies is to apply such methodologies to hPSC-derived populations for the purposes of studying human disease. Such experiments could potentially provide a detailed, transcriptome-level view of human disease states in the relevant tissue types. For diseases such as PKD, such an unbiased, global analysis may reveal specific pathways critical to pathophysiology that have been previously overlooked. However, iPSCs from different patients may be too heterogenous to provide meaningful comparisons at the genome scale (see above section 8, Making Meaningful Comparisons Between iPSCs from Different Patients). Genome-edited or RNAi-treated hPSCs are likely to be less variable and provide a more useful starting point for these experiments.
11. iPSC research moves from dish to clinic
The field of iPSC research has important translational potential for improving patient health, which has fueled its expansion and visibility. One area in which iPSCs can benefit disease research is in the discovery and testing of new drugs. As iPSC-based disease phenotypes and biomarkers are discovered and characterized, they raise the possibility of performing screens for therapeutic compounds that ameliorate iPSC phenotypes in vitro. Such an approach would guide the selection of candidate therapeutics for testing in pre-clinical animal models and ultimately in human patients. Recently, a screen of ~5,000 small molecules identified a GSK3β inhibitor, kenpaullone, as consistently enhancing survival in motor neurons derived from mouse ESCs.33 The authors then performed a small-scale “clinical trial in vitro” to demonstrate that the compound was also effective at sustaining motor neurons produced from a small cohort of hPSCs, including iPSCs from two patients with amyotrophic lateral sclerosis.33 The advantages of hPSCs – reproducibility, species specificity, and high throughput – make them an attractive model system for biotechnology and pharmaceutical screening applications. What is now required at the basic research level is identification of disease-specific phenotypes in iPSCs which may be useful for performing therapeutic screens. For the kidney, this will require disease modeling efforts with iPSCs representing a broad spectrum of kidney disorders, with the goal of identifying cellular phenotypes that inform our understanding of the molecular pathophysiology underlying each disease. This information can then be used to guide the development of therapeutics that target the root causes of disease at the molecular level. Such early-stage interventions would complement and enhance current clinical strategies, which primarily target late-stage consequences of kidney dysfunction. Collaborations between iPSC researchers and laboratories studying specific kidney diseases will be essential for this effort to succeed.
An attractive feature of iPSCs is that they may someday be utilized to produce patient-specific tissue which is both transplantable and immunocompatible with the original patient.8–12 The need for this is clear for kidney disease, in which ~15% of grafts suffer acute rejection, and the remainder require life-long immunosuppression to slow chronic rejection. Differentiated tissues derived from iPSCs are predicted to be essentially 100% immunocompatible with the original patient, based on mouse studies.8,9 Transplantation experiments in several organs, including the heart, liver, and pancreas, suggest that hPSC-derived somatic tissue can engraft and function in animal models.10–12 The therapeutic potential of iPSCs for autologous transplantation is currently being evaluated in Japan in a small clinical trial for wet age-related macular degeneration (AMD), using retinal pigmented epithelial (RPE) cells derived from iPSCs.108 This pilot safety study follows on the heels of an ongoing, high-profile clinical trial testing the safety of ESC-derived RPEs for AMD and Stargardt’s disease, sponsored by Ocata Therapeutics Inc. (OCAT; formerly Advanced Cell Technology Inc.).109,110
Practical experience has also revealed significant barriers to iPSC-based therapeutics, which have limited their transition from bench to bedside. One financial consideration is that Dr. Yamanaka and Kyoto University (iPS Academia Japan, Inc.) hold an international patent on iPSCs. Whereas academic institutions may utilize iPSC technology cost-free for noncommercial uses, for-profit entities must license iPSC technology for fees and royalties, typically in the range of tens of thousands of dollars per year. According to the licensing agency (http://ips-cell.net/e/), over 100 entities have signed agreements, suggesting that the licensing per se does not constitute a major barrier to iPSC product development. A more significant concern, strategically, is that each individual iPSC-based therapeutic needs to be developed on-demand and in a patient-specific manner, to achieve immunocompatibility. Development of such therapeutics would require close to a year under ideal circumstances – and this would need to be done for every patient (Table 3; see above section 8, Making Meaningful Comparisons Between iPSCs from Different Patients). Furthermore, success is not guaranteed. For instance, in the aforementioned iPSC-based clinical trial in Japan, the second patient’s iPSCs have reportedly failed to pass the study’s rigorous quality control standards, possibly due to a mutation. While the scientific details have not yet been made public, this quality control issue has reportedly resulted in a temporary halt to iPSC-derived RPE implantation in that trial, according to the RIKEN research institute, where the trial is being conducted (http://www.riken-ibri. jp/AMD). This case highlights an important safety issue: because iPSCs are expected to be 100% immunocompatible with patients, they and their derivatives will be invisible to the immune system after implantation. It is therefore critical that any implant be scrutinized carefully for oncogenic potential, lest the graft result in an autologous tumor and potentially cause more harm than good.
