Abstract
Late Embryogenesis Abundant (LEA) proteins are an ubiquitous group of polypeptides that were first described to accumulate during plant seed dehydration, at the later stages of embryogenesis. Since then they have also been recorded in vegetative plant tissues experiencing water limitation and in anhydrobiotic bacteria and invertebrates and, thereby, correlated with the acquisition of desiccation tolerance. This study provides the first comprehensive study about the LEA gene family in sweet orange (Citrus sinensis L. Osb.), the most important and widely grown fruit crop around the world. A surprisingly high number (72) of genes encoding C. sinensis LEAs (CsLEAs) were identified and classified into seven groups (LEA_1, LEA_2, LEA_3 and LEA_4, LEA_5, DEHYDRIN and SMP) based on their predicted amino acid sequences and also on their phylogenetic relationships with the complete set of Arabidopsis thaliana LEA proteins (AtLEAs). Approximately 60% of the CsLEAs identified in this study belongs to the unusual LEA_2 group of more hydrophobic LEA proteins, while the other LEA groups contained a relatively small number of members typically hydrophilic. A correlation between gene structure and motif composition was observed within each LEA group. Investigation of their chromosomal localizations revealed that the CsLEAs were non-randomly distributed across all nine chromosomes and that 33% of all CsLEAs are segmentally or tandemly duplicated genes. Analysis of the upstream sequences required for transcription revealed the presence of various stress-responsive cis-acting regulatory elements in the promoter regions of CsLEAs, including ABRE, DRE/CRT, MYBS and LTRE. Expression analysis using both RNA-seq data and quantitative real-time RT-PCR (qPCR) revealed that the CsLEA genes are widely expressed in various tissues, and that many genes containing the ABRE promoter sequence are induced by drought, salt and PEG. These results provide a useful reference for further exploration of the CsLEAs functions and applications on crop improvement.
Introduction
Late Embryogenesis Abundant (LEA) proteins have been found to accumulate in tissues of plants exposed to stresses that include dehydration or during some stages of plant development involving water limitation, such as seed, pollen grain, shoot and root development [1]. Their name is related to the fact that the originally described proteins were observed to accumulate at high levels during the later stages of embryo development [2,3]. Since the orthodox seeds acquire the ability to withstand and tolerate desiccation at this stage of development, LEA proteins have been associated with dehydration tolerance [4,5].
LEAs are widely distributed proteins in the plant kingdom, from algae to angiosperms, and they are also found in anhydrobiotic invertebrates and in some bacterial species [1]. They are mainly composed of hydrophilic amino acids arranged in repeated sequences, forming a highly hydrophilic structure and with thermal stability [5,6]. Analyses of their amino acid sequences have separated LEA proteins into seven different groups, each containing distinctive motifs [1]. LEAs appear located in many cell types and in varying concentrations. Inside the cell they appear predominantly, but not exclusively, in the cytosol [7]. Other locations include chloroplast, mitochondria, protein and lipid bodies, plasmodesmata and nucleus [8].
The importance of LEA proteins has been inferred from their abundance and expression patterns, as well as their overexpression in transgenic plants, since the in vivo activities for most of them remain unknown. LEA genes have been shown to be significantly induced by abiotic stresses, such as cold, drought and salinity, and their overexpression in transgenic plants has resulted in increased tolerance to such abiotic stresses [9–14]. For these reasons, it has been postulated that there is a positive correlation between the expression of LEAs and abiotic stress tolerance in plants [15,16]. The observations that silencing of one or two of the three LEA_4 proteins in Arabidopsis thaliana is sufficient to cause water deficit susceptibility [17] and that a DEHYDRIN gene co-segregated with chilling tolerance during cowpea (Vigna unguiculata L.) seedling emergence [18] also reinforce this interpretation.
The possible functions of LEA proteins have been demonstrated by in vitro experiments, where those LEAs from groups 2, 3 and 4 were observed to prevent the inactivation of enzymes like lactate dehydrogenase (LDH) and malate dehydrogenase (MDH) upon different levels of dehydration [19–21]. Similar protective properties of LEA proteins were also observed during in vitro freeze-thaw assays and attributed to the presence of the K-segment in LEA_2 and a conserved region in LEA_4 proteins [21,22]. Some LEA and LEA-like proteins have been also shown to avoid protein aggregation in in vitro dehydration assays [19,23]. This protective activity is thought to be carried out by direct protein-protein interactions [20,24]. LEA proteins have also another interesting functional property related to its supposed function as membrane stabilizers under stress conditions [18,25–27]. Additional functions have been suggested for LEA proteins, including ion sequestration, particularly for LEA_2 and LEA_4, where histidine-containing motifs seem to bind divalent cations [28], and oxidant scavenger [28,29].
Citrus are economically important fruit crops cultivated in many tropical and subtropical areas of the world, where they are constantly exposed to a range of environmental stresses that include drought, high salinity, and extreme temperature. For this reason, efforts are required to improve their tolerance to abiotic stresses, particularly by transferring genes related to salt and drought tolerance, such as those acting in osmotic adjustment or membrane stabilization, to citrus rootstocks [30]. Although the complete set of LEA protein encoding genes has been characterized in Arabidopsis [31], the characterization of citrus LEAs is still rudimentary. A cDNA clone homologue to the cotton LEA5 gene, named here as CsLEA49, was isolated from an ovule-derived cell suspension of ‘Shamouti’ sweet orange (Citrus sinensis L. Osbeck) and shown to be expressed in NaCl-treated cell suspension and leaves, as well as in seedlings exposed to drought and heat stress [32]. A cDNA clone encoding a DEHYDRIN (orthologue of CsLEA65) was also isolated from the epicarp of ‘Satsuma’ mandarin (Citrus unshiu) and observed to be expressed in different tissues and in leaves exposed to cold stress [22]. Overexpression of this gene in tobacco has enhanced the cold tolerance and inhibited the cold-induced lipid peroxidation in the transgenic plants [33]. Another cDNA encoding an orthologue DEHYDRIN was isolated from the flavedo of Fortune mandarin (Citrus clementina Hort. Ex Tanaka x Citrus reticulata Blanco) and found to be constitutively expressed in the fruit flavedo and highly induced in leaves exposed to cold and water stresses [34]. The purified protein was also observed to confer in vitro protection against freezing and dehydration inactivation for LDH and MDH enzymes [34].
