Abstract
Theileria spp. are tick-transmitted, intracellular apicomplexan protozoan parasites infecting a wide range of animals. As there is very limited information on the prevalence of Theileria spp. in the Caribbean we used the recently described genus-specific pan-Theileria FRET-qPCR to identify infected animals in the region and a standard 18S rRNA gene PCR and sequencing to determine the species involved. We found Theileria spp. in 9% of the convenience samples of animals (n = 752) studied from five Caribbean islands. Donkeys (20.0%: 5/25) were most commonly infected, followed by sheep (17.4%, 25/144), cattle (6.8%; 22/325), goats (5.0%; 12/238), and horses (5.0%; 1/20). Six species of Theileria were identified: T. equi (donkeys, cattle, goats, and sheep), Theileria sp. OT3 (sheep and goats), Theileria sp. NG-2013a (cattle), Theileria sp. YW-2014 (donkeys), Theileria sp. B15a (goats), and Babesia vulpes or a closely related organism (sheep and goats). Only T. equi has been previously reported in the Caribbean. Our findings expand the known host ranges of Theileria spp. and the known distribution of the organisms around the world.
1. Background
Theileria spp. are tick-transmitted, intracellular apicomplexan protozoan parasites infecting leukocytes and erythrocytes of a wide range of animals [1, 2]. The organisms have been described in all livestock species and can cause significant economic losses to farmers. They are transmitted by a variety of ixodid ticks of the genera Rhipicephalus, Hyalomma, Amblyomma, and Haemaphysalis [3]. Infections with some Theileria spp. can result in fever, anemia, hemoglobinuria, and death in severe cases, but many species are benign and cause minor or no signs. Animals that recover from acute or primary infections usually remain persistently infected and may act as reservoirs for tick vectors [4, 5]. Infected animals are found particularly in tropical and subtropical regions in Africa, the Middle East, Southern Europe, and Asia [6–11].
There is little information on infectious agents in livestock in the Caribbean although animal production is an important source of income for many people in the region. In the case of Theileria spp., morphological and serological evidence has been presented that T. mutans and T. velifera, both benign species transmitted by Amblyomma spp., occur in cattle on Guadeloupe [12]. Also, an organism with the morphology of T. mutans was seen in a blood smear from a bovine on Martinique [13]. In Trinidad, T. equi (previously Babesia equi) has been demonstrated in horses with a specific nested 18S rRNA PCR [14, 15] and a serosurvey has provided supporting evidence for its presence [16].
While there are many tests to detect Theileria spp. in animals, their specificity varies, as does their usefulness in finding the full spectrum of organisms present in an area. Microscopic detection of parasites can be difficult with low parasitemia and does not readily allow differentiation of species [2]. Serological studies, although sensitive and relatively easy to perform, are not specific as there is cross-reactivity between Theileria spp. [17]. Although molecular techniques have been described, many are for specific species which limits their usefulness in surveys. Reverse line blotting (RLB) assays enable the simultaneous identification of multiple species [18, 19], but they are cumbersome and time demanding to perform and identifying stringent species-specific oligonucleotide sequences can be challenging [20]. Recently, a sensitive genus-specific pan-Theileria FRET-qPCR has been described that detects the recognized Theileria spp. of domestic animals in a single reaction (Table 1) [21]. To provide further data on Theileria spp. in the Caribbean, we used the pan-Theileria FRET-qPCR to screen livestock from five islands for evidence of infection. Further, we used a standard 18S rRNA PCR and gene sequencing on positive reactors to identify the Theileria spp. involved. The results of this survey are described below.
Table 1.
Alignment of the nucleotides used in the primers and probes of the pan-Theileria FRET-qPCR used in this study.
| Forward primer | LCRed-640 | 6-FAM | Reverse primer | |
|---|---|---|---|---|
| T AGTGACAAGAAATAACAATACGGGGC- -TT | GTCTTGTAATTGGAATGATGGGAATT | AAACCTCTTCCAGAGTATCAATTGG | AGTTAAAAAGCTCGTAGTTGAATTTCTGCTG | |
| T. orientalis | ...........................- -.. | .......................... | ......................... | ............................... |
| T. buffeli | ...........................- -.C | .......................... | ......................... | ............................... |
| T. annulata | ........................... - - .. | .......................... | ......................... | ............................... |
| T. sergenti | ........................... - - .. | .......................... | ......................... | ............................... |
| T. luwenshuni | ........................... - - .. | .......................... | ......................... | ............................... |
| T. velifera | ........................... - - .. | .C........................ | ......................... | ..............................A |
| T. ovis | ........................... - - .. | .......................... | ......................... | ............................... |
| T. parva | ........................... - - .. | .......................... | ......................... | ............................... |
| T. uilenbergi | ........................... - - .. | .......................... | ......................... | .............................C. |
| T. lestoquardi | ........................... - - .. | .......................... | ......................... | ............................... |
| T. equi | ......................A.... - - .. | .......................... | .....C................... | ............................