Table 3.
Comparison of iPSCs and ESCs as potential cell therapeutics.
| SELF-RENEWING AND PLURIPOTENT? | DERIVATION TIME (WKS) | DIFFERENTIATION TIME (WKS) | LICENSING FEE ($/YR) | OFF-THE-SHELF? | IMMUNOCOMPATIBILITY/IMMUNOSUPPRESSION | RISKS | ETHICAL ISSUES | |
|---|---|---|---|---|---|---|---|---|
| iPSC | Yes | 24 | 2–10 | 50,000 + | No | Autologous/no | Tumorigenicity | Privacy |
| ESC | Yes | 12 | 2–10 | Free | Yes | Allogeneic/yes | Rejection, graft v. host | Embryo destruction |
ESCs represent an alternative to iPSCs as a source of human tissue. ESCs feature self-renewal and pluripotency characteristics similar to iPSCs, but with key logistical and strategical differences (Table 3). Financially, ESCs may be more attractive to companies than iPSCs. The patentability of ESCs is disputed, and key patents are expected to expire in 2015, twenty years after their filing. The safety and efficacy of ESC-derived RPE grafts is currently being evaluated in the Ocata Therapeutics clinical trial for AMD and Stargardt’s disease. In a test group of 18 patients, the only safety issues identified appeared to be related to immunosuppression and surgery, not the presence of the ESCs themselves. In an encouraging sign, several patients exhibited improvements in visual acuity after RPE-implantation, which were not observed in untreated control patients.109,110 Of course, for any therapy to be adopted as a clinical standard, it would need to be tested against the existing clinical gold standard, which has not yet been done. From an ethical standpoint, it is worth noting that ESC derivation typically requires the destruction of a human blastocyst-stage embryo, which may be ethically unacceptable to some patients. To avoid this ethical issue, Ocata is utilizing ESCs derived from individual blastomere cells, which can be procured without causing harm to the embryo.111 The ESCs used in this trial are furthermore not HLA-matched, taking advantage of the eye as an immunotolerant organ. For other organs, allogeneic transplants from ESCs will require human leukocyte (HLA) matching between donor and recipient to encourage successful engraftment. To this end, an allogeneic, HLA-matched library of hPSCs has been proposed in hopes of providing an ‘off-the-shelf’ source of cell and tissue grafts.112 In contrast to ESCs, iPSCs are typically derived from living, adult humans, for whom the major ethical consideration is privacy. In the case of the RIKEN trial, the study leaders have disclosed that allogeneic iPSCs are being considered for use in the second patient, instead of the autologous iPSCs that failed quality control. This unexpected development will provide an interesting side-by-side comparison of autologous and allogeneic iPSCs. It is possible that genome editing in ESCs or iPSCs might also enable generation of a ‘universal donor’ hPSC with increased graft immunocompatibility.113,114 For the kidney, improved immunocompatibility, however achieved, might offer a key advantage over the existing paradigm of allogeneic, HLA-matched organ transplant (see Fig. 2).
