Abstract
In fish, oocyte meiotic maturation is regulated by 17α, 20β-dihydroxy-progesterone through cAMP. To study the role of cAMP response element binding protein (CREB) in meiotic maturation, we cloned and characterized the expression pattern of CREBs from two fish models, the Nile tilapia and catfish. In the Nile tilapia three different CREBs were identified where in CREB1 was found in many tissues including gonads with abundant expression in testis. CREB2, few amino acids shorter than CREB1, was expressed in several tissues with abundant expression in ovary. In addition, a 3’UTR variant form, CREB3 was exclusively found in ovary. During natural 14-day ovarian cycle of the Nile tilapia, CREB1 expression was stable throughout vitellogenesis with a sharp decrease on the day of spawning. In contrast, CREB2 remain unchanged throughout the ovarian cycle, however elevated in 11-day full-grown immature ovarian follicle and after hCG-induction. Interestingly, CREB3 expression was induced three folds on the day of spawning as well as during hCG-induced oocyte maturation. Based on the synergistic expression pattern, CREB1 is likely to control oocyte growth, whereas CREB 2 and 3 contribute to oocyte maturation in tilapia and the latter seems to be critical. In catfish, a single form of CREB showed a maximum expression during spawning phase and hCG-induced maturation both in vivo and in vitro augmented CREB expression. These results suggest that spatial and temporal expression of CREBs seems to be important for final oocyte maturation and may also regulate oocyte growth in fish.
Introduction
Oogenesis in teleosts in its broadest sense comprises of two phases, oocyte growth (vitellogenesis) and final maturation (resumption of meiosis) that is regulated by follicle stimulating hormone (FSH) and luteinizing hormone (LH) respectively [1]. In certain cases, LH regulates both the processes with very less role for FSH [2,3]. It is well established that, estradiol 17β (E2) produced in ovarian granulosa cells by the enzyme ovarian cytochrome P450 aromatase (Cyp19a1a), largely controls vitellogenesis [1]. On the other hand, oocyte maturation is promoted [1] by the maturation inducing steroid, 17α, 20β-dihydroxyprogesterone (17α, 20β-DP), produced in ovarian granulosa cells by 20β-hydroxysteroid dehydrogenase (20β-HSD). Plasma E2 levels rise gradually throughout oocyte growth and decreases suddenly with the onset of oocyte maturation while 17α, 20β-DP levels stays basal during oocyte growth and increases sharply with maturation [4]. This shift in steroidogenesis seems to be a critical step during oocyte maturation in several teleost species [1,5].
Molecular mechanisms governing shift in steroidogenesis were studied in greater details in tilapia and to some extent in catfish model [1,5,6]. In tilapia, consistent with plasma steroid levels, a gradual increase in expression and activity of Cyp19a1a throughout vitellogenesis and a diminished or undetectable activity and expression in post-vitellogenic ovarian follicles/during meiotic maturation was found [7]. A down-regulation of Cyp19a1a expression was noticed when post-vitellogenic follicles of tilapia were incubated with hCG, in vitro [7]. Further studies demonstrated that AD4BP/SF-1 and FOXL-2 regulate the expression Cyp19a1a [7–9]. An increase in 20β-HSD mRNA and/or activity is known in ayu, catfish and trout [10–12] during oocyte maturation, whereas in tilapia, 20β-HSD expression was found to be basal in vitellogenic follicles, undetectable in post-vitellogenic follicles and a rise in expression was observed during oocyte maturation [13]. Based on these studies, it is suggested that shift in steroidogenesis is governed by subjugation of Cyp19a1a expression and an increase in 20β-HSD expression [1,5].
Transcriptional regulation of Cyp19a1a is relatively well studied [7,8,14]. However, 20β-HSD promoter was not analyzed explicitly except for a study from our laboratory [15]. We have previously reported the characterization of Cyp19a1a promoter and its regulation by Ad4BP/SF-1 [7] and 20β-HSD promoter by cAMP responsive elements [15]. Since, expression of both 20β-HSD and Cyp19a1a in granulosa cells is modulated by gonadotropins via cAMP, understanding how cAMP controls the up-regulation of 20β-HSD and down regulation of Cyp19a1a during steroidogenic shift is interesting. cAMP responsive elements have been identified on both 20β-HSD [15] and Cyp19a1a promoter motifs [7,14], hence transcriptional regulation of these genes during shift in steroidogenesis could possibly occur by spatial and temporal expression of CREBs. Present study is intended to test the possibilities of how CREB’s regulate the shift in steroidogenesis vis-à-vis final oocyte maturation by characterizing the expression pattern. We have identified multiple forms of CREBs from gonads of Nile tilapia (Oreochromis niloticus) and a single form of CREB from air-breathing catfish (Clarias gariepinus) and characterized their expression patterns during natural and gonadotropin (hCG) induced final oocyte maturation. To our knowledge, this is the first report to implicate a pivotal role for CREBs in meiotic maturation in any lower vertebrates.
