Abstract
The Epstein-Barr virus (EBV)-positive subtype of gastric adenocarcinoma is conventionally identified by in situ hybridization (ISH) for viral nucleic acids, but next-generation sequencing represents a potential alternative. We therefore determined normalized EBV read counts by whole genome, whole exome, mRNA and miRNA sequencing for 295 fresh-frozen gastric tumor samples. Formalin-fixed, paraffin-embedded tissue sections were retrieved for ISH confirmation of 13 high-EBV and 11 low-EBV cases. In pairwise comparisons, individual samples were either concordantly high or concordantly low by all genomic methods for which data were available. Empiric cut-offs of sequencing counts identified 26 (9%) tumors as EBV-positive. EBV-positivity or negativity by molecular testing was confirmed by EBER-ISH in all but one tumor evaluated by both approaches (kappa=0.91). EBV-positive gastric tumors may be accurately identified by quantifying viral sequences in genomic data. Simultaneous analyses of human and viral DNA, mRNA and miRNA could streamline tumor profiling for clinical care and research.
Keywords: Stomach cancer, Molecular subtypes, EBV, TCGA
INTRODUCTION
Epstein-Barr virus (EBV) is a recognized carcinogenic agent for several malignancies, accounting for about 200,000 new cancer cases annually worldwide.[1] Approximately 9% of gastric adenocarcinomas have latent EBV infection in every tumor cell.[2] In viral-positive tumors, the nucleic acids typically present as monoclonal episomes with uniform terminal repeats, indicating infection was present at the time of transformation in the clonal progenitor cell.[3] EBV-positive adenocarcinoma cases differ from other gastric cancers, exhibiting distinct epidemiological (e.g., male predominance, post-gastrectomy), pathological (e.g., preferentially non-antral anatomic subsites) and clinical (e.g., better survival) features.[2, 4, 5] Based on a comprehensive molecular analysis of 295 gastric adenocarcinomas performed by The Cancer Genome Atlas (TCGA), EBV-positivity was identified to mark one of four molecularly distinct subtypes of this disease. EBV-positive tumors were characterized by extreme DNA CpG island hypermethylation phenotype (CIMP), frequent PIK3CA mutation, absence of TP53 mutation, and recurrent amplifications of the chromosome 9 locus containing JAK2, CD274/PD-L1, and PDCD1LG2/PD-L2.[6]
EBV is almost ubiquitous in the human population, primarily maintained as a latent infection in a subset of B-lymphocytes comprising roughly 10−5 peripheral blood mononuclear cells. Detection of EBV in tumor tissue is therefore needed to implicate the infection in gastric carcinogenesis. However, the tissue inflammation often present in gastric cancer may lead to infiltration of EBV-infected leukocytes as a non-specific source of viral sequences. Conventionally, EBV is localized to particular cells within tumor tissue by in situ hybridization (ISH) for EBV-encoded small RNA (EBER) types-1 and -2, abundant untranslated transcription products of unknown function.[7] This assay is considered to be the “gold standard” for assigning EBV status based on its high sensitivity and specificity, as long as adequate quantity and integrity of lesional tissue are available.[8] Importantly, in situ analyses can determine whether virions are localized within tumor cells or a different tissue compartment.
Massive parallel sequencing methodologies offer an alternative approach for detecting nucleic acids originating from infectious agents. In the current study, we determine assay cut-offs for distinguishing EBV-positive gastric cancer from other molecular subtypes in sequencing data from TCGA and evaluate agreement among four genomic technologies as well as with conventional EBER-ISH.
METHODS
EBV sequences in nucleic acid extracts of 295 fresh-frozen gastric adenocarcinoma samples from TCGA were determined by whole genome (n=77), whole exome (n=263), mRNA (n=237) and miRNA (n=293) sequencing and normalized to corresponding human sequence counts, as previously reported.[6] Briefly, DNA or RNA sequence reads matching EBV were identified by the PathSeq [9] or BioBloom [10] algorithms, respectively. Viral DNA abundance was normalized to human sequences by dividing #reads mapped to the microbe by #reads mapped to human in the sample/average # reads mapped to human in the sample cohort/4.857, the latter constant representing the ratio of the genome size of EBV to the average of all viruses. RNA counts were normalized by millions of total reads sequenced as the #reads mapped to the microbe*106/#chastity passed reads. Tumor EBV status was provisionally classified based on detection of high or low normalized viral read counts by at least two sequencing platforms. All patients provided informed consent, and local Institutional Review Boards approved tissue collection.