For therapeutic transplantation of hPSC-derived kidney grafts to become a reality, it will be necessary to demonstrate that the implanted cells are functional in animals. Early studies have suggested that transplanted fetal kidney cells may confer benefit on mouse hosts suffering from renal insufficiency.27,115 As the effects were modest, additional studies are required to clearly demonstrate this, first with fetal cells, and later with hPSC-derived NPCs. In parallel, studies demonstrating physiological function of hPSC-derived tissues in vitro can provide important preliminary validation of proposed therapeutics. For genetic diseases, it will be important to genetically correct the disease prior to transplantation, as demonstrated in a mouse model of sickle cell anemia.116 As DNA sequencing may not correctly predict a disease-causing mutation,34 the ideal assay would be rescue of a disease-specific phenotype in cultured lines in vitro prior to implantation into human patients. Development of phenotyping assays is therefore an important step not only for disease modeling in vitro but also for the longer term goal of transplantation of corrected organs into patients.
V. Conclusion and Outlook
Surveying the field, the use of iPSCs for disease modeling has only scratched the surface of what is possible and what will hopefully be achieved in the future. Many more iPSC lines have been produced than have actually been utilized in disease modeling experiments. This reflects the fact that considerably more labor and rigor is required to identify a phenotype in iPSCs than to produce them. Nevertheless, once lines are available and growing in culture, quantitative data can be obtained rapidly. The rate-limiting step is development of an assay/biomarker that represents the disease process and can distinguish between mutant and non-mutant iPSCs.
For kidney disease, iPSCs present opportunities for human disease modeling which are not currently available using any other system. In particular, iPSCs can form well-differentiated human epithelia in culture, including tubular epithelial and podocyte-like cell populations. Nephron-like organoids have been derived from iPSCs by several groups, but the extent to which these cells truly represent the kidney, and at what developmental stage, are critical questions which disease modeling and nephrotoxicity approaches may play a role in answering. Advancements in the arena of functionality in vitro will foster more strategic and informed approaches for transplantation of iPSC-derived tissues and development of safe and efficacious therapeutics.
Most iPSC research has focused on a relatively small subset of somatic lineages and a correspondingly limited subset of diseases. There is fertile ground here for the study of other lineages and many more diseases. In order for iPSCs to produce clinically useful information, it is important to invest and capitalize on this technology now and translate it into novel biological and clinical insights. As iPSC technologies become more mainstream, they will find their way out of laboratories specializing in iPSCs and into laboratories specializing in specific diseases. Collaborations between stem cell scientists, geneticists, clinicians, and pathologists will result in more specific, rapid, and effective application of iPSC technologies for the kidney and other organs.
Acknowledgments
We thank Joseph Bonventre, Jing Zhou, Theodore Steinman, Albert Lam, Hongxia Fu, Todd Valerius, Vishal Vaidya, Vicki Kelley, Stuart Shankland, Jonathan Himmelfarb, Akio Kobayashi, Neal Paragas, and Kelly McNagny for helpful discussions. We also acknowledge the authors of many excellent studies that we were unable to cite due to space limitations.
Footnotes
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2,151 words, excluding any confidential comments to the academic editor.
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
FUNDING: This work was supported by the National Institutes of Health (DK102826 and NCATS Loan Repayment Program) and the National Kidney Foundation (Young Investigator Grant). The author confirms that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: The author has a patent pending for methods of generating intermediate mesoderm cells from human pluripotent stem cells. He also has a patent pending for three dimensional differentiation of epiblast spheroids into kidney tubular organoids modeling human microphysiology, toxicology and morphogenesis.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the manuscript, revised the manuscript, and approved the final version: BSF. The author reviewed and approved of the final manuscript.