The recent completion and publication of the draft genome sequences of sweet orange [35–37] now allows the identification and characterization of the complete repertoire of LEAs in citrus. Therefore, in this study we have carried out a genome-wide analysis of LEA protein encoding genes in the sweet orange genome in order to characterize their sequences, evolutionary relationships, putative functions and expression patterns in various tissues and in response to different abiotic stresses.
Materials and Methods
Plant material and stress treatments
For the drought stress experiment, two-year-old sweet orange [C. sinensis (L.) Osbeck var. Westin] plants grafted on the drought-tolerant citrus rootstock Rangpur lime (C. limonia Osbeck) were grown in plastic pots of 45L, containing a mixture of soil and sand (ratio 3:1), under optimal conditions (irrigated with tap water twice a week and fertilized weekly) in a greenhouse (25±4°C, 16 h of light and RH oscillating between 80 and 95%) for 90 days. After that, the pots were covered with aluminum foil to prevent water loss by evaporation, and a set of 10 plants was randomized over the experimental area and subjected to the following treatments: (i) control treatment, in which five plants were maintained at leaf predawn water potential values of -0.2 to -0.4 MPa by daily irrigation and (ii) drought treatment, in which the other five plants were exposed to a progressive soil water deficit until their leaves reach predawn water potential values of -1.5 MPa. The leaves were then harvested and immediately frozen in liquid nitrogen and stored at -80°C until use. The leaf predawn water potential was recorded on the third fully expanded mature leaf from the apex of each plant, between 2 AM and 4 AM, using a Scholander-type pressure pump (m670, Pms Instrument Co., Albany, USA).
For the salt and osmotic stress experiments, sweet orange [C. sinensis (L.) Osbeck var. Valencia] seeds were in vitro germinated as previously described [38]. Twenty-day-old seedlings of nucellar origin were selected, based on their uniformity, and then transferred to MS medium alone (control) or containing 150 mM NaCl (Merck, Darmstadt, Germany) or 25% polyethylene glycol 6000 (PEG-6000) (Merck, Darmstadt, Germany). Each treatment consisted of fifteen biological replicates. Leaves and roots were harvested 20 days after the treatments and immediately frozen in liquid nitrogen and stored at -80°C until use.
Identification and sequence analysis of LEA protein encoding genes in the sweet orange genome
The HMM (Hidden Markov Model) profiles of the PFAM (http://pfam.sanger.ac.uk/; [39]) motifs PF03760 (LEA1), PF03168 (LEA2), PF03242 (LEA3), PF02987 (LEA4), PF00477 (LEA5), PF00257 (DEHYDRIN), and PF04927 (SMP) were used as keywords to search the sweet orange genome sequence database (http://www.phytozome.org/citrus). The sweet orange genome was also queried by the 51 Arabidopsis LEA protein sequences downloaded from TAIR (http://www.arabidopsis.org), using the TBLASTN tool [40]. Comparison of the sizes of the different LEA groups in other species of higher plants was performed by searching their well-annotated genomes available at Phytozome (www.phytozome.net/), using the same strategy as outlined for sweet orange.
The molecular weight (MW) and GRAVY (grand average of hydropathy) of CsLEA proteins were predicted by the PROTPARAM tool (http://web.expasy.org/protparam/) [41]. The subcellular localization of the proteins was predicted by the WoLF PSORT tool available at http://www.genscript.com/psort/wolf_psort.html [42]. The exon-intron structure of the sweet orange LEA genes was analyzed using the sweet orange gene models annotated in Phytozome.
Multiple sequence alignment of the deduced amino acid sequences of CsLEA proteins were performed using the default parameters of ClustalX 2 [43] and the dendrogram was constructed by the neighbor joining (NJ) method [44] and bootstrap analysis (1,000 replications) in MEGA 6 program [45]. The protein motif analysis was conducted using the program MEME/MAST (http://meme.nbcr.net/meme/) [46].
Analysis of promoter regions and chromosomal locations of CsLEA genes
To identify the presence of the stress-responsive cis-acting regulatory elements ABRE (ABA-responsive element; ACGTG), DRE/CRT (dehydration responsive element/C-repeat; G/ACCGAC), MYBS (MYB binding site; TAACTG) and LTRE (low-temperature-responsive element; CCGAC) in their promoters, the one kb upstream region from the translation start site of the CsLEA genes was analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) [47]. The physical location of each CsLEA was determined by confirming the starting position of all genes on each chromosome, using BLASTN searching against the local database of the C. sinensis Annotation Project (CAP; http://citrus.hzau.edu.cn/orange/). MapChart software was used to plot the gene loci on the sweet orange chromosomes [48].
Expression analysis of CsLEAs
RNA-Seq data were downloaded from CAP [36] and used to generate the expression patterns of CsLEAs in different tissues, namely callus, flower, leaf and fruit (flesh tissue). The heatmap was generated using the Cluster 3.0 software.
Quantitative real-time RT-PCR (qPCR) analysis was used to measure the expression changes of CsLEAs in response to different abiotic stresses. Total RNA isolation and cDNA synthesis were performed as described previously [38]. qPCR primers were designed in order to avoid the conserved regions, by using the Primer 3 tool (http://bioinfo.ut.eeprimer3-0.4.0). GAPC2 (Glyceraldehyde-3-phosphate dehydrogenase C2) and UPL7 (ubiquitin protein ligase 7) were used as internal reference genes to normalize expression among the different samples [49]. Primer sequences are shown in detail in S1 Table. The qPCR reactions were run on a Stratagene Mx3005P real-time PCR System (Agilent Technologies, Santa Clara, CA, USA), following the manufacturers' instructions. The reactions were performed in triplicate, containing 10 μl (10 ng) of the cDNA sample, 10 nM of each forward and reverse primers, 12.5 μl Maxima® SYBR Green/ROX qPCR Master Mix (2X) (Fermentas, Maryland, USA) and sterile Milli-Q water for a final volume of 25 μl. The thermal cycling conditions were: 10 min at 95°C and 40 cycles of 15 s at 95°C, 30s at 60°C and 30s at 72°C. Data were obtained from a pool of three biological replicates that were individually validated.