For comparison to conventional determination of EBV status, we retrieved formalin-fixed, paraffin-embedded (FFPE) tissue sections from a matched tumor block for 13 high-EBV cases and 11 low-EBV cases selected at random from the same tissue source. EBER-ISH was performed at the University of North Carolina. Briefly, three adjacent sections were stained by hematoxylin and eosin and by ISH for EBER and for oligodT control RNA to confirm RNA integrity, with inclusion of known EBER-positive and -negative tumors as external controls. Hybridization was performed using the Leica Bond system with 5 minutes of protease digestion and 2 hours of probe hybridization. A tumor was interpreted as EBV-negative if EBER staining was undetected or only localized to benign-appearing lymphoid cells, and EBV-positive if EBER staining was localized to the nucleus of malignant epithelial cells, as previously described [11]. Cases with unsatisfactory or indeterminate results were re-tested using additional sections from the same block. Histopathologic examinations and ISH were performed under code such that laboratory personnel did not have access to results of molecular testing.
Relative frequencies of log-transformed EBV read counts were graphed as probability density functions using z-scores normalized by subtracting mean counts and dividing by standard deviations. Scatterplots were used to compare sample measurements and cutoffs selected empirically to optimize concordance across assay platforms. Spearman rank correlations between read counts on different platforms were calculated, combined and separately for EBV-positive and negative tumors, with p-values less than 0.05 considered statistically significant. Sensitivity, specificity and kappa statistics were calculated for conventional EBER-ISH as compared to genomic-assigned EBV status. All statistical analyses were performed using StataSE v13 (College Station, TX).
RESULTS
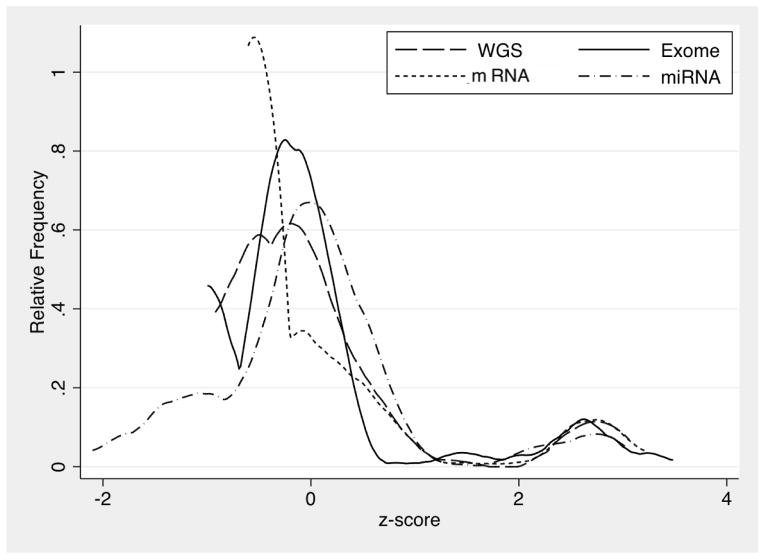
By each of the four methods of whole genome, whole exome, mRNA or miRNA massive parallel sequencing, numbers of normalized EBV reads across individual samples were bimodally distributed, with distinct separation of a minority of tumors having substantially higher counts. For each platform, the two modes of log-transformed values were separated by approximately three standard deviations (Figure 1).
Figure 1.
Probability density plots of normalized EBV read counts in gastric cancer tissues by whole genome (WGS; n=77), exome (n=263), mRNA (n=237) and miRNA (n=293) sequencing.
