Abstract
Background
Prostate cancer is a one of the most common malignant diseases in men worldwide. Now it is a challenge to identify patients at higher risk for relapse and progression after surgery, and more novel prognostic biomarkers are needed. The aim of this study was to investigate the clinical significance of protocadherin17 (PCDH17) methylation in serum and its predictive value for biochemical recurrence (BCR) after radical prostatectomy.
Material/Methods
We evaluated the methylation status of PCDH17 in serum samples of 167 early-stage prostate cancer patients and 44 patients with benign prostatic hyperplasia (BPH) using methylation-specific PCR (MSP), and then evaluated the relationship between PCDH17 methylation and clinicopathologic features. Kaplan-Meier survival analysis and Cox analysis were used to evaluate its predictive value for BCR.
Results
The ratio of PCDH17 methylation in prostate cancer patients was higher than in patients with BPH. Moreover, PCDH17 methylation was significantly associated with advanced pathological stage, higher Gleason score, higher preoperative PSA levels, and BCR. Kaplan-Meier survival analysis indicated that patients with methylated PCDH17 had shorter BCR-free survival time compared to patients with unmethylated PCDH17. Cox regression analysis indicated that PCDH17 methylation was an independent predictive factor for the BCR of patients after radical prostatectomy.
Conclusions
PCDH17 methylation in serum is a frequent event in early-stage prostate cancer, and it is an independent predictor of BCR after radical prostatectomy.
MeSH Keywords: Biological Markers, DNA Methylation, Prostatic Neoplasms
Background
Prostate cancer is a common disease, especially in Western countries [1]. It is estimated that there will be 220 800 new cases and 27 540 deaths in the USA in 2015 [2]. Simultaneously, with changing lifestyles, the growing proportion of elderly people in the population, and improvement in awareness and disease surveillance, the morbidity of prostate cancer has increased in Eastern countries in the past few decades [3,4]. Now, with the widespread use of prostate-specific antigen (PSA) test and biopsy, more prostate cancers can be diagnosed at an early stage and treated by radical prostatectomy [5]. Unfortunately, approximately 30% of prostate cancer patients will suffer biochemical recurrence (BCR) after initial curative surgery, ultimately leading to death [6]. However, prostate cancer is a heterogeneous disease; it is difficult to distinguish indolent cases from aggressive ones. Although preoperative serum PSA, Gleason score, and TNM stages are routinely used in clinical practice as prognostic indicators, their accuracy is limited [7–9]. Thus, novel biomarkers are needed to improve the accuracy of prognosis.
Like many other tumors, the initiation and progression of prostate cancer arises from progressive genetic and epigenetic alterations [10]. Epigenetic modifications include chromation remodeling, nucleosome occupancy, histone modifications, and DNA methylation. In particular, DNA methylation is the most well-studied and well-described epigenetic change in human cancers [11]. Aberrant DNA methylation, particularly in gene promoter regions, results in transcriptional inactivation and silencing of tumor suppressor genes in tumors, and has been linked to a wide variety of important molecular pathways. Thus, DNA methylation is one of the most reliable sources for novel biomarker discovery [12–15]. Studies have indentified some new specific DNA methylation sites as biomarkers of prostate cancer. For example, aberrant methylation of some important genes, such as GSTP1, LINE-1, HSPB1, CCND2, and DPYS, are potential prognostic biomarkers of prostate cancer [16–19].
Protocadherin17 (PCDH17), a member of the cadherin superfamily, functions as a tumor suppressor in several human caners, including prostate cancer [20–23]. The human PCDH17 gene locates on chromosome 13q21.2, and is often inactivated by aberrant hypermethylation in tumors [20–24]. Our previous study has indicated that PCDH17 methylation is a frequent event in prostate cancer tissues, and it is correlated with malignant behaviors and poor outcomes of patients with prostate cancer [22]. DNA methylation can be detected in tumor tissues, as well as in body fluids, especially in serum [23–25]. DNA fragments are frequently and abundantly detected in serum of patients with malignant tumors, and the detection of serum tumor-related methylated genes may be used as a potential biomarker in human cancers [26–28].
To the best of our knowledge, this is the first study to investigate the clinical significance of PCDH17 methylation in serum of patients with prostate cancer. In the current study we analyzed the methylation status of PCDH17 in preoperative serum of prostate cancer patients, and then correlated it with clinical and pathological parameters, as well as patient outcome, in order to develop a potential prognostic biomarker for use at the time of diagnosis.