Results and Discussion
LEA protein encoding genes in the sweet orange genome
Existing annotation in the sweet orange genome sequence database at Phytozome and BLAST searches using the 51 amino acid sequences of the complete set of A. thaliana LEA proteins (AtLEAs) as query sequences have resulted in the identification of a total of 72 different LEA encoding protein genes in C. sinensis (CsLEAs), which were distributed in seven distinct groups (LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, DEHYDRIN and SMP) (S2 Table). The sweet orange genome was observed to contain 4 LEA_1, 43 LEA_2, 5 LEA_3, 7 LEA_4, 3 LEA_5, 6 DEHYDRIN and 4 SMP proteins encoding genes. This number is far higher than the numbers of LEA genes previously reported in the genomes of rice (34) [50], Arabidopsis (51) [31], Chinese plum (30) [51], poplar (53) [52], tomato (27) [53] and potato (29) [54].
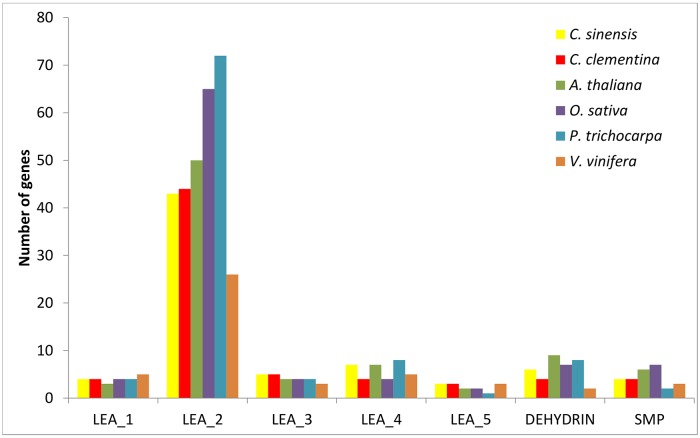
In order to compare the numbers of LEA genes within each group among the different species of higher plants, we searched the well-annotated genomes of Clementine mandarin (Citrus clementina), Arabidopsis (A. thaliana), rice (Oryza sativa), poplar (Populus trichocarpa) and grapevine (Vitis vinifera) available at Phytozome, using the same strategy as outlined for sweet orange. The analysis revealed that main differences occur in the LEA_2 group (Fig 1). The abundance of LEA_2 genes is lowest in grapevine (26) and especially higher in rice (65) and poplar (72). More significantly, such a large number of LEA_2 members have not been described in the previously investigated genomes of Arabidopsis [31], rice [50] and poplar [52]. This result may be explained in part by the improved annotation of the higher plant genomes available at Phytozome (v10.2) and also by the fact that LEA_2 is an unusual group composed of 'atypical' LEA proteins because of their more hydrophobic character. These findings suggest that the LEA protein family in higher plants may be larger and much more complex than previously described. On the other hand, minor variations were observed in the other sweet orange LEA groups, which showed a similar number of members in the analyzed higher plant genomes. This may indicate that the LEA_2 group has evolved later in higher plants.
Fig 1. Comparison of the size of the different LEA gene groups in sweet orange (Citrus sinensis), Clementine mandarin (Citrus clementina), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), poplar (Populus trichocarpa) and grapevine (Vitis vinifera).
Phylogenetic analysis and characteristics of CsLEA proteins
All the CsLEA proteins were aligned by the NJ method in ClustalX2. The resulting dendrogram shows that they were clustered into seven groups, helping to confirm or clarify their classification as proposed in the present study (Fig 2). In the latter case, two LEA protein encoding genes (CsLEA53 and CsLEA54) with no significant Pfam hits were aligned in the LEA_4 group and, hence, classified in this group. Conversely, CsLEA42 and CsLEA65 did not cluster with their respective LEA groups, despite containing the Pfam motifs that are characteristic of their groups. It may be due to the fact that CsLEA42 and CsLEA65 are, respectively, C-terminally extended and N-terminally truncated proteins compared to the other members of their respective groups.
Fig 2. Phylogenetic comparison of the complete set of 72 different LEA genes (CsLEAs) encoded in the sweet orange genome.
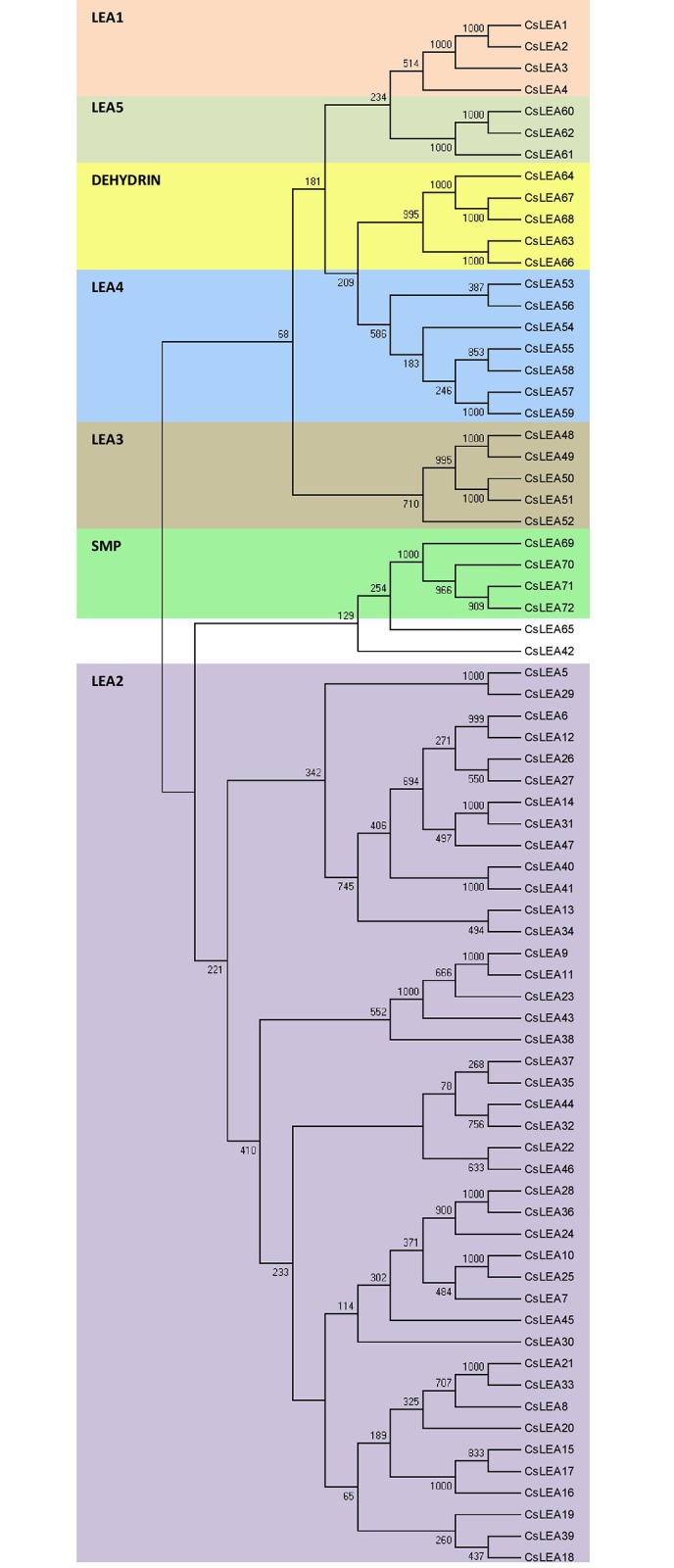
The different LEA groups are indicated by different colors. Sequence alignment was performed using ClustalX2 and the phylogenetic tree was generated using Bootstrap NJ tree (1,000 resamplings) method and MEGA program (v6.0.5).