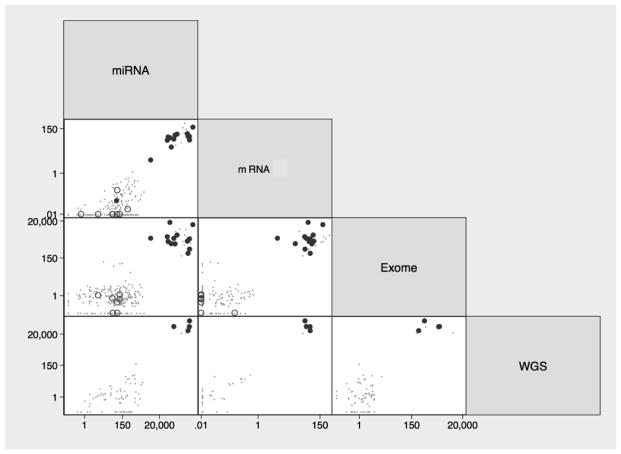
Pairwise comparisons of the four sequencing platforms indicated that individual samples were either consistently high or consistently low by all genomic methods for which data were available. Log-log scatterplots of the 295 TCGA samples were confined to upper right and lower left quadrants only (Figure 2).
Figure 2.
Pairwise comparisons of normalized EBV read counts in gastric cancer tissues by whole genome (WGS), exome, mRNA and miRNA sequencing. Solid circles represent EBER-positive tumors (n=14), open circles represent EBER-negative tumors (n=9) and dots indicate TCGA tumors not tested by in situ hybridization (n=272).
Overall quantitative counts were moderately correlated (all p-values < .001), with Spearman rank correlation coefficients (Rho) ranging from 0.2 to 0.8 (Table 1). Stratified by EBV status, counts were less correlated. Among EBV-positive tumors, the only significant correlation among the four genomic platforms was between miRNA and mRNA (rho=0.6). Among EBV-negative tumors, four of the six pairwise correlations were significant, with higher correlation between mRNA and whole genome (rho=0.6) and lower correlations for the other three comparisons (rho<0.3).
TABLE 1.
Spearman coefficients (Rho), numbers of observations (n) and significance levels (p) for rank correlations of normalized EBV-specific read counts in gastric cancers analyzed by whole genome (WGS), exome, mRNA and miRNA sequencing
| All tumors combined
|
EBV-negative tumors
|
EBV-positive tumors
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WGS | Exome | mRNA | miRNA | WGS | Exome | mRNA | miRNA | WGS | Exome | mRNA | miRNA | ||
| WGS | Rho | 1 | 1 | 1 | |||||||||
| n | 77 | 70 | 7 | ||||||||||
| p | |||||||||||||
|
| |||||||||||||
| Exome | Rho | 0.4 | 1 | 0.2 | 1 | 0.3 | 1 | ||||||
| n | 75 | 263 | 68 | 237 | 7 | 26 | |||||||
| p | 0.001 | 0.2 | 0.6 | ||||||||||
|
| |||||||||||||
| mRNA | Rho | 0.8 | 0.4 | 1 | 0.6 | 0.2 | 1 | −0.09 | 0.3 | 1 | |||
| n | 40 | 210 | 237 | 34 | 186 | 213 | 6 | 24 | 24 | ||||
| p | 0.000 | 0.000 | 0.000 | 0.03 | 0.9 | 0.2 | |||||||
|
| |||||||||||||
| miRNA | Rho | 0.5 | 0.2 | 0.5 | 1 | 0.3 | −0.01 | 0.3 | 1 | 0.3 | −0.2 | 0.6 | 1 |
| n | 77 | 261 | 235 | 293 | 70 | 237 | 213 | 269 | 7 | 24 | 22 | 24 | |
| p | 0.000 | 0.000 | 0.000 | 0.02 | 0.9 | 0.000 | 0.5 | 0.4 | 0.01 | ||||
By comparing distributions of the genomic data, empiric cut-offs were defined as 1000 normalized EBV reads for whole genome sequencing, 100 for exome, 4 for mRNA, and 5000 for miRNA for perfect concordance in identifying 26 (9%) EBV-positive samples among the 295 TCGA tumors analyzed (Table 2).