Materials and Methods
Animals
The Nile tilapia has a fortnight spawning cycle and is an excellent model to study oocyte growth and maturation events. The average ovarian cycle is of 14–18 days within which the ovarian follicles undergo different developmental stages. Ovarian follicle begins early vitellogenesis (1–4 days) a day after ovulation and passes through mid vitellogenic stage at about 5–7 days and becomes full grown immature follicle by 8 – 11th day. Spawning usually occurs on 14th day and in some cases it may extend up to 18th day [7,13]. The Nile tilapias were reared and maintained as described earlier and ovarian follicles at different stages of natural ovarian cycle were collected after careful observation and utilized for gene expression analysis [13]. Experiments involving hCG-induced oocyte maturation are done as described earlier using 11 day ovarian follicles isolated from tilapia which has 14-day spawning cycle tested at least for three consecutive durations as explained earlier [13].
Air-breathing Catfish, Clarias gariepinus has an annual ovarian cycle. Preparatory phase (January–April) in which vitellogenesis begins and passes through mid vitellogenic stage. In pre-spawning phase (May–June) ovarian follicles develop through late vitellogenic stage and become full-grown immature ovarian follicles ready to be spawned. The spawning phase lasts typically in July–October that matches with Monsoon season in the South India and we observed an extended spawning period that overlaps with post-spawning phase of catfish in the North Indian region. Ovary in the months of November–December is generally in resting phase. Catfish used in this study were purchased locally from farmers. They were maintained under normal photoperiod and ambient temperature conditions in aquarium tanks during acclimation and experimentation. Feeding as well as rearing of catfish and the hCG-induced oocyte maturation experiments are performed as described earlier [13]. All the experiments conducted on the Nile tilapia and catfish were following general guidelines of Institutional Animal Ethical Committee (IAEC, National Institute of Basic Biology, Japan and University of Hyderabad, India respectively). An approval was not required for the edible fish sacrifice.
Cloning and sequence analysis of CREB cDNAs
RT-PCR amplification of partial cDNAs homologous to CREB
Based on nucleotide sequences of vertebrate CREB’s, sets of degenerate primers were (Table 1) designed and used in RT-PCR amplification of ovarian first-strand cDNA preparation. The amplicons were cloned in pGEM-T easy (Promega) vector and subsequently sequenced. The similarity of cloned sequences was assessed by performing BLAST analysis.
Table 1. Oligonucleotide sequences used in cloning and expression analysis of tilapia and catfish CREBs.