REFERENCES
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998 Nov 6;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30;131(5):8, 61–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Bock C, Kiskinis E, Verstappen G, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011 Feb 4;144(3):439–52. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008 Jan 10;451(7175):141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007 Dec 21;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 7.Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009 Dec 3;462(7273):595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guha P, Morgan JW, Mostoslavsky G, Rodrigues NP, Boyd AS. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013 Apr 4;12(4):407–12. doi: 10.1016/j.stem.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Araki R, Uda M, Hoki Y, et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013 Feb 7;494(7435):100–4. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 10.Chong JJ, Yang X, Don CW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014 Jun 12;510(7504):273–7. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013 Jul 25;499(7459):481–4. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 12.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nature biotechnology. 2008 Apr;26(4):443–52. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 13.Rao M. iPSC crowdsourcing: a model for obtaining large panels of stem cell lines for screening. Cell Stem Cell. 2013 Oct 3;13(4):389–91. doi: 10.1016/j.stem.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells–opportunities for disease modelling and drug discovery. Nature reviews Drug discovery. 2011 Dec;10(12):915–29. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 15.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008 Sep 5;134(5):877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boland MJ, Hazen JL, Nazor KL, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009 Sep 3;461(7260):91–4. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 17.Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009 Sep 3;461(7260):86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 18.Taguchi A, Kaku Y, Ohmori T, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014 Jan 2;14(1):53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Takasato M, Er PX, Chiu HS, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015 Oct 22;526(7574):564–8. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 20.Freedman BS, Brooks CR, Lam AQ, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nature communications. 2015 Oct 23;6(8715):1–13. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nature biotechnology. 2015 Nov;33(11):1193–200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faa G, Gerosa C, Fanni D, et al. Marked interindividual variability in renal maturation of preterm infants: lessons from autopsy. The journal of maternal-fetal & neonatal medicine : the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2010 Oct;23(Suppl 3):129–33. doi: 10.3109/14767058.2010.510646. [DOI] [PubMed] [Google Scholar]
- 23.Hartman HA, Lai HL, Patterson LT. Cessation of renal morphogenesis in mice. Dev Biol. 2007 Oct 15;310(2):379–87. doi: 10.1016/j.ydbio.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphreys BD, Valerius MT, Kobayashi A, et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008 Mar 6;2(3):284–91. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Rinkevich Y, Montoro DT, Contreras-Trujillo H, et al. In vivo clonal analysis reveals lineage–restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell reports. 2014 May 22;7(4):1270–83. doi: 10.1016/j.celrep.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barak H, Huh SH, Chen S, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell. 2012 Jun 12;22(6):1191–207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harari-Steinberg O, Metsuyanim S, Omer D, et al. Identification of human nephron progenitors capable of generation of kidney structures and functional repair of chronic renal disease. EMBO molecular medicine. 2013 Oct;5(10):1556–8. doi: 10.1002/emmm.201201584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009 Dec 4;5(6):584–95. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. The European Polycystic Kidney Disease Consortium. Cell. 1994 Aug 26;78(4):725. [PubMed] [Google Scholar]
- 30.Mochizuki T, Wu G, Hayashi T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996 May 31;272(5266):1339–42. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Fan X, Basora N, et al. Late onset of renal and hepatic cysts in Pkd1-targeted heterozygotes. Nat Genet. 1999 Feb;21(2):160–1. doi: 10.1038/5944. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Tian X, Nishimura S, et al. Trans-heterozygous Pkd1 and Pkd2 mutations modify expression of polycystic kidney disease. Hum Mol Genet. 2002 Aug 1;11(16):1845–54. doi: 10.1093/hmg/11.16.1845. [DOI] [PubMed] [Google Scholar]