TABLE 2.
Distributions of normalized EBV-specific read counts by whole genome (WGS), exome, mRNA and miRNA sequencing in gastric cancers with (N=23) and without (N=272) EBER-ISH confirmation of EBV status
| EBV status | WGS | Exome | mRNA | miRNA | ||||
|---|---|---|---|---|---|---|---|---|
| cases | #reads | cases | #reads | cases | #reads | cases | #reads | |
| EBER-ISH-positive* | 4 | 36,000–170,000 | 13 | 280–17,000 | 13 | 5–180 | 13 | 6000–1,500,000 |
| presumptive positive | 3 | 25,000–68,000 | 13 | 210–7400 | 11 | 25–290 | 11 | 140,000–1,100,000 |
| EBER-ISH-negative | 0 | - | 7 | 0–1 | 9 | 0–1 | 9 | 1–580 |
| presumptive negative | 70 | 0–193 | 230 | 0–93 | 204 | 0–1 | 260 | 0–2200 |
omits one case with 0 mRNA reads and 60 miRNA reads (see Results for details)
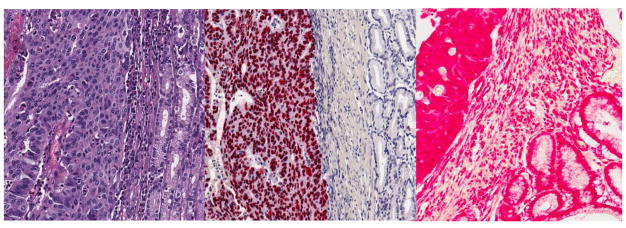
In blinded evaluations of 24 gastric cancer tissues, 13 cases exhibited distinct EBER localization to tumor cells (Figure 3); initial assay results were equivocal for a fourteenth case that on re-testing was classified as EBER-positive with some background staining. Nine tumors were clearly EBER-ISH negative. One case was unclassifiable because sampled fixed tissue did not contain any tumor cells in two separately evaluated sections.
Figure 3.
Representative photomicrographs of an EBV-positive gastric cancer tumor stained with hematoxylin and eosin (left panel), EBER-ISH (center panel) and RNA preservation control (right panel).
For the 23 tumors with EBV status determined by both genomic and conventional approaches, agreement was observed in all but one case (Figure 2). The sole exception was the tumor with initially equivocal EBER-ISH results, reclassified as positive; this case was EBV-negative by both mRNA and miRNA sequencing and was classified as microsatellite instability-type gastric cancer by DNA methylation and other genomic data. Assuming greater accuracy of the molecular assignments, EBER-ISH was 100% sensitive and 90% specific with a kappa statistic of 0.91, representing 96% observed agreement between conventional and molecular assignment of EBV status.
DISCUSSION
The current study capitalizes on TCGA data on a large set of gastric cancer specimens collected under standardized conditions with detailed annotation, and subjected to multiple analytical platforms. Four different next-generation sequencing methods had perfect concordance classifying EBV status for gastric cancer tissues. The accepted standard technique of EBER-ISH had excellent agreement to the genomic classification, with one presumed false-positive in the presence of background hybridization.
Our data suggest that next generation sequencing platforms may provide an accurate replacement for conventional ISH. However, there are several potential hurdles to practical implementation for routine use. Quantitation of viral sequences may vary due to differences in specimen processing, assay protocols and inherent batch-to-batch fluctuation.[12] The specific cut-offs generated for this sample set may not be applicable to other cases and testing laboratories need to determine their own criteria for establishing EBV-positivity. Furthermore, these excellent genomic results were obtained on frozen tissues of optimal nucleic acid quality; replication is needed on a wider variety of sample types, including fixed tissues.
The robust detectability of EBV by whole exome sequencing was unexpected. Our target enrichment platform (Agilent SureSelect Human All Exon) utilized 120-nucleotide RNA baits designed to capture all human exons with relative exclusion of other DNA sequences. Nevertheless, there were sufficient off-target reads to detect at least some portion of the viral genome in every EBV-positive gastric tumor. An alternative strategy pursued for TCGA analysis of esophageal cancer is to supplement the exome capture library with 120-mer probes specifically designed to cover cancer-related viruses, based on spacing, GC-content, repeat content and lack of similarity to human sequence (Michael McLellan, personal communication).