| Primer | Sequence 5' to 3' | Purpose |
|---|---|---|
| DF1 | CATMTATCAGACYAGCASSGGSCA | For amplification of partial CREB cDNA from Tilapia and catfish. |
| DR1 | CYTTCTTCTTCCTGCGACACTC | For amplification of partial CREB cDNA from Tilapia and catfish. |
| NT CREB1-F | GGCACAGATTGCTACTTTGG | For RT-PCR of Tilapia CREB 1 |
| NT CREB1-R | CAGGTGTGGCAGCAGCAGC | For RT-PCR of Tilapia CREB 1 |
| NT CREB2-F | GGAGTACGTGAAGTGTCTGGAG | For RT-PCR of Tilapia CREB 2 |
| NT CREB2-R | CTGATGGTTGATTTCAAATTGCTC | For RT-PCR of Tilapia CREB 2 |
| NT CREB3-F | CTGGGTAAATCTACCGCTCATC | For RT-PCR of Tilapia CREB 3 |
| NT CREB3-R | GAACATTTGTTTGTTTTAATATATG | For RT-PCR of Tilapia CREB 3 |
| CF GSP-R1 | AGTTTGCAGCCCTTGCACGCCGTC | For 5' RACE of catfish CREB |
| CF GSP-R2 | CTGGATGGCTCCACCCTGTGTGAT | For 5' RACE of catfish CREB |
| CF GSP-F1 | CGCCTCATGAAGAACAGGGAAGC | For 3' RACE of catfish CREB |
| CF GSP-F2 | AGGGAAGCGGCCCGAGAGTGTCGC | For 3' RACE of catfish CREB |
| CF qRT-F | CGTCCTTCTTACAGGAAGATCC | For Real-time RT-PCR of catfish CREB |
| CF qRT-R | TCTCTGAGCTGTATTTGGCACG | For Real-time RT-PCR of catfish CREB |
cDNA library construction and screening
cDNA libraries from tilapia ovary and testis were constructed as described earlier [13]. In brief, 5 μg of poly (A)+ RNA was isolated from ovarian follicles and a λl-ZAP library was constructed. The library was packaged into UNI-ZAP XR using Gigapack II gold packaging extract as per the manufacturer instructions. Partial cDNAs cloned (described in the above section) were labeled with α-P32 using random hexa-nucleotide kit. Libraries were screened with the probe under high stringency conditions for three rounds. After three rounds of screening, positive clones were rescued as pBluescript phagemids. Subsequently, positive clones were sequenced bi-directionally and the sequence analysis was performed using LaserGene software (release 3.05:DNASTAR, Madson, WI, USA).
5’ and 3’ Rapid Amplification of cDNA Ends (RACE)
To clone CREB’s from catfish ovary, 5’ and 3’ RACE, as described earlier [13], was carried out using the gene specific primers as listed in Table 1.
Northern blot analysis
About 5 μg of poly (A)+ RNA was obtained from ovarian follicles of different stages and mature testis as described earlier [13]. The RNA was resolved on 1.2% denaturing formaldehyde-agarose gel and capillary transferred onto nylon membranes. cDNA fragments for single partial cDNA CREB (common for all forms of CREB), as well as catfish CREB were radiolabelled and used as probes separately for each blot. The blots were stripped and re-hybridized with β-actin probe that served as internal control. Signals were detected using a Fuji BSA2000 Phosphorimager. The signals were quantified by densitometry using NIH image J software.
Semi-quantitative RT-PCR
A semi-quantitative RT-PCR analysis as described by Kwon et al.[16] was employed to study the tissue distribution pattern and expression of CREBs’ during natural ovarian cycle in tilapia. Gene specific primers used in PCR reactions were shown in Table 1. qPCR was not performed as the variations among all the three CREBs are mostly in UTR regions.
qRT-PCR
Expression of CREB in catfish during different stages of reproductive cycle and hCG-induced oocyte maturation were analyzed by quantitative RT-PCR as described earlier [12].Total RNA was isolated from different stages of ovary as well as during hCG-induction, in vivo and in vitro. First strand cDNA was synthesized using random hexamers and Superscript III cDNA synthesis kit (Invitrogen) with 5 μg of total RNA. The cDNA template was used in qPCR reaction with CREB and β-actin primers (Table 1) and amplifications and fluorescence detection were performed on ABIPrism 7500 (Applied Biosystems) real-time PCR machine under the manufacturer’s universal thermal cycling conditions. Cycle threshold (CT) values were recorded during exponential phase of PCR amplification, the expression of CREB was normalized to that of β-actin (ΔCT = CREB CT - β-actin CT) and abundance of CREB mRNA was calculated using the formula 2-ΔΔCT. All the RT-PCR data was presented as mean ± SEM and statistical analysis was performed using Graphpad Prism software. Differences between groups were analyzed by ANOVA following Kruskal-Wallis’ test and P values ≤0.05 were considered significant.
Results
Cloning and sequence analysis of CREBs
A partial cDNA of 405 nt was obtained from the ovarian follicles using a set of degenerate primers by RT-PCR. The identity of the cloned cDNA was confirmed by BLAST analysis. This partial CREB cDNA was used as a probe for the extensive screening of tilapia ovarian and testicular cDNA libraries. After three rounds of cDNA library screening, several positive clones were identified and sequenced. Sequence analysis revealed multiple forms of CREBs. CREB1 cDNA, isolated from testis cDNA library was 1278 nt long with an ORF of 990 nt encoding for a protein of 330 amino acids. CREB2, cloned from tilapia ovary cDNA library was 2826 nt in length with an ORF of 954 nt encoding for a protein of 318 amino acids. Though they differ in few amino acids at N-terminal, they both have the characteristic kinase inducible and DNA binding zinc finger domains. However, CREB2 has a very long 3' UTR (1753 nt) relative to CREB1. In addition to CREB2, in the ovary a 3’UTR variant form was identified and designated as CREB3. CREB3 was 2757 nt in length with an ORF same as CREB2, but has few nucleotides shorter than 3' UTR of CREB2 (1684nt).