Analysis of the physicochemical properties showed that the CsLEA proteins have a molecular weight ranging from 8.2 to 59.4 kDa, with the smaller proteins belonging to DEHYDRIN and LEA_3 groups (~8.5 kDa) and the largest proteins belonging to the LEA_4 group (59.4 kDa) (S2 Table). GRAVY values indicated that the CsLEA proteins are quite hydrophilic, except for the LEA_2 group that contains hydrophobic proteins. Similar characteristics have also been reported for LEA proteins of Arabidopsis [31], Chinese plum [51], poplar [52] and tomato [53], indicating that they are evolutionary conserved proteins in higher plants. The prediction of subcellular localization indicated that LEA_1 and DEHYDRIN proteins are exclusively located in the nucleus, with most members of LEA_5 and SMP and some members of LEA_2 and LEA_4 groups also targeted to this compartment (S2 Table). The majority of LEA_2 proteins are located in the cytoplasm (35.7%) or chloroplast (30.9%), with some of them also targeted to endoplasmic reticulum (RE) and mitochondrion. Most LEA_3 proteins are located in the chloroplast, while those from LEA_4 group have a more diverse localization, including chloroplast and mitochondrion (S2 Table). Thus, the CsLEA proteins can be present in all subcellular compartments, as reported for Arabidopsis [31] and tomato [53] LEAs; however, whether they have different functions in the different compartments, these functions need to be further determined.
Motif analysis of the predicted CsLEA proteins by the MEME program showed that members of each LEA group contain all the conserved motifs (Fig 3) that have been previously identified in other plant species, including Chinese plum [51], potato [54], tomato [53] and Arabidopsis [31]. For instance, an important conserved motif in the group DEHYDRIN is a repetitive 15-mer motif, EKKGIMDKIKEKLPG, called K-segment because of its richness in lysine (K) residues [1]. A particular protective role was described for this segment [20]. These results suggest that the CsLEAs are functional LEA proteins that play group-specific functions. Besides, the conserved motifs observed within each LEA group indicate that their members were likely originated from gene expansion within the groups.
Fig 3. Conserved motifs in the different groups of sweet orange LEA proteins (CsLEAs).
The conserved motifs were obtained using the MEME program.
Genomic organization of CsLEAs
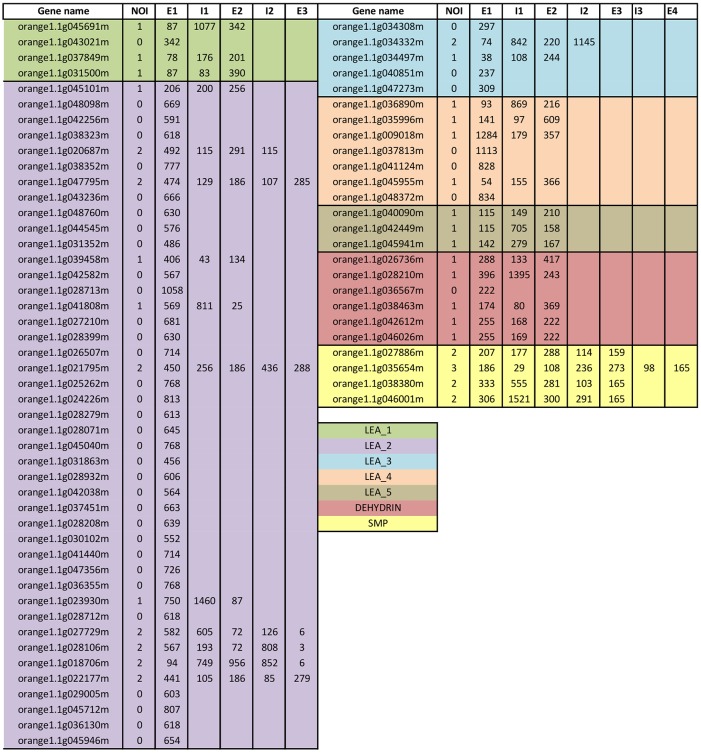
Analysis of the exon-intron structure of all 72 CsLEA genes was carried out using the sweet orange gene models annotated in Phytozome. The number and size of the exons, but not of the introns, were usually conserved within each LEA group (Fig 4). Most CsLEA genes contain no or few introns, with 56% of them having no intron, 43% with one or two introns, and only one gene (CsLEA70) of SMP group showing three introns (Fig 4). Similar exon-intron organization were also reported in LEA genes of Chinese plum [51], poplar [52], tomato [53] and potato [54]. It has been argued that genes related to the stress response usually contain few introns [55]. A proposed hypothesis to explain such observation is that introns may have a deleterious effect on gene expression, since they can delay transcript production due to the various steps of splicing and processing into mature mRNA, besides the additional energetic cost caused by the increased transcript length [55].