EBV-positive gastric cancer tissues have much higher levels of viral miRNA as compared to EBV-negative tumors.[13] Viral-derived miRNAs may also be detected in various body fluids[14] and levels in blood plasma have been evaluated as diagnostic and prognostic markers for nasopharyngeal carcinoma, the second most frequent EBV-associated malignancy.[15, 16] Circulating blood levels of EBV miRNA warrant investigation as a potential non-invasive test for EBV-positive gastric cancer when tumor tissue is inadequate or unavailable for direct assessment.
EBV-positive gastric cancers exhibit a restricted transcription pattern of viral genes, with most of the highly expressed sequences encoded in the BamH1A gene region of the genome.[6, 17] These transcripts and their protein products are candidate targets for functional studies to explore mechanisms of viral carcinogenesis. Elucidating the viral contribution to gastric cancer pathophysiology could lead to novel strategies for prevention and treatment, with possible extension to other EBV-related malignancies.
The recognition of EBV-positive gastric cancer as a distinct entity has informed scientific understanding of gastric carcinogenesis. Increasing availability of massive parallel sequencing will facilitate routine identification of these tumors for clinical translation of important research findings.
Acknowledgments
The authors thank Michael Button, Asterand USA, Hark Kim, National Cancer Center of Korea, Trey Kohl, ILSBio, Troy Shelton, International Genomics Consortium and Maciej Wiznerowicz, Greater Poland Cancer Center for their invaluable assistance in providing patient specimens for this study.
FINANCIAL SUPPORT: This work was supported by the Intramural Research Program and the following grants from the United States National Institutes of Health: 5U24CA143799, 5U24CA143835, 5U24CA143840, 5U24CA143843, 5U24CA143845, 5U24CA143848, 5U24CA143858, 5U24CA143866, 5U24CA143867, 5U24CA143882, 5U24CA143883, 5U24CA144025, U54HG003067, U54HG003079, U54HG003273 and P30CA16672.
References
- 1.Cohen JI, Fauci AS, Varmus H, et al. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med. 2011;3(107):107fs7. doi: 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy G, Pfeiffer R, Camargo MC, et al. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137(3):824–33. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 4.Camargo MC, Murphy G, Koriyama C, et al. Determinants of Epstein-Barr virus-positive gastric cancer: an international pooled analysis. Br J Cancer. 2011;105(1):38–43. doi: 10.1038/bjc.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camargo MC, Kim WH, Chiaravalli AM, et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63(2):236–43. doi: 10.1136/gutjnl-2013-304531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Network TCGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe JG, Shu MD. Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell. 1989;57(5):825–34. doi: 10.1016/0092-8674(89)90797-6. [DOI] [PubMed] [Google Scholar]
- 8.Gulley ML, Tang W. Laboratory assays for Epstein-Barr virus-related disease. J Mol Diagn. 2008;10(4):279–92. doi: 10.2353/jmoldx.2008.080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostic AD, Ojesina AI, Pedamallu CS, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011;29(5):393–6. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloom BH. Space/time trade-offs in hash coding with allowable errors. Communications ACM. 1970;13(7):422–6. [Google Scholar]
- 11.Ryan JL, Morgan DR, Dominguez RL, et al. High levels of Epstein-Barr virus DNA in latently infected gastric adenocarcinoma. Lab Invest. 2009;89(1):80–90. doi: 10.1038/labinvest.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.‘t Hoen PA, Friedlander MR, Almlof J, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31(11):1015–22. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 13.Kim do N, Chae HS, Oh ST, et al. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Virol. 2007;81(2):1033–6. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–56. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 15.Chan JY, Gao W, Ho WK, et al. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 2012;32(8):3201–10. [PubMed] [Google Scholar]
- 16.Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136(5):E301–12. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 17.Strong MJ, Xu G, Coco J, et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 2013;9(5):e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]