Abstract
We have examined whether dissociation of the histone octamer is required for elongation of RNA transcripts through arrays of nucleosome cores in vitro. Control or dimethyl suberimidate-crosslinked histone octamers were reconstituted onto supercoiled, closed circular pT207-18 DNA, which contains tandemly repeated 207-base-pair (bp) 5S rDNA nucleosome positioning sequences inserted between the T7 and SP6 transcription promoters of pGEM-3Z. Double label transcription experiments showed that there was little or no effect of extensive crosslinking of the histone octamers on transcription initiation and elongation by T7 RNA polymerase in vitro. Continuous regularly spaced linear arrays of either crosslinked or control nucleosome cores were obtained by digesting reconstituted nucleosomal pT207-18 templates with Dra I, a site that is protected from digestion by the presence of positioned nucleosome cores in the 207-pb sequence. After in vitro transcription with T7 RNA polymerase, an RNA ladder with 207-nucleotide spacing was obtained from templates reconstituted both with crosslinked and with control histone octamers, demonstrating clearly that neither partial nor complete dissociation of the histone octamer is essential for transcription elongation through arrays of nucleosome cores in vitro.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Dong F., Hansen J. C., van Holde K. E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould H. J., Cowling G. J., Harborne N. R., Allan J. An examination of models for chromatin transcription. Nucleic Acids Res. 1980 Nov 25;8(22):5255–5266. doi: 10.1093/nar/8.22.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C., Ausio J., Stanik V. H., van Holde K. E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989 Nov 14;28(23):9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991 Apr;5(4):683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov N., Tsaneva I., Einbinder E., Tsanev R. In vitro transcription through nucleosomes by T7 RNA polymerase. EMBO J. 1992 May;11(5):1941–1947. doi: 10.1002/j.1460-2075.1992.tb05247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Transcription-induced nucleosome 'splitting': an underlying structure for DNase I sensitive chromatin. EMBO J. 1991 Mar;10(3):607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. On the displacement of histones from DNA by transcription. Cell. 1988 Dec 2;55(5):743–744. doi: 10.1016/0092-8674(88)90128-6. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5 S rDNA. J Mol Biol. 1991 Jul 5;220(1):89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- Morse R. H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989 Aug;8(8):2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton V. G., Imai B. S., Yau P., Bradbury E. M. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989 May 5;57(3):449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- O'Neill T. E., Roberge M., Bradbury E. M. Nucleosome arrays inhibit both initiation and elongation of transcripts by bacteriophage T7 RNA polymerase. J Mol Biol. 1992 Jan 5;223(1):67–78. doi: 10.1016/0022-2836(92)90716-w. [DOI] [PubMed] [Google Scholar]
- Pfaffle P., Gerlach V., Bunzel L., Jackson V. In vitro evidence that transcription-induced stress causes nucleosome dissolution and regeneration. J Biol Chem. 1990 Oct 5;265(28):16830–16840. [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
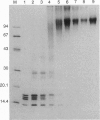
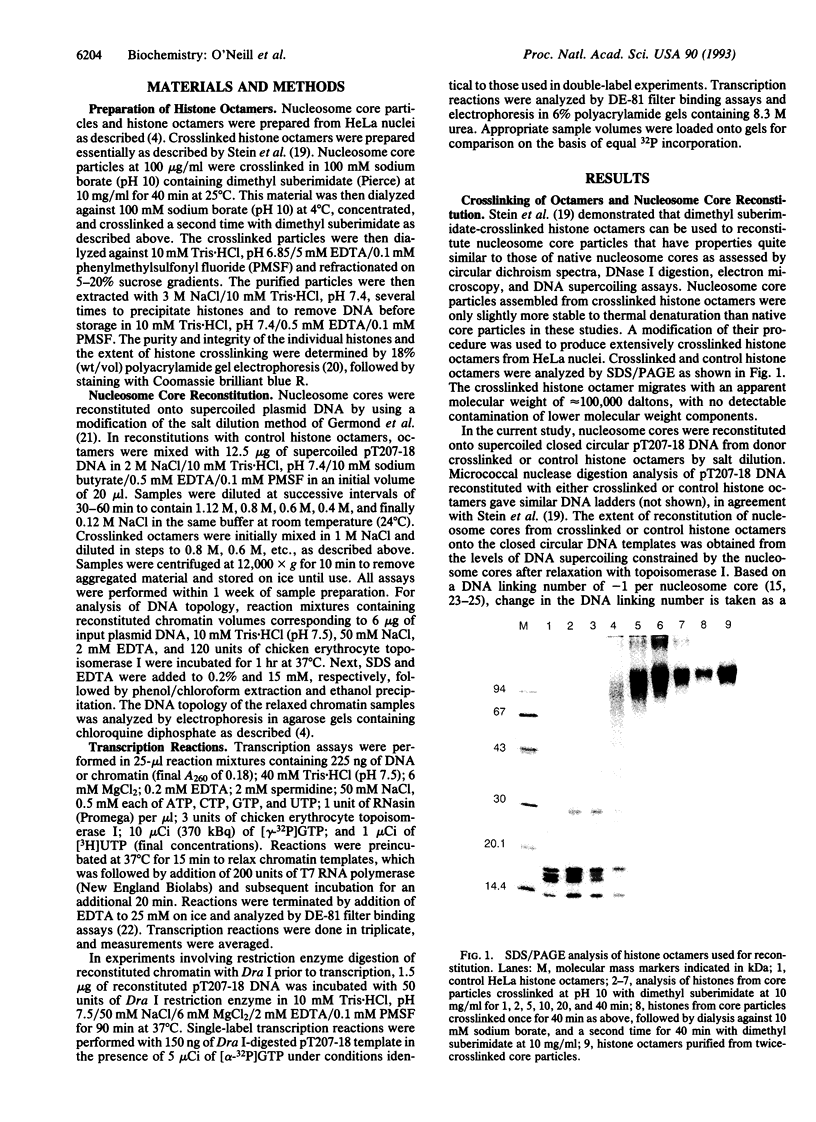
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. Structural changes in nucleosomes during transcription: strip, split or flip? Trends Genet. 1991 Jun;7(6):175–177. doi: 10.1016/0168-9525(91)90429-t. [DOI] [PubMed] [Google Scholar]
- Thomas J. O. Chemical cross-linking of histones. Methods Enzymol. 1989;170:549–571. doi: 10.1016/0076-6879(89)70064-1. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Studies on the mechanism of transcription of nucleosomal complexes. Eur J Biochem. 1980 Jan;103(2):219–226. doi: 10.1111/j.1432-1033.1980.tb04306.x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
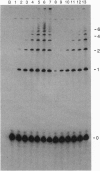
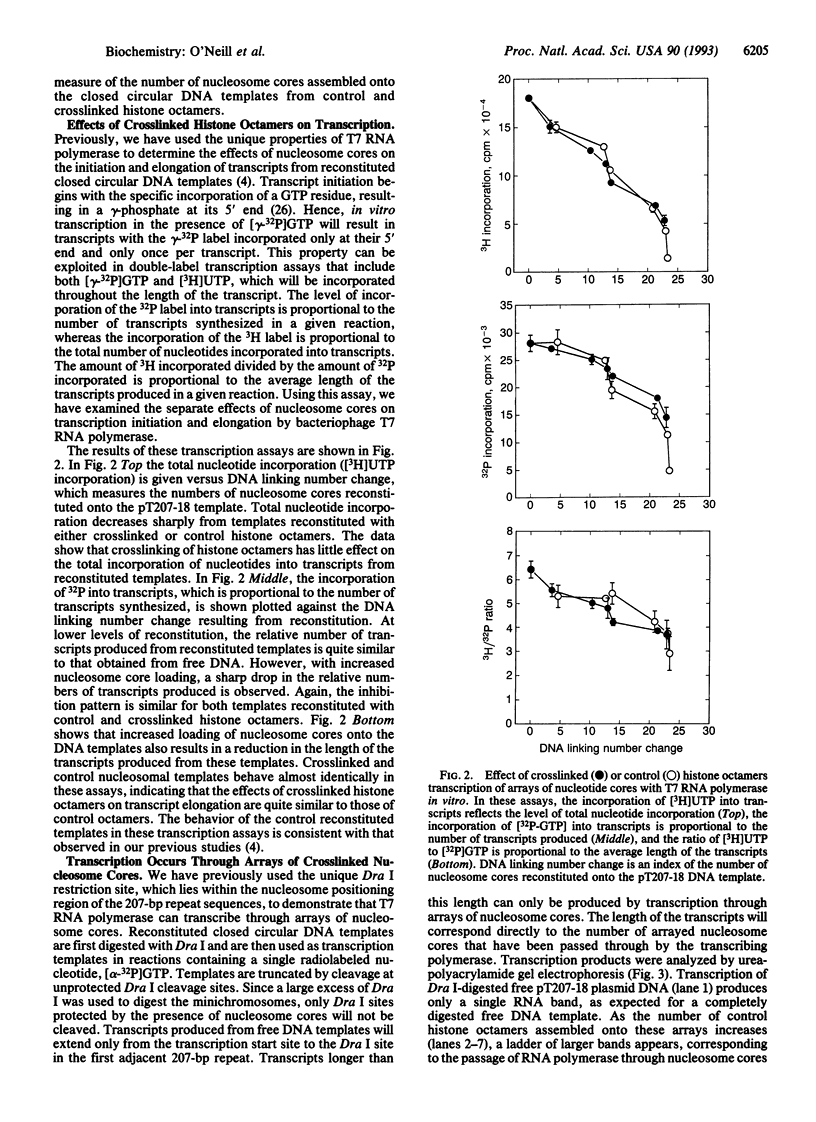
- van Holde K. E., Lohr D. E., Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992 Feb 15;267(5):2837–2840. [PubMed] [Google Scholar]