- 33.Yang YM, Gupta SK, Kim KJ, et al. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013 Jun 6;12(6):713–26. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010 Apr 16;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 35.Freedman BS, Lam AQ, Sundsbak JL, et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J Am Soc Nephrol. 2013 Oct;24(10):1571–86. doi: 10.1681/ASN.2012111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thatava T, Armstrong AS, De Lamo JG, et al. Successful disease-specific induced pluripotent stem cell generation from patients with kidney transplantation. Stem cell research & therapy. 2011;2(6):48. doi: 10.1186/scrt89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Y, Nivet E, Sancho-Martinez I, et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol. 2013 Dec;15(12):1507–15. doi: 10.1038/ncb2872. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Luo R, Xu Y, et al. Generation of systemic lupus erythematosus- specific induced pluripotent stem cells from urine. Rheumatology international. 2013 Aug;33(8):2127–34. doi: 10.1007/s00296-013-2704-5. [DOI] [PubMed] [Google Scholar]
- 39.Tropel P, Tournois J, Come J, et al. High-efficiency derivation of human embryonic stem cell lines following pre-implantation genetic diagnosis. In Vitro Cell Dev Biol Anim. 2010 Apr;46(3–4):376–85. doi: 10.1007/s11626-010-9300-8. [DOI] [PubMed] [Google Scholar]
- 40.Teo AK, Windmueller R, Johansson BB, et al. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J Biol Chem. 2013 Feb 22;288(8):5353–6. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang L, Hua H, Foo K, et al. beta-cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes. 2014 Mar;63(3):923–33. doi: 10.2337/db13-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu S, Kanekura K, Hara T, et al. A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proc Natl Acad Sci U S A. 2014 Dec 9;111(49):E5292–5301. doi: 10.1073/pnas.1421055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen WB, Huang JR, Yu XQ, Lin XC, Dai Y. Identification of microRNAs and their target genes in Alport syndrome using deep sequencing of iPSCs samples. Journal of Zhejiang University Science B. 2015 Mar;16(3):235–50. doi: 10.1631/jzus.B1400272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nauli SM, Rossetti S, Kolb RJ, et al. Loss of polycystin-1 in human cyst-lining epithelia leads to ciliary dysfunction. J Am Soc Nephrol. 2006 Apr;17(4):1015–1025. doi: 10.1681/ASN.2005080830. [DOI] [PubMed] [Google Scholar]
- 45.Xu C, Rossetti S, Jiang L, et al. Human ADPKD primary cyst epithelial cells with a novel, single codon deletion in the PKD1 gene exhibit defective ciliary polycystin localization and loss of flow-induced Ca2+ signaling. Am J Physiol Renal Physiol. 2007 Mar;292(3):F930–945. doi: 10.1152/ajprenal.00285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiprilov EN, Awan A, Desprat R, et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008 Mar 10;180(5):897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H, Xu H, Yao Q, et al. Ciliary membrane proteins traffic through the Golgi via a Rabep1/GGA1/Arl3-dependent mechanism. Nature communications. 2014;5:5482. doi: 10.1038/ncomms6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gainullin VG, Hopp K, Ward CJ, Hommerding CJ, Harris PC. Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J Clin Invest. 2015 Jan 9; doi: 10.1172/JCI76972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng L, Okuhara D, Yu Z, et al. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006 Apr 1;119(Pt 7):1383–95. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- 50.Proulx-Bonneau S, Annabi B. The primary cilium as a biomarker in the hypoxic adaptation of bone marrow-derived mesenchymal stromal cells: a role for the secreted frizzled-related proteins. Biomarker insights. 2011;6:107–18. doi: 10.4137/BMI.S8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossetti S, Consugar MB, Chapman AB, et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2007 Jul;18(7):2143–60. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 52.Maehr R, Chen S, Snitow M, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A. 2009 Sep 15;106(37):15768–73. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohmine S, Squillace KA, Hartjes KA, et al. Reprogrammed keratinocytes from elderly type 2 diabetes patients suppress senescence genes to acquire induced pluripotency. Aging. 2012 Jan;4(1):60–73. doi: 10.18632/aging.100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kudva YC, Ohmine S, Greder LV, et al. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem cells translational medicine. 2012 Jun;1(6):451–61. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua H, Shang L, Martinez H, et al. iPSC-derived beta cells model diabetes due to glucokinase deficiency. J Clin Invest. 2013 Jul;123(7):3146–53. doi: 10.1172/JCI67638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Thompson CJ, Charlton J, Walford S, et al. Vasopressin secretion in the DIDMOAD (Wolfram) syndrome. The Quarterly journal of medicine. 1989 Apr;71(264):333–45. [PubMed] [Google Scholar]
- 57.Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical pharmacology and therapeutics. 2001 Mar;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 58.Lin Z, Will Y. Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicol Sci. 2012 Mar;126(1):114–27. doi: 10.1093/toxsci/kfr339. [DOI] [PubMed] [Google Scholar]