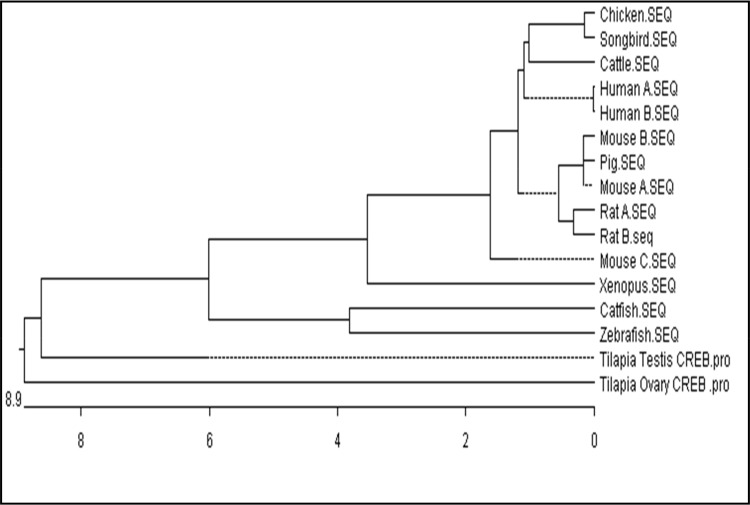
In order to extend our studies in catfish, we intended to study the expression pattern of CREBs in catfish. Using the same set of degenerate primers, a partial cDNA of 405 nt that was quite similar to tilapia CREB1 was obtained from the ovarian follicles of catfish. This partial cDNA was used to isolate and identify CREBs in catfish. However, using 5' and 3' RACE strategies, a single CREB that is similar to tilapia CREB1 was isolated. The catfish CREB was 1398 nt with an ORF of 975 bp encoding a protein of 324 amino acids. All CREB nucleotide sequences are submitted to GenBank. An amino acid sequence alignment and a phylogenic tree are shown in Figs 1 and 2. The putative proteins from both tilapia and catfish are highly conserved encompassing signature domains such as kinase inducible and zinc finger regions.
Fig 1. Clustal W multiple alignment of tilapia and catfish CREB deduced amino acid sequences.
Fig 2. Cladogram showing phylogenetic analysis of the Nile tilapia and catfish CREBs with other vertebrate analogs.
(Accession no.: Human A NM_004379; Human B NM_134442; Mouse C NM_001037726; Xenopus NM_001086603;Mouse A NM_009952; Mouse B NM_133828; Rat A NM_ 134443; Rat B NM_031017; Chicken NM_ 204450; Zebrafish NM_ 200909; Pig NM_001099929; Cattle NM_174285; Songbird NM_001048256).
Tissue expression pattern of CREBs
In order to detect the different CREB mRNAs cloned from Nile tilapia, Northern blot analysis was performed using a partial CREB cDNA fragment. Northern blot analysis detected a single transcript of ~1.3 kb (CREB1) in testis, whereas in ovary, two transcripts of ~2.75 (CREB 3) and ~2.85 (CREB 2) kb were detected thus confirming our cloning analysis (Fig 3A and 3B). Further, CREB2 showed a very strong signal in ovarian follicles in three different stages with a slight increase in full-grown immature follicles while CREB3 signal was relatively faint but expression was abundant during spawning phase (day) (Fig 3A and 3B). Owing to high sequence similarity, the probe in Northern blot could detect CREB1 only in testis, but not in ovarian follicles. This high degree of sequence similarity also posed a limitation to design real-time quantitative PCR assay. Therefore, we had employed semi-quantitative RT-PCR to analyze the expression of Nile tilapia CREBs in different tissues as well as during oocyte maturation.
Fig 3. Northern blot analysis of CREBs.