Fig 4. Exon-intron structure of the 72 sweet orange LEA genes (CsLEAs).
NOI denotes the number of introns, E the exon and I the intron. Numbers on the E and I columns indicate, respectively, the base pair length of the exonic and intronic sequences.
The positions of most CsLEA genes were mapped on the sweet orange chromosomes by homology searches against the full-length sweet orange genome assembly available at the C. sinensis Annotation Project (CAP) (S3 Table). Except for eight CsLEA genes that could not be exactly located on any chromosome (ChrUN) because of an incomplete physical map for sweet orange, all the loci were precisely mapped on the sweet orange chromosomes (Fig 5 and S4 Table). CsLEAs can be found on every chromosome, indicating a wide distribution of the gene family on sweet orange genome. However, the density of these loci was variable across the nine chromosomes of sweet orange. The largest number of genes was located on chromosomes 1 and 6 (12 genes each), followed by chromosome 2 (10 genes), chromosome 5 (8 genes), chromosome 8 (7 genes), chromosome 4 (5 genes), chromosomes 3 (4 genes) and chromosomes 7 and 9 (3 genes each). Members of the group LEA_1 were distributed on 2 (chr 1 and chr 7), LEA_2 on 8 (chr 1, chr 2, chr 3, chr 4, chr 5, chr 6, chr 8 and chr 9), LEA_3 on 3 (chr 1, chr 5 and chr 8), LEA_4 on 4 (chr 1, chr 2, chr 3 and chr 6), LEA_5 on 2 (chr 6 and chr 7), DEHYDRIN on 4 (chr 1, chr 2, chr 3 and chr 8) and SMP on 3 (chr 3, chr 5 and chr 9) different chromosomes. Interestingly, some closely related CsLEA isoforms showed significant reciprocal best BLAST hits to a single gene on CAP database and, thereby, they were mapped on the same chromosome positions (see S4 Table and Fig 5). These results might be due to the different sequencing depth and assembly quality between the sweet orange genomes available at the Phytozome and CAP databases.
Fig 5. Chromosomal locations of CsLEAs.
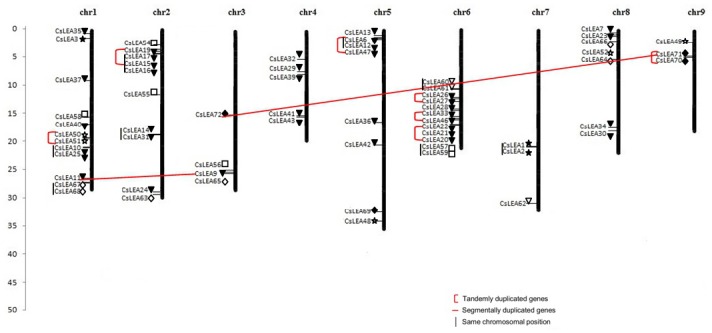
The chromosomal position of each CsLEA was mapped according to the Citrus sinensis Annotation Project (CAP). The CsLEA5/29 pair of segmentally duplicated genes is not indicated in the Figure since CsLEA5 did not map on any sweet orange chromosome (see S4 Table). The scale is in Mb. LEA_1 (closed star), LEA_2 (closed triangle), LEA_3 (open star), LEA_4 (open square), LEA_5 (open triangle), DEHYDRIN (open diamond) and SMP (closed diamond).
Segmental duplication, tandem duplication and transposition events are the main drivers of gene family expansion. To investigate potential gene duplications in sweet orange, segmental duplications and tandem duplications were identified. We detected 12 pairs of paralogous CsLEA genes based on phylogenetic analysis. Among them, three pairs of paralogous genes were putative segmental duplication events, according to criteria of Gu et al. [56]: the length of aligned sequence covers >80% of the longer gene and the similarity of the aligned region is >70%. These were the LEA_2 genes CsLEA5/29 (80.4% similarity) and CsLEA9/11 (74.6% similarity), and the SMP genes CsLEA71/72 (71.3% similarity). The other paralogous genes were putative tandem duplications according to the criteria of Hanada et al. [57], where tandem duplicates are genes in any gene pair, tandem 1 (T1) and tandem 2 (T2), that (1) belong to the same gene family, (2) are located within 100 kb each other, and (3) are separated by 10 or fewer nonhomologous spacer genes. These were the LEA_3 genes CsLEA50/51 on chromosome 1, the LEA_2 genes CsLEA15/16/17/19 on chromosome 2, CsLEA6/12/47 on chromosome 5, CsLEA26/27, CsLEA33/46 and CsLEA20/21/22 on chromosome 6, and the SMP genes CsLEA70/71 on chromosome 9 (S3 Table and Fig 5). Thus, these results suggest that segmental duplication and tandem duplication events have contributed to the expansion of the LEA gene family in sweet orange, especially in the large LEA_2 group, as proposed for the LEA genes of Arabidopsis [31], rice [50], Chinese plum [51], poplar [52], tomato [53] and potato [54].
Stress-responsive cis-acting regulatory elements in the CsLEA promoters
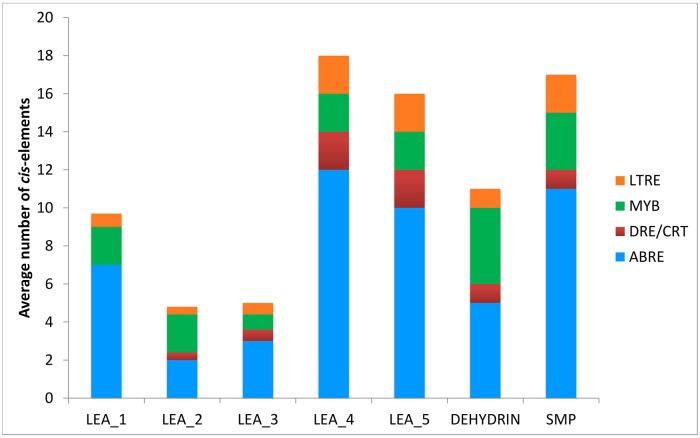
Analysis of the promoter region of all CsLEA genes identified the presence of various stress responsive cis-acting regulatory elements, including ABRE, DRE/CRT, MYBS and LTRE. These stress-responsive elements were relatively abundant in the promoters of the CsLEA genes, especially ABREs (Fig 6), indicating that LEA proteins may play an important role in abiotic stress response and tolerance in sweet orange. Variations in the average number of these promoter elements were observed among the different LEA groups (Fig 6). CsLEA genes from LEA_4, LEA_5 and SMP groups contained the highest average number of each of the four investigated stress-responsive elements, while those from LEA_2 and LEA_3 the lowest. Similar results, with the predominance of ABRE elements, have been reported for LEA genes in Arabidopsis [31], Chinese plum [51] and tomato [53]. ABRE is the major cis-acting element involved in ABA signaling during seed maturation and dormancy and also in response to abiotic stresses, while DRE/CRT and LTRE are major cis-acting regulatory elements involved in the ABA-independent gene expression in response to dehydration (DRE/CRT) and cold (DRE/CRT and LTRE) [58]. MYBS is another well known cis-acting regulatory element involved in the ABA-dependent signaling pathway in response to abiotic stresses, such as drought, salt and cold [59].