- 59.Andersson H, Steel D, Asp J, et al. Assaying cardiac biomarkers for toxicity testing using biosensing and cardiomyocytes derived from human embryonic stem cells. Journal of biotechnology. 2010 Oct 1;150(1):175–81. doi: 10.1016/j.jbiotec.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 60.Carlson C, Koonce C, Aoyama N, et al. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. Journal of biomolecular screening. 2013 Dec;18(10):1203–11. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 61.Whitemarsh RC, Strathman MJ, Chase LG, et al. Novel application of human neurons derived from induced pluripotent stem cells for highly sensitive botulinum neurotoxin detection. Toxicological sciences: an official journal of the Society of Toxicology. 2012 Apr;126(2):426–35. doi: 10.1093/toxsci/kfr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kondo T, Asai M, Tsukita K, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell stem cell. 2013 Apr 4;12(4):487–96. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 63.Ichimura T, Bonventre JV, Bailly V, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998 Feb 13;273(7):4135–42. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 64.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008 May;118(5):1657–68. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishra J, Dent C, Tarabishi R, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. 2005 Apr 2–8;365(9466):1231–8. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- 66.Paragas N, Nickolas TL, Wyatt C, et al. Urinary NGAL marks cystic disease in HIV-associated nephropathy. J Am Soc Nephrol. 2009 Aug;20(8):1687–92. doi: 10.1681/ASN.2009010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Somers EB, O’Shannessy DJ. Folate receptor alpha, mesothelin and megakaryocyte potentiating factor as potential serum markers of chronic kidney disease. Biomarker insights. 2014;9:29–37. doi: 10.4137/BMI.S15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lam AQ, Freedman BS, Morizane R, Lerou PH, Valerius MT, Bonventre JV. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J Am Soc Nephrol. 2014 Jun;25(6):1211–25. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takasato M, Er PX, Becroft M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014 Jan;16(1):118–26. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 70.Mae S, Shono A, Shiota F, et al. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nature communications. 2013;4:1367. doi: 10.1038/ncomms2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lam AQ, Freedman BS, Bonventre JV. Directed Differentiation of Pluripotent Stem Cells to Kidney Cells. Seminars in nephrology. 2014 Jul;34(4):445–61. doi: 10.1016/j.semnephrol.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nature reviews Molecular cell biology. 2014 Oct;15(10):647–64. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boletta A, Qian F, Onuchic LF, et al. Polycystin-1, the gene product of PKD1, induces resistance to apoptosis and spontaneous tubulogenesis in MDCK cells. Molecular cell. 2000 Nov;6(5):1267–73. doi: 10.1016/s1097-2765(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 74.Carone FA, Nakamura S, Bacallao R, Nelson WJ, Khokha M, Kanwar YS. Impaired tubulogenesis of cyst-derived cells from autosomal dominant polycystic kidneys. Kidney international. 1995 Mar;47(3):861–68. doi: 10.1038/ki.1995.129. [DOI] [PubMed] [Google Scholar]
- 75.Wilson PD, Schrier RW, Breckon RD, Gabow PA. A new method for studying human polycystic kidney disease epithelia in culture. Kidney international. 1986 Sep;30(3):371–8. doi: 10.1038/ki.1986.194. [DOI] [PubMed] [Google Scholar]
- 76.Saleem MA, O’Hare MJ, Reiser J, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002 Mar;13(3):630–8. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 77.Barua M, Shieh E, Schlondorff J, Genovese G, Kaplan BS, Pollak MR. Exome sequencing and in vitro studies identified podocalyxin as a candidate gene for focal and segmental glomerulosclerosis. Kidney international. 2014 Jan;85(1):124–33. doi: 10.1038/ki.2013.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kavoura E, Gakiopoulou H, Paraskevakou H, et al. Immunohistochemical evaluation of podocalyxin expression in glomerulopathies associated with nephrotic syndrome. Human pathology. 2011 Feb;42(2):227–35. doi: 10.1016/j.humpath.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 79.Doyonnas R, Kershaw DB, Duhme C, et al. Anuria, omphalocele, and perinatal lethality in mice lacking the CD34-related protein podocalyxin. The Journal of experimental medicine. 2001 Jul 2;194(1):13–27. doi: 10.1084/jem.194.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Self M, Lagutin OV, Bowling B, et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006 Nov 1;25(21):5214–28. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi A, Valerius MT, Mugford JW, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008 Aug 7;3(2):169–81. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliver G, Wehr R, Jenkins NA, et al. Homeobox genes and connective tissue patterning. Development. 1995 Mar;121(3):693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 83.Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008 Mar;26(3):313–15. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 84.Boulting GL, Kiskinis E, Croft GF, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011 Mar;29(3):279–86. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009 May 8;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nature methods. 2011 May;8(5):409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 87.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy. Series B, Physical and biological sciences. 2009;85(8):348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou T, Benda C, Duzinger S, et al. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011 Jul;22(7):1221–28. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi SM, Liu H, Chaudhari P, et al. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011 Aug 18;118(7):1801–5. doi: 10.1182/blood-2011-03-340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song B, Niclis JC, Alikhan MA, et al. Generation of induced pluripotent stem cells from human kidney mesangial cells. J Am Soc Nephrol. 2011 Jul;22(7):1213–20. doi: 10.1681/ASN.2010101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aasen T, Raya A, Barrero MJ, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008 Nov;26(11):1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 92.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010 Sep 16;467(7313):285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohi Y, Qin H, Hong C, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011 May;13(5):541–49. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009 Nov;27(11):1033–7. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 95.Sridharan R, Tchieu J, Mason MJ, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009 Jan 23;136(2):364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li M, Suzuki K, Kim NY, Liu GH, Izpisua Belmonte JC. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J Biol Chem. 2014 Feb 21;289(8):4594–9. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Disease models & mechanisms. 2013 Jul-Aug;6(4):896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013 Apr 4;12(4):393–4. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bellin M, Casini S, Davis RP, et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013 Dec 11;32(24):3161–75. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu GH, Suzuki K, Qu J, et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011 Jun 3;8(6):688–94. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yusa K, Rashid ST, Strick-Marchand H, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011 Oct 20;478(7369):391–4. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li LB, Chang KH, Wang PR, Hirata RK, Papayannopoulou T, Russell DW. Trisomy correction in Down syndrome induced pluripotent stem cells. Cell Stem Cell. 2012 Nov 2;11(5):615–9. doi: 10.1016/j.stem.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005 Mar;23(3):299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 105.Tokuda S, Higashi T, Furuse M. ZO-1 knockout by TALEN-mediated gene targeting in MDCK cells: involvement of ZO-1 in the regulation of cytoskeleton and cell shape. PLoS One. 2014;9(8):e104994. doi: 10.1371/journal.pone.0104994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014 Jan 3;343(6166):84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gifford CA, Ziller MJ, Gu H, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013 May 23;153(5):1149–63. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kamao H, Mandai M, Okamoto S, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem cell reports. 2014 Feb 11;2(2):205–18. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schwartz SD, Hubschman JP, Heilwell G, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012 Feb 25;379(9817):713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 110.Schwartz SD, Regillo CD, Lam BL, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015 Feb 7;385(9967):509–16. doi: 10.1016/S0140-6736(14)61376-3. [DOI] [PubMed] [Google Scholar]
- 111.Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R. Human embryonic stem cell lines derived from single blastomeres. Nature. 2006 Nov 23;444(7118):481–5. doi: 10.1038/nature05142. [DOI] [PubMed] [Google Scholar]
- 112.Turner M, Leslie S, Martin NG, et al. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013 Oct 3;13(4):382–4. doi: 10.1016/j.stem.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 113.Riolobos L, Hirata RK, Turtle CJ, et al. HLA engineering of human pluripotent stem cells. Mol Ther. 2013 Jun;21(6):1232–41. doi: 10.1038/mt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rong Z, Wang M, Hu Z, et al. An effective approach to prevent immune rejection of human ESC-derived allografts. Cell Stem Cell. 2014 Jan 2;14(1):121–30. doi: 10.1016/j.stem.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rogers SA, Hammerman MR. Prolongation of life in anephric rats following de novo renal organogenesis. Organogenesis. 2004 Jul;1(1):22–5. doi: 10.4161/org.1.1.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007 Dec 21;318(5858):1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]