Material and Methods
Patients and serum samples
We recruited a total of 167 consecutive patients with prostate cancer undergoing radical prostatectomy (open surgery) and 44 patients with benign prostatic hyperplasia (BPH) undergoing transurethral resection of the prostate with histologically verified nonmalignant prostate tissues in the Third Hospital of Hebei Medical University between March 2006 and March 2010. None of the patients had received any kind of anticancer therapy before surgery. Prostatectomy specimens were reviewed by 2 senior pathologists to confirm the diagnosis and Gleason score. Relevant clinical data, such as age, serum PSA level before surgery, pathologic stage, and follow-up data, were recorded (Table 1). Whole peripheral blood samples (10 ml) were collected before surgery from all the patients. Clotting of serum samples was allowed for 60 min before centrifugation at 1800× gravity per minute for 10 min, and the supernatants were stored at −80°C. BCR was defined as the period between radical prostatectomy and the measurement of 2 successive values of serum PSA level ≥0.2 ng/ml [29]. All participants gave written informed consent according to the institutional guidelines before inclusion into the study. This study was performed according to the Declaration of Helsinki and was approved by the Ethics Committee of the Third Hospital of Hebei Medical University (No. HMU20050712X).
Table 1.
The clinicopathologic features of patients with prostate cancer (n=167), and the correlations between PCDH17 methylation and these features.
| Features | Variables | No. | M (%) | U (%) | P |
|---|---|---|---|---|---|
| Age (years) | ≤70 | 89 | 48 (53.9) | 41 (46.1) | 0.410 |
| >70 | 78 | 47 (60.3) | 31 (39.7) | ||
| Preoperative PSA (ng/ml) | ≤10 | 64 | 29 (45.3) | 35 (54.7) | 0.017 |
| >10 | 103 | 66 (64.1) | 37 (35.9) | ||
| Pathologic stage | T1 | 85 | 43 (50.6) | 42 (49.4) | 0.033 |
| T2 | 57 | 32 (56.1) | 25 (43.9) | ||
| T3 | 25 | 20 (80.0) | 5 (20.0) | ||
| Gleason score | ≤6 | 43 | 19 (44.2) | 24 (55.8) | 0.025 |
| 7 | 72 | 39 (54.2) | 33 (45.8) | ||
| ≥8 | 52 | 37 (71.2) | 15 (28.8) | ||
| Lymph node status | N0 | 149 | 82 (55.0) | 67 (45.0) | 0.164 |
| N1 | 18 | 13 (72.2) | 5 (27.8) | ||
| Surgical margin status | Negative | 156 | 88 (59.1) | 68 (40.9) | 0.640 |
| Positive | 11 | 7 (63.6) | 4 (36.4) | ||
| Recurrence | No | 120 | 55 (45.8) | 65 (54.2) | <0.001 |
| Yes | 47 | 40 (85.1) | 7 (14.9) |
M – methylation; U – unmethylation.
DNA extraction, bisulfite treatment, and methylation-specific PCR (MSP)
DNA was extracted from 1 ml of archived serum from each patient, using the QIAmp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. The isolated DNA was modified with bisulfite using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA, USA) and standard protocol, as described previously [21–24].The methylation status of the PCDH17 was detected using MSP, as described previously [21–24]. The primers were used as previously reported. Sequences were unmethylated: forward 5′-AGATTATTGGGTGTTGTAGTTT-3′ and reverse 5′-AACCCTAACACAACATACACA-3′; and methylated: forward 5′-GATTATCGGGTGTCGTAGTTC-3′ and reverse 5′-CCCTAACGCAACGTACGCG-3′ [21–24]. The PCR amplification was carried out as previously reported [21–24]. In vitro methylated DNA and unmethylated DNA (New England Biolabs, Beverly, MA, USA) were used as methylation and unmethylation positive control, as described previously. The MSP products were separated in 2% agarose gel, stained with ethidium bromide, and visualized under ultraviolet illumination for analysis. The product was defined as methylation-positive when methylated allele was present in the methylated DNA lane or both in the methylated and unmethylated DNA lanes, and was defined as methylation-negative when a band was present only in the unmethylated DNA lane, as reported previously [21–24].
Statistical analysis
The difference in PCDH17 methylation between prostate cancer patients and controls was evaluated using Fisher’s exact test. The associations between PCDH17 methylation and clinicopathologic features were evaluated by chi square test. For BCR-free survival analysis, Kaplan-Meier survival analysis was used and the differences in survival were analyzed using the log-rank test. Univariate and multivariate Cox regression analysis was used to evaluate the predictive effect of PCDH17 methylation in prostate cancer [30]. The a priori sample size was calculated and power analysis was performed. The statistical analyses were performed using SPSS 13.0 software. P<0.05 was considered to be statistically significant.