Fig 6. Average number of the cis-elements ABRE (ACGTG), DRE/CRT (G/ACCGAC), MYBS (TAACTG) and LTRE (CCGAC) in promoter region of sweet orange LEA genes from each LEA group.
The cis-elements were analyzed in the 1 kb upstream promoter region of translation start site using the PLACE database.
Expression patterns of CsLEAs in different tissues
In order to investigate the expression patterns of CsLEAs in different tissues, RNA-seq data were downloaded from CAP [36]. The heatmap generated demonstrates that all the 72 CsLEA genes are expressed in one or more of the major sweet orange tissues, namely callus, flower, leaf and fruit (Fig 7). The clustering revealed five main clades. CsLEA genes from clade 1 corresponded to family members from LEA_1, LEA_2 and LEA_3 groups displaying the highest expression in callus, flower and/or leaf tissues, while those from clade 2 included LEA_2, LEA_3 and DEHYDRIN members displaying the highest expression in flower, leaf and/or fruit. Clade 3 and clade 4 included genes from all the seven CsLEA groups with high expression, respectively, in callus and leaf or callus and fruit. Clade 5 corresponded to genes from LEA_2, LEA_3, LEA_5 and DEHYDRIN groups with high expression in fruit. Some CsLEAs showed preferential expression in a specific tissue, such as CsLEA8/11/28/51/58 in callus, CsLEA3/42 in flower and CsLEA36/63 in fruit (Fig 7). It suggests their involvement in developmental processes that are specific to these organs. In tomato, all the SlLEA genes were also observed to be expressed in at least one of the ten tissues tested, with some members of the LEA_2, LEA_3 and DEHYDRIN groups showing higher expression in all the tissues [53]. On the other hand, the expression of most Chinese plum PmLEAs (22/30) were higher in flower, with the expression of LEA_2 and DEHYDRIN members higher than that of the other groups [51].
Fig 7. Heatmap of expression of the 72 CsLEA genes in different tissues of sweet orange.
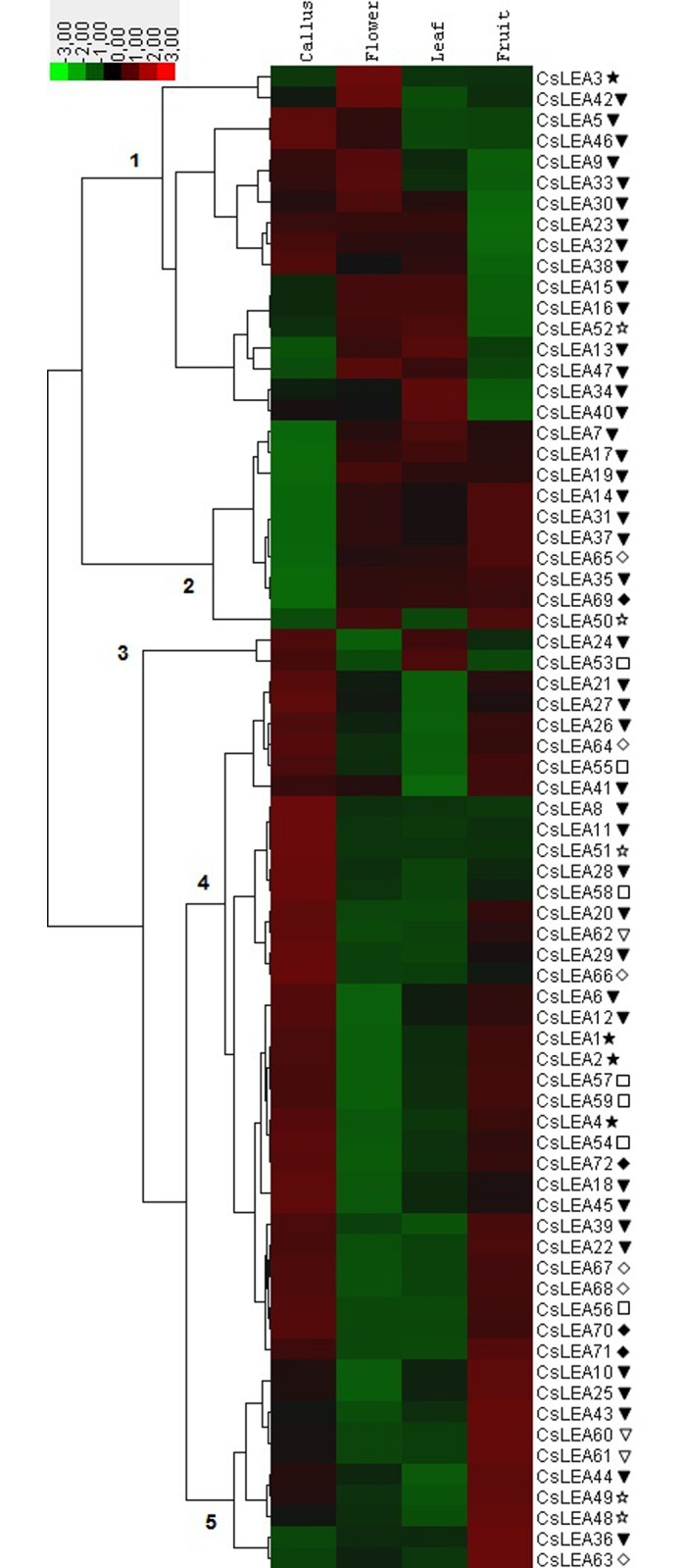
The heatmap was generated using Cluster 3.0 software. The color scale shown represents RPKM-normalized log2-transformed counts. LEA_1 (closed star), LEA_2 (closed triangle), LEA_3 (open star), LEA_4 (open square), LEA_5 (open triangle), DEHYDRIN (open diamond) and SMP (closed diamond).