(A) Expression of CREBs in tilapia ovary and testis (VOF; Vitellogenic Ovarian Follicle; FIO, Full grown Immature Ovarian Follicle; MOF, Mature Ovarian follicle). (B) Representative densitometric analysis of CREB2 and CREB3 of tilapia. Bars depicting same letter are significantly different from each other (mean ±SEM of two independent experiments). (C) Northern blot analysis of CREBs catfish ovary (P, Preparatory; PS, Pre-spawning; S, Spawning). Note: only one form of CREB (~ 1.3 kb) in tilapia testis [CREB 1] while two forms of CREB (~2.85 [CREB 2] and ~2.75 kb [CREB 3]) in tilapia ovary.
Semi-quantitative RT-PCR analysis showed that CREB1 and CREB2 transcripts were detected in most of the tissues analyzed and the expression was predominant in brain, gonads, kidney and spleen (Fig 4). There was no appreciable difference in CREB1 and CREB2 expression pattern between male and female tilapia. Interestingly, expression of CREB3 was exclusively found in ovary and ovarian follicles, while it was undetectable in all other tissues analyzed in both sexes (Fig 4).
Fig 4. Tissue distribution pattern of tilapia CREBs as determined by semi-quantitative RT-PCR.
A plasmid DNA of CREB partial cDNA was used as positive control (PC) and PCR reaction without RT was used as negative control (NC). (Ma, Marker; B, Brain; A, Adrenal; H, Heart; S, Spleen; L, Lung; I, Intestine; K, Kidney; M, Muscle; T, Testis; O, Ovary; F, Ovarian follicle).
In contrast to multiple forms of CREBs in the Nile tilapia, only a single transcript of CREB was detected in catfish ovary by Northern blot analysis (Fig 3C).Tissue expression pattern of catfish CREB was similar to tilapia CREB1 (data not shown).
Expression of tilapia CREBs in natural ovarian cycle and hCG-induced maturation
During natural ovarian cycle of the Nile tilapia, CREB1 expression remained stable throughout vitellogenesis and expression was down-regulated on the day of spawning. In contrast, CREB2 expression was unchanged throughout the cycle with a minor elevation on day 14. Intriguingly, CREB3 expression was found to be the same from 0 to 8 days after spawning, then decreased in full-grown immature follicles (day 11) and a dramatic increase on day 14 (spawning) was noticed. Taken together these results shows, diminished expression of CREB1 and a great augment in expression of CREB3 and a minor elevation of CREB 2 during spawning were observed in natural ovarian cycle of tilapia (Fig 5A), indirectly support their role in oocyte growth and meiotic maturation.
Fig 5. Expression of tilapia CREBs during: (A) natural ovarian cycle and (B) hCG induced -oocyte maturation as determined by RT-PCR and Northern Blot respectively. A plasmid DNA with CREB partial cDNA was used as positive control (PC) and PCR reaction without RT was used as negative control (NC) (Ma, Marker). (C) Densitometric analysis of CREB2 and CREB3 (mean ±SEM of two independent experiments).
Further we sought to determine the expression of CREB2 and CREB3 following hCG- induced oocyte maturation. Northern blot analysis demonstrated that expression of CREB2 increased about 1.5 folds 1 hour after hCG-injection. CREB3 expression before hCG-injection was very low and its expression induced 3 folds 1 hour after hCG-injection (Fig 5B and 5C). As a control, tilapias were also injected with physiological saline, which showed slight decrease in CREB2 and CREB3 expression (Fig 5B and 5C). These results strongly support the expression pattern of CREB2 and CREB3 during natural ovarian cycle and confirmed the overexpression of CREB3 and to some extent CREB2 during spawning (on day 14) in the Nile tilapia.
Expression of catfish CREB in natural ovarian cycle and hCG-induced maturation
In catfish reproductive cycle, CREB expression was low in pre-spawning and regressed/resting phases and a maximum expression was noticed during spawning phase (Fig 6A). In catfish, both in vivo and in vitro hCG-induced oocyte maturation, CREB mRNA levels augmented rapidly (Fig 6B and 6C).
Fig 6. (A) Expression of catfish CREB during natural ovarian cycle (A) and hCG induced oocyte maturation in vivo (B) and in vitro (C) as determined by real-time RT-PCR.