Results
PCDH17 methylation in serum samples
We first examined the methylation status of PCDH17 in serum samples of early-stage prostate cancer patients and patients with BPH. The results indicated that PCDH17 methylation occurred in 95 prostate cancer patients (56.9%) and in none of the patients with BPH, and the difference was statistically significant (P<0.001). A representative MSP result is shown in Figure 1.
Figure 1.
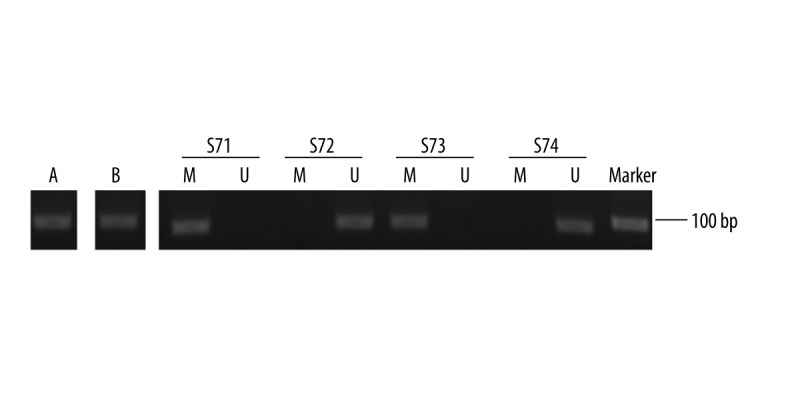
Representative MSP results for PCDH17 methylation in serum of patients with prostate cancer. A – methylation-positive control; B – unmethylation-positive control; S – serum sample from patients; S71 and 73 – exhibited methylated PCDH17; S72 and 74 – exhibited unmethylated PCDH17.
Correlations between PCDH17 methylation and clinicopathologic features
Subsequently, we correlated the methylation status of PCDH17 in serum with the clinicopathologic features of patients with prostate cancer. Aberrant methylation of PCDH17 was significantly associated with advanced pathological stage (P=0.033), higher Gleason score (P=0.025), higher preoperative PSA levels (P=0.017), and BCR (P<0.001). There were no statistically significant associations between PCDH17 methylation and age, lymph node status, or surgical margin status. These findings are presented in Table 1.
PCDH17 methylation predicts BCR after radical prostatectomy
The follow-up data was obtained from all the prostate cancer patients. The patients were followed up at between 7 and 60 months (median 60 months). BCR was detected in 47 (28.1%) patients during the follow-up period. To investigate the association of PCDH17 methylation with BCR, patients were divided into a methylated PCDH17 group and an unmethylated PCDH17 group. BCR-free survival curves were established according to PCDH17 methylation status. Kaplan-Meier survival analysis and log-rank test suggested that patients with methylated PCDH17 had a significantly shorter BCR-free survival time than patients with unmethylated PCDH17 (Figure 2, x2=19.458, P<0.001, log-rank test). Moreover, univariate regression analysis indicated that higher preoperative PSA level, higher Gleason score, and PCDH17 methylation were significantly associated with poor BCR-free survival. All factors determined to be significant in the univariate analysis were tested with multivariate analysis for association with BCR-free survival. Multivariate analysis demonstrated that higher Gleason score and PCDH17 methylation were independently associated with shorter BCR-free survival. These results are shown in Table 2.
Figure 2.
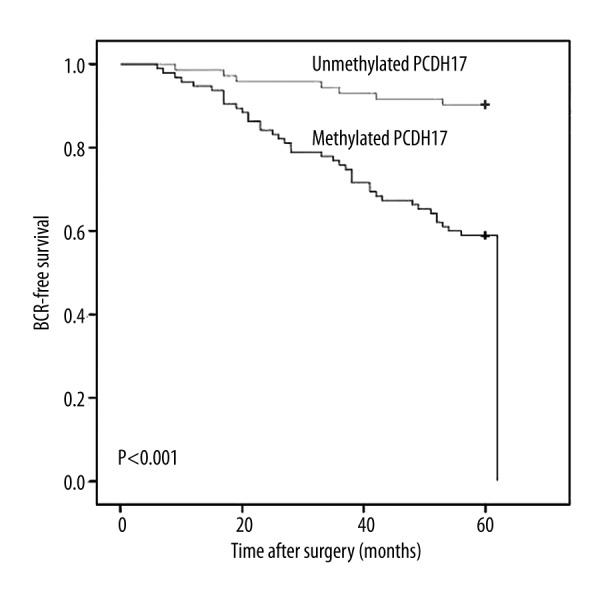
Associations between PCDH17 methylation and BCR-free survival of patients after radical prostatectomy. Patients with methylated PCDH17 showed significantly shorter BCR-free survival than those with unmethylated PCDH17 (P<0.001, log-rank test).