Expression patterns of CsLEAs under abiotic stresses
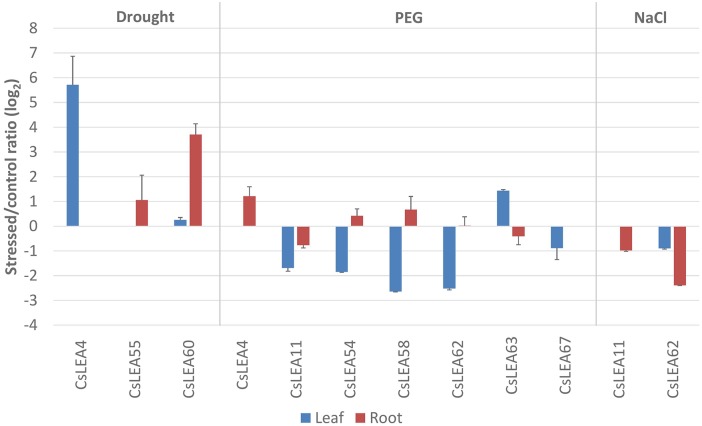
To identify CsLEA genes with a potential role in abiotic stress response, the expression patterns of 17 candidate genes were investigated by qPCR analysis in sweet orange plants exposed to drought, PEG and salinity. These genes were selected based on the presence of 10 or more ABRE elements in their promoter region (S5 Table). Of the 17 CsLEA genes analyzed, nine of them were observed to have their expression changed (log2 fold change of ≥1.0 or ≤-1.0 as cutoff threshold between stressed and control plants) in response to at least one stress condition and tissue analyzed (Fig 8). Analysis of expression of CsLEA genes under drought treatment showed an induction of mRNA expression of CsLEA4 in leaf and CsLEA55 and CsLEA60 in roots (Fig 8). Osmotic stress caused by PEG treatment induced the expression of CsLEA63 in leaf and CsLEA4 in roots, while this treatment downregulated the expression of CsLEA11, CsLEA54, CsLEA58, CsLEA62 and CsLEA67 in leaf. On the other hand, salt treatment caused a downregulation of CsLEA11 and CsLEA62 in roots. Taken together, these data suggest that the differentially expressed CsLEA genes belonging to the LEA_1, LEA_2, LEA_4, LEA_5 and DEHYDRIN groups may play a role in the adaptation of sweet orange to the tested stress conditions. Members of these LEA groups have been found to respond to abiotic stresses like drought and salt in different plant species. For example, members of the LEA_1, LEA_3, LEA_4 and DEHYDRIN groups were observed to be induced by drought stress in Arabidopsis, while some members of LEA_3, LEA_4, LEA_5 and DEHYDRIN groups were salt-induced [31]. Five members of the LEA_1, LEA_2, LEA_4 and DEHYDRIN groups were upregulated by drought and salt stresses in tomato [53]. In rice, LEA genes of the groups LEA_2, LEA_3 and DEHYDRIN exhibited strong response to osmotic stress (PEG), salt and ABA [50].
Fig 8. Expression profiles of the differentially expressed CsLEA genes in response to drought, PEG and salt (NaCl) stresses.
Ratios (log2) of relative mRNA levels between stressed and control plants in leaves and roots, as measured by qPCR. GAPC2 was used as an endogenous control. The bars show means ± SE from three biological replicates.
Conclusions
In the present study we have identified and characterized for the first time the whole repertoire of LEA encoding genes in the sweet orange genome. The results indicate that LEA constitute a large family of proteins in sweet orange, exhibiting a diversity of sequences, motif composition, gene structure, chromosomal locations and expression patterns. Segmental and tandem duplication events are proposed to be the main contributors to the expansion and functional diversification of the LEA gene family in sweet orange. The future efforts to elucidate their functional role, as well as to explore their potential on the genetic improvement of abiotic stress tolerance in citrus, should greatly benefit from the presented comprehensive analysis.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
The chromosomal position of each CsLEA was mapped according to the C. sinensis Annotation Project (CAP).
(DOCX)
(DOCX)
Acknowledgments
The authors are grateful to Dr. Eduardo A. Girardi (Embrapa Cassava & Fruits, Cruz das Almas, Bahia, Brazil) for kindly providing the grafted citrus plant material used in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by research grants from CNPq (Brasília, Brazil), CAPES (Brasília, Brazil) and FAPESP (São Paulo, Brazil). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Battaglia M, Covarrubias AA. Late Embryogenesis Abundant (LEA) proteins in legumes. Front Plant Science. 2013;25–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dure L, Greenway SC, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry. 1981;20: 4162–4168. [DOI] [PubMed] [Google Scholar]
- 3. Dure L, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination: XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiology. 1981;68:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cumming AC. LEA proteins In: Shewry P, Casey R, editors. Seed Proteins. Boston: Kluwer Acad; 1999. pp. 753–780. [Google Scholar]
- 5. Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. [DOI] [PubMed] [Google Scholar]
- 6. Hong-Bo S, Zong-Suo L, Ming-An S. LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids and Surfaces B: Biointerfaces. 2005;45:131–135. [DOI] [PubMed] [Google Scholar]
- 7. Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47: 377–403. [DOI] [PubMed] [Google Scholar]
- 8. Rorat T. Plant dehydrins—Tissue location, structure and function. Cel Mol Bio Lett. 2006;11: 536–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Babu RC, Zhang J, Blum A, Ho THD, Wu R, Nguyen HT. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 2004;166: 855–862. [Google Scholar]
- 10. Kim H-S, Lee JH, Kim JJ, Kim C-H, Jun S-S, Hong Y-N. Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum . Gene. 2005;344:115–123. [DOI] [PubMed] [Google Scholar]
- 11. Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol. 2006;17: 113–122. [DOI] [PubMed] [Google Scholar]
- 12. Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. Plos One. 2012;7(9): e45117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Checker VG, Chhibbar AK, Khurana P. Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Research. 2012;21: 939–957. 10.1007/s11248-011-9577-8 [DOI] [PubMed] [Google Scholar]
- 14. Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, et al. ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 2013;54:944–959. 10.1093/pcp/pct047 [DOI] [PubMed] [Google Scholar]
- 15. Reddy PS, Reddy GM, Pandey P, Chandrasekhar K, Reddy MK. Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: protection against abiotic stresses. Mol Biol Rep. 2012;39:7163–7174. 10.1007/s11033-012-1548-5 [DOI] [PubMed] [Google Scholar]
- 16. Mizoi J, Yamaguchi-Shinozaki K. molecular approaches to improve rice abiotic stress tolerance. Rice Protocols—Methods in Molecular Biology. 2013;956:269–283. [DOI] [PubMed] [Google Scholar]
- 17. Olvera-carrillo Y, Campos F, Reyes L, Garciarrubio A, Covarrubias AA. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010;154:373–390. 10.1104/pp.110.158964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ismail AM, Hall AE, Close TJ. Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 1999; 120:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;188:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reyes JL, Rodrigo MJ, Colmenero-Flores JM, Gil JV, Garay-Arroyo A, Campos F, et al. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 2005;28: 709–718. [Google Scholar]
- 21. Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, et al. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008;31: 1781–1790. 10.1111/j.1365-3040.2008.01879.x [DOI] [PubMed] [Google Scholar]
- 22. Hara M, Wakasugi Y, Ikoma Y, Yano M, Ogawa K, Kuboi T. cDNA sequence and expression of a cold-responsive gene in Citrus unshiu . Biosci Biotechnol Biochem. 1999;63: 433–437. [DOI] [PubMed] [Google Scholar]
- 23. Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci USA. 2007;104: 18073–18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olvera-carrillo Y, Reyes JL, Covarrubias AA. Versatile players in the plant adaptation to water limiting environments. Plant Signal Behav. 2011;6:586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koag MC, Fenton RD, Wilkens S, Close TJ. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003;131: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009;150: 1503–1514. 10.1104/pp.109.136697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kosová K, Vitámvás P, Prášil IT. The role of dehydrins in plant responses to cold. Biol Plant. 2007;51:601–617. [Google Scholar]
- 28. Hara M, Fujinaga M, Kuboi T. Metal binding by citrus dehydrin with histidine-rich domains. J Exp Bot. 2005;56:2695–2703. [DOI] [PubMed] [Google Scholar]
- 29. Liu GB, Xu H, Zhang L, Zheng YZ. Fe binding properties of two soybean (Glycine max L.) LEA4 proteins associated with antioxidant activity. Plant Cell Physiol. 2011;52: 994–1002. 10.1093/pcp/pcr052 [DOI] [PubMed] [Google Scholar]
- 30. Gong X-Q, Liu J-H. Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tiss Organ Cult. 2013;113: 137–147. [Google Scholar]
- 31. Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118 10.1186/1471-2164-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naot D, Ben-Hayyim G, Eshdat Y, Holland D. Drought, heat and salt stress induces the expression of a citrus homologue of an atypical late-embryogenesis Lea5 gene. Plant Mol Biol. 1995;27: 619–622. [DOI] [PubMed] [Google Scholar]
- 33. Hara M, Terashima S, Fukaya T, Kuboi T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta. 2003;217: 290–293. [DOI] [PubMed] [Google Scholar]
- 34. Sanchez-Ballesta MT, Rodrigo MJ, Lafuente MT, Granell A, Zacarias L. Dehydrin from Citrus, which confers in vitro dehydration tolerance and freezing protection activity, is constitutive and highly expressed in the flavedo of fruit but responsive to cold and water stress in leaves. J Agric Food Chem. 2004;2: 1950–1957. [DOI] [PubMed] [Google Scholar]
- 35. Xu Q, Chen L, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 2013;45: 59–66. 10.1038/ng.2472 [DOI] [PubMed] [Google Scholar]
- 36. Wang J, Chen D, Lei Y, Chang JW, Hao BH, Xing F, et al. Citrus sinensis Annotation Project (CAP): A comprehensive database for sweet orange genome. PLoS One. 2014;9(1): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biothecnol. 2014;32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Oliveira TM, Cidade LC, Gesteira AS, Coelho-Filho MA, Soares-Filho WS, Costa MGC. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet Genomes. 2011;7: 1123–1134. [Google Scholar]
- 39. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2011;40(D1): D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36: D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson JD, Higgens DG, Gibson TJ. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4: 406–425. [DOI] [PubMed] [Google Scholar]
- 45. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database:1999. Nucleic Acids Research. 1999;27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. [DOI] [PubMed] [Google Scholar]
- 49. Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One. 2012;7(2): e31263 10.1371/journal.pone.0031263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, Zhu H, Jin G, Liu H, Wu W, Zhu J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007;172: 414–420. [Google Scholar]
- 51. Du D, Zhang Q, Cheng T. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep. 2013;400: 1937–1946. [DOI] [PubMed] [Google Scholar]
- 52. Lan T, Gao J, Zeng Q. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes. 2013;9: 253–264. [Google Scholar]
- 53. Cao J, Li X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta. 2015;241: 757–772. 10.1007/s00425-014-2215-y [DOI] [PubMed] [Google Scholar]
- 54. Charfeddine S, Saïdi MN, Charfeddine M, Gargouri-Bouzid R. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol Biol Rep. 2015;42: 1163–1174. 10.1007/s11033-015-3853-2 [DOI] [PubMed] [Google Scholar]
- 55. Jeffares DC, Penkett CJ, Bahler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24: 375–378. 10.1016/j.tig.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 56. Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19: 256–262. [DOI] [PubMed] [Google Scholar]
- 57. Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148: 993–1003. 10.1104/pp.108.122457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress- responsive promoters. Trends Plant Sci. 2005;10: 88–94. [DOI] [PubMed] [Google Scholar]
- 59. Li C, Ng CK-Y, Fan L-M. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2015;114: 80–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
The chromosomal position of each CsLEA was mapped according to the C. sinensis Annotation Project (CAP).
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.