Data are mean±SEM of two independent experiments (n = 3). Bars depicting same letter are significantly different from each other and bars depicting * are statistically significant compared to 0 hours. A plasmid DNA with CREB partial cDNA was used as positive control and PCR reaction without RT was used as negative control.
Discussion
cAMP mediates communication between cells in many biological processes such as cellular homeostasis, cell proliferation and death, neuronal plasticity, long term memory, steroid hormone and glucose metabolism etc., [17]. cAMP employs the transcription factor CREB which is activated by phosphorylation and modulates gene expression by binding to cAMP responsive elements on gene promoters. cAMP has multiple roles in fish oocytes. For example, cAMP is known to be critical for the synthesis of both E2 and 17α, 20β-DP. Prophase I arrest maintenance requires a state of decreased intra-oocyte cAMP. On the other hand, gonadotropins, in large part, mediate their actions on several steroidogenic enzyme genes through cAMP [18–20]. Together, the identification of cAMP response elements on both 20β-HSD and Cyp19a1a promoters added an enigma to our understanding of molecular mechanism of shift in steroidogenesis in fish oocytes [1,5,6]. We speculated that CREBs’ might temporally regulate the different stages of oocyte maturation [5,6]. Surprisingly, we identified multiple forms of CREB in tilapia and a single form of CREB in catfish.
CREB1 and CREB2 are ubiquitously expressed in several tissues in addition to ovary and testis. Stable expression of CREB2 throughout of tilapia ovarian cycle with an elevation during spawning indicates its possible role in maintaining basal level of steroidogenic enzymes in particular 20β-HSD and to some extent Cyp19a1a. This notion is documented well by the observation of CREB2 overexpression following hCG-induction. In mammals, CREB and SF-1 bind to Cyp19a1a gene in co-operative manner to mediate cAMP action in granulosa cells [21–23]. Although, putative cAMP responsive elements were identified on fish Cyp19a1a promoters, either a direct role or interactions with other factors remain elusive. However, CREB1 expression pattern correlates well with Cyp19a1a expression and possibly involve in down regulation of Cyp19a1a expression during steroidogenic shift [7,8]. CREB3 seems to play a very prominent role in the up regulation of 20β-HSD expression vis-à-vis final oocyte maturation. This contention is supported by i) the exclusive presence of CREB3 in ovary and ovarian follicles, ii) synergistic expression pattern of CREB3 with 20β-HSD in natural ovarian cycle, iii) over expression of CREB3 preceding 20β-HSD during hCG-induced oocyte maturation. Therefore, these results strongly support that shift in steroidogenesis at transcriptional level is possibly regulated by multiple forms of CREBs in tilapia. It seems CREB1 regulates Cyp19a1a while CREB3 as well as CREB2 regulates 20β-HSD expression. However, we could not be able to provide the direct evidence to this notion, due to the lack of fish specific CREBs recombinants and future studies shall address this fact. Nevertheless, using natural and hCG-induced oocyte maturation with two fish different models and identification of CRE motif [15] (in fish 20β-HSD promoter supports this concept strongly.
To extend these observations in an annual breeder, catfish, similar studies were conducted. As opposed to tilapia, a single form of CREB that is homologous to tilapia CREB1/2 was identified. In spite of repeated attempts we could not able to get the variant form of CREB in ovary. Unlike tilapia, we did not use testicular tissue for cloning CREB. Expression pattern of CREB in catfish correlate more or less with 20β-HSD expression and enzyme activity in natural and hCG-induced oocyte maturation. Thus, we presume that CREB might regulate both Cyp19a1a and 20β-HSD as the two events of ovarian cycle, vitellogenesis and maturation are widely spaced in catfish as opposed to the short time in tilapia. Another explanation could be the interaction of multiple transcription factors to modulate expression of these two enzymes.
In conclusion, multiple forms of CREBs were identified in tilapia ovary for the first time in any lower vertebrates. Based on their expression patterns, CREB1 could be involved in regulating Cyp19a1a and an alternatively spliced (UTR variant) CREB3 is exclusively expressed in ovary playing a major role in shift in steroidogenesis by targeting 20β-HSD. Further, hCG-induced over expression of CREB3 as well as CREB2 demonstrate the robust action of these correlates to promote final oocyte maturation by transcriptionally regulating [15] 20β-HSD expression and function [6]. In catfish, a single form of CREB1 was identified that may probably implicated in final oocyte maturation and to some extent in vitellogenesis.