Table 2.
The prognostic value of PCDH17 methylation for the BCR-free survival in univariate and multivariate Cox regression analysis.
| Varivale | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Exp (B) | 95% CI | P | Exp (B) | 95% CI | P | |
| Age | 1.003 | 0.693–1.537 | 0.874 | 1.425 | 0.819–6.305 | 0.067 |
| PSA | 2.537 | 1.049–3.641 | 0.003 | |||
| Pathologic Stage | 1.104 | 0.921–2.466 | 0.571 | |||
| Gleason score | 3.237 | 1.369–5.824 | <0.001 | 1.875 | 1..43–4.382 | 0.001 |
| Lymph node status | 1.136 | 0.798–2.594 | 0.542 | |||
| Surgical margin status | 1.078 | 0.896–1.763 | 0.755 | |||
| PCDH17 methylation | 4.331 | 1.783–6.583 | <0.001 | 3.167 | 1.534–5.073 | <0.001 |
Discussion
Prostate cancer is a heterogeneous disease with outcome difficult to predict. The major challenge in prostate cancer management is to distinguish aggressive tumors from indolent ones at the time of diagnosis, due to the limited accuracy of currently available predictive factors, such as serum PSA, Gleason score, and tumor stage [31]. Therefore, there is an urgent need to develop sensitive, non-invasive, and cost-effective early prognosis biomarkers.
Aberrant DNA methylation results in the silencing of tumor suppressor genes, contributing to cancer initiation, progression, invasion, and metastasis, and thus could be a potential candidate for inclusion into improved prognostic factors, either alone or in combination with other molecular markers [32]. It has been shown that DNA methylation can not only be detected in tissue samples, but also in body fluids, such as blood, urine, and cerebrospinal fluid, allowing for development of non-invasive biomarkers [13,32–34]. It is known that the circulating DNA in serum results from apoptosis or necrosis of tumor cells and originates from neoplastic cells, and the circulating DNA has a methylation pattern similar to that in the primary tumor. Previous studies have shown that detection of aberrant tumor-related DNA methylation in serum is feasible and may be useful for development of clinically relevant prognostic biomarkers for human cancers [13,32–34].
In the present study, we detected the methylation status of PCDH17 in serum samples of early-stage prostate cancer patients and patients with BPH by using MSP. PCDH17 has been reported to be hypermethylated in urologic cancers [35]. Our data indicate that the frequency of PCDH17 methylation in serum of prostate cancer patients was 56.9%, but no methylation of PCDH17 was found in the controls. The difference of PCDH17 methylation in serum between prostate cancer patients and controls was significant, in accordance with a previous study in prostate cancer tissues [22]. Because DNA methylation plays an important role in the initiation and progression of tumors, we further explored the clinicopathological significance and prognostic value of PCDH17 methylation in prostate cancer patients. According to our findings, PCDH17 methylation was significantly associated with advanced pathological stage, higher Gleason score, and higher preoperative PSA levels. These features are all considered as risk factors for the prognosis of prostate cancer. Thus, PCDH17 methylation in serum may be associated with poor outcome of prostate cancer patients. Moreover, PCDH17 methylation was more frequent in patients with BCR than in patients without BCR. Our findings are similar to those of previous studies in renal cell carcinoma and bladder cancer [21,23,24]. These results suggest that PCDH17 methylation in serum may be a useful predictor for BCR of prostate cancer after surgery.
Subsequently, the prostate cancer patients were divided into 2 groups according to the methylation status of PCDH17 in serum: a methylated PCDH17 group and an unmethylated PCDH17 group. Then, the BCR-free survival curves of the patients were established and the log-rank test indicated that patients with methylated PCDH17 had worse BCR-free survival than patients without methylated PCDH17. Furthermore, univariate Cox regression analysis suggested that PCDH17 methylation is an important risk factor of BCR, and multivariate Cox regression analysis verified that PCDH17 methylation is an independent predictor for BCR after surgery. Our findings are similar to those of a previous study in bladder cancer [24]. The results indicate that PCDH17 methylation in serum has significant independent prognostic value for prediction of BCR after radical prostatectomy, at the time of diagnosis, beyond routine clinicopathologic predictors. Patients with methylated PCDH17 in serum have increased risk of BCR after surgery; therefore, adjuvant radiotherapy or chemotherapy should be performed after surgery in order to prevent tumor recurrence and progression.
Our study has some limitations. Our cohort consisted of only 167 patients with prostate cancer, and this study only identified 1 candidate marker. Thus, prospective studies are needed to identify more DNA methylations that could have higher predictive capability as a panel. The use of BCR as a clinical endpoint is also a limitation of our study, and other clinical endpoints, such as cancer-specific survival and overall survival, should be included in future studies. Given the long disease course of prostate cancer, this would require large cohorts with decades of follow-up [31].
Conclusions
Our findings suggest that PCDH17 methylation in serum is a frequent event in early-stage prostate cancer, and is associated with malignant clinicopathologic factors of prostate cancer. Furthermore, PCDH17 methylation in serum is an independent predictor of BCR after radical prostatectomy. Our study supports a further role for PCDH17 methylation in serum in identifying patients at higher risk for BCR who may be benefit from more aggressive therapies after surgery.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare in relation to this article.
Source of support: This study was supported by Xuzhou Medical Talented Youth Project (No: 2014007), Xuzhou Science and Technology Project (No: KC14SH015), Jiangsu University Clinical Fund (No: JLY20140109), Jiangsu Province Health and Family Planning Fund (No: Q201514), and Jiangsu Province “six talents peak” Project (No: 2014-WSW-066)
References
- 1.Kapoor A. What’s New in Prostate Cancer Research? Highlights of GU-ASCO 2015. Can Urol Assoc J. 2015;9(5–6 Suppl 3):S148–53. doi: 10.5489/cuaj.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Basu S, Majumder S, Bhowal A, et al. A study of molecular signals deregulating mismatch repair genes in prostate cancer compared to benign prostatic hyperplasia. PLoS One. 2015;10(5):e0125560. doi: 10.1371/journal.pone.0125560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Zhang G, Wei M, et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein. Cancer Biol Ther. 2014;15(1):108–18. doi: 10.4161/cbt.26722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidenreich A, Bastian PJ, Bellmunt J, et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2014;65(2):467–79. doi: 10.1016/j.eururo.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Strand SH, Orntoft TF, Sorensen KD. Prognostic DNA methylation markers for prostate cancer. Int J Mol Sci. 2014;15(9):16544–76. doi: 10.3390/ijms150916544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grishina YA, Khmelevsky EV. Radiotherapy for local recurrences of prostate cancer after radical prostatectomy. Vopr Onkol. 2015;61(1):7–13. [PubMed] [Google Scholar]
- 8.Filson CP, Marks LS, Litwin MS. Expectant management for men with early stage prostate cancer. Cancer J Clin. 2015;65(4):265–82. doi: 10.3322/caac.21278. [DOI] [PubMed] [Google Scholar]
- 9.Esfahani M, Ataei N, Panjehpour M. Biomarkers for evaluation of prostate cancer prognosis. Asian Pac J Cancer Prev. 2015;16(7):2601–11. doi: 10.7314/apjcp.2015.16.7.2601. [DOI] [PubMed] [Google Scholar]
- 10.Adjakly M, Ngollo M, Dagdemir A, et al. Prostate cancer: The main risk and protective factors-Epigenetic modifications. Ann Endocrinol (Paris) 2015;76(1):25–41. doi: 10.1016/j.ando.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 12.Paska AV, Hudler P. Aberrant methylation patterns in cancer: A clinical view. Biochem Med (Zagreb) 2015;25(2):161–76. doi: 10.11613/BM.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahara T, Arisawa T. DNA methylation as a molecular biomarker in gastric cancer. Epigenomics. 2015;7(3):475–86. doi: 10.2217/epi.15.4. [DOI] [PubMed] [Google Scholar]