Acknowledgments
BS and CCS are the recipients of the Japan Society for the Promotion of Science, which is acknowledged. We also thank the support from DST-FIST of the Department of Animal Biology and School of Life Sciences. GS acknowledges junior and senior research fellowships from Council of Scientific and Industrial Research, India. BS is a recipient of TATA innovation fellowship, Department of Biotechnology, Government of India, which is duly acknowledged.
Data Availability
All relevant data are within the paper.
Funding Statement
Grants-in-aid for Research from CREST (Core Research for Evolutional Science and Technology), JST (Japan Science and Technology Corporation), Research for the Future (JSPS-RFTF 96L00401) and Priority Areas (07283104 and 14042267) from the Ministry of Education, Science, Culture, and Sports, Japan, and Bio Design Program from the Ministry of Agriculture, Forestry and Fisheries, Japan to YN is acknowledged. A Grant-in-aid (SP/SO/AS-49/2003) from the Department of Science and Technology (DST), India, to BS is acknowledged. BS and CCS are the recipients of the Japan Society for the Promotion of Science, which is acknowledged. The authors also thank the support from DST-FIST of the Department of Animal Biology and School of Life Sciences. GS acknowledges junior and senior research fellowships from Council of Scientific and Industrial Research, India. BS is a recipient of TATA innovation fellowship, Department of Biotechnology, Government of India, which is duly acknowledged.
References
- 1. Nagahama Y, Yamashita M (2008) Regulation of oocyte maturation in fish. Dev Growth Differ 50 Suppl 1: S195–219. 10.1111/j.1440-169X.2008.01019.x [DOI] [PubMed] [Google Scholar]
- 2. Gen K, Okuzawa K, Senthilkumaran B, Tanaka H, Moriyama S, Kagawa H (2000) Unique expression of gonadotropin-I and -II subunit genes in male and female red seabream (Pagrus major) during sexual maturation. Biol Reprod 63: 308–319. [DOI] [PubMed] [Google Scholar]
- 3. Koide Y, Noso T, Schouten G, Peute J, Zandbergen MA, Bogerd J, et al. (1992) Maturational gonadotropin from the African catfish, Clarias gariepinus: purification, characterization, localization, and biological activity. Gen Comp Endocrinol 87: 327–341. [DOI] [PubMed] [Google Scholar]
- 4. Senthilkumaran B, Joy KP (2001) Periovulatory changes in catfish ovarian oestradiol-17beta, oestrogen-2-hydroxylase and catechol-O-methyltransferase during GnRH analogue-induced ovulation and in vitro induction of oocyte maturation by catecholoestrogens. J Endocrinol 168: 239–247. [DOI] [PubMed] [Google Scholar]
- 5. Senthilkumaran B, Yoshikuni M, Nagahama Y (2004) A shift in steroidogenesis occurring in ovarian follicles prior to oocyte maturation. Mol Cell Endocrinol 215: 11–18. [DOI] [PubMed] [Google Scholar]
- 6. Senthilkumaran B (2011) Recent advances in meiotic maturation and ovulation: comparing mammals and pisces. Frontiers in Bioscience-Landmark 16: 1898–1914. [DOI] [PubMed] [Google Scholar]
- 7. Yoshiura Y, Senthilkumaran B, Watanabe M, Oba Y, Kobayashi T, Nagahama Y (2003) Synergistic expression of Ad4BP/SF-1 and cytochrome P-450 aromatase (ovarian type) in the ovary of Nile tilapia, Oreochromis niloticus, during vitellogenesis suggests transcriptional interaction. Biol Reprod 68: 1545–1553. [DOI] [PubMed] [Google Scholar]
- 8. Watanabe M, Tanaka M, Kobayashi D, Yoshiura Y, Oba Y, Nagahama Y (1999) Medaka (Oryzias latipes) FTZ-F1 potentially regulates the transcription of P-450 aromatase in ovarian follicles: cDNA cloning and functional characterization. Mol Cell Endocrinol 149: 221–228. [DOI] [PubMed] [Google Scholar]
- 9. Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, Sakai F, et al. (2007) Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol 21: 712–725. [DOI] [PubMed] [Google Scholar]