- 14.Shiovitz S, Grady WM. Molecular markers predictive of chemotherapy response in colorectal cancer. Curr Gastroenterol Rep. 2015;17(2):431. doi: 10.1007/s11894-015-0431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanwal R, Gupta K, Gupta S. Cancer epigenetics: an introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, Jiao H, Zhang X, et al. Correlation between the expression of DNMT1, and GSTP1 and APC, and the methylation status ofGSTP1 and APC in association with their clinical significance in prostate cancer. Mol Med Rep. 2015;12(1):141–46. doi: 10.3892/mmr.2015.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasiljević N, Ahmad AS, Thorat MA, et al. DNA methylation gene-based models indicating independent poor outcome in prostate cancer. BMC Cancer. 2014;14:655. doi: 10.1186/1471-2407-14-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry KH, Moore LE, Liao LM, et al. Prospective study of DNA methylation at LINE-1 and Alu in peripheral blood and the risk ofprostate cancer. Prostate. 2015;75(15):1718–25. doi: 10.1002/pros.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Shen Y, Dai D, et al. Meta-analyses of methylation markers for prostate cancer. Tumour Biol. 2014;35(10):10449–55. doi: 10.1007/s13277-014-2300-7. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Yasuda S, Tanaka H, et al. Non-clustered protocadherin. Cell Adh Migr. 2011;5(2):97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YL, Gui SL, Guo H, et al. Protocadherin17 promoter methylation is a potential predictive biomarker in clear cell renal cell carcinoma. Med Sci Monit. 2015;21:2870–76. doi: 10.12659/MSM.895603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin YL, Xie PG, Wang L, et al. Aberrant methylation of protocadherin 17 and its clinical significance in patients with prostate cancer after radical prostatectomy. Med Sci Monit. 2014;20:1376–82. doi: 10.12659/MSM.891247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XB, Lin YL, Li ZG, et al. Protocadherin 17 promoter methylation in tumour tissue from patients with bladder transitional cell carcinoma. J Int Med Res. 2014;42(2):292–99. doi: 10.1177/0300060513504364. [DOI] [PubMed] [Google Scholar]
- 24.Luo ZG, Li ZG, Gui SL, et al. Protocadherin-17 promoter methylation in serum-derived DNA is associated with poor prognosis of bladder cancer. J Int Med Res. 2014;42(1):35–41. doi: 10.1177/0300060513504705. [DOI] [PubMed] [Google Scholar]
- 25.Severi G, Southey MC, English DR, et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat. 2014;148(3):665–73. doi: 10.1007/s10549-014-3209-y. [DOI] [PubMed] [Google Scholar]
- 26.Raja UM, Gopal G, Rajkumar T. Intragenic DNA methylation concomitant with repression of ATP4B and ATP4A gene expression in gastric canceris a potential serum biomarker. Asian Pac J Cancer Prev. 2012;13(11):5563–68. doi: 10.7314/apjcp.2012.13.11.5563. [DOI] [PubMed] [Google Scholar]
- 27.Jing F, Jun L, Yong Z, et al. Multigene methylation in serum of sporadic Chinese female breast cancer patients as a prognostic biomarker. Oncology. 2008;75(1–2):60–66. doi: 10.1159/000155145. [DOI] [PubMed] [Google Scholar]
- 28.How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications. Biochimie. 2012;94(11):2314–37. doi: 10.1016/j.biochi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Carter HB. American Urological Association (AUA) guideline on prostate cancer detection: process and rationale. BJU Int. 2013;112(5):543–47. doi: 10.1111/bju.12318. [DOI] [PubMed] [Google Scholar]
- 30.Niu WB, Gui SL, Lin YL, et al. Promoter methylation of protocadherin8 is an independent prognostic factor for biochemical recurrence of early-stage prostate cancer. Med Sci Monit. 2014;20:2584–89. doi: 10.12659/MSM.893083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristensen H, Haldrup C, Strand S, et al. Hypermethylation of the GABRE~miR-452~miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2014;20(8):2169–81. doi: 10.1158/1078-0432.CCR-13-2642. [DOI] [PubMed] [Google Scholar]
- 32.Strand SH, Orntoft TF, Sorensen KD. Prognostic DNA methylation markers for prostate cancer. Int J Mol Sci. 2014;15(9):16544–76. doi: 10.3390/ijms150916544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol. 2015;36(1):7–19. doi: 10.1007/s13277-014-2758-3. [DOI] [PubMed] [Google Scholar]
- 34.Gao T, He B, Pan Y, et al. The association of retinoic acid receptor beta2(RARβ2) methylation status and prostate cancerrisk: A systematic review and meta-analysis. PLoS One. 2013;8(5):e62950. doi: 10.1371/journal.pone.0062950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa VL, Henrique R, Danielsen SA, et al. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6(9):1120–30. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]