- 10. Guan G, Tanaka M, Todo T, Young G, Yoshikuni M, Nagahama Y (1999) Cloning and expression of two carbonyl reductase-like 20beta-hydroxysteroid dehydrogenase cDNAs in ovarian follicles of rainbow trout (Oncorhynchus mykiss). Biochem Biophys Res Commun 255: 123–128. [DOI] [PubMed] [Google Scholar]
- 11. Tanaka M, Nakajin S, Kobayashi D, Fukada S, Guan G, Todo T, et al. (2002) Teleost ovarian carbonyl reductase-like 20beta-hydroxysteroid dehydrogenase: potential role in the production of maturation-inducing hormone during final oocyte maturation. Biol Reprod 66: 1498–1504. [DOI] [PubMed] [Google Scholar]
- 12. Sreenivasulu G, Senthilkumaran B (2009) New evidences for the involvement of 20beta-hydroxysteroid dehydrogenase in final oocyte maturation of air-breathing catfish. Gen Comp Endocrinol 163: 259–269. 10.1016/j.ygcen.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 13. Senthilkumaran B, Sudhakumari CC, Chang XT, Kobayashi T, Oba Y, Guan G, et al. (2002) Ovarian carbonyl reductase-like 20beta-hydroxysteroid dehydrogenase shows distinct surge in messenger RNA expression during natural and gonadotropin-induced meiotic maturation in nile tilapia. Biol Reprod 67: 1080–1086. [DOI] [PubMed] [Google Scholar]
- 14. Huang W, Zhou L, Li Z, Gui JF (2009) Expression pattern, cellular localization and promoter activity analysis of ovarian aromatase (Cyp19a1a) in protogynous hermaphrodite red-spotted grouper. Mol Cell Endocrinol 307: 224–236. 10.1016/j.mce.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 15. Sreenivasulu G, Senthilkumaran B, Sudhakumari CC, Guan G, Oba Y, Kagawa H, et al. (2012) 20beta-hydroxysteroid dehydrogenase gene promoter: potential role for cyclic AMP and xenobiotic responsive elements. Gene 509: 68–76. 10.1016/j.gene.2012.07.017 [DOI] [PubMed] [Google Scholar]
- 16. Kwon JY, McAndrew BJ, Penman DJ (2001) Cloning of brain aromatase gene and expression of brain and ovarian aromatase genes during sexual differentiation in genetic male and female Nile tilapia Oreochromis niloticus. Mol Reprod Dev 59: 359–370. [DOI] [PubMed] [Google Scholar]
- 17. Sands WA, Palmer TM (2008) Regulating gene transcription in response to cyclic AMP elevation. Cell Signal 20: 460–466. [DOI] [PubMed] [Google Scholar]
- 18. Kanamori A, Nagahama Y (1988) Involvement of 3',5'-cyclic adenosine monophosphate in the control of follicular steroidogenesis of amago salmon (Oncorhynchus rhodurus). Gen Comp Endocrinol 72: 39–53. [DOI] [PubMed] [Google Scholar]
- 19. Nagahama Y (1997) 17 alpha,20 beta-dihydroxy-4-pregnen-3-one, a maturation-inducing hormone in fish oocytes: mechanisms of synthesis and action. Steroids 62: 190–196. [DOI] [PubMed] [Google Scholar]
- 20. Haider S (2003) Cyclic AMP level and phosphodiesterase activity during 17alpha,20beta-dihydroxy-4-pregnen-3-one induction and theophylline inhibition of oocyte maturation in the catfish, Clarias batrachus. Comp Biochem Physiol A Mol Integr Physiol 134: 267–274. [DOI] [PubMed] [Google Scholar]
- 21. Carlone DL, Richards JS (1997) Evidence that functional interactions of CREB and SF-1 mediate hormone regulated expression of the aromatase gene in granulosa cells and constitutive expression in R2C cells. J Steroid Biochem Mol Biol 61: 223–231. [PubMed] [Google Scholar]
- 22. Young M, McPhaul MJ (1998) A steroidogenic factor-1-binding site and cyclic adenosine 3',5'-monophosphate response element-like elements are required for the activity of the rat aromatase promoter in rat Leydig tumor cell lines. Endocrinology 139: 5082–5093. [DOI] [PubMed] [Google Scholar]
- 23. Fitzpatrick SL, Carlone DL, Robker RL, Richards JS (1997) Expression of aromatase in the ovary: down-regulation of mRNA by the ovulatory luteinizing hormone surge. Steroids 62: 197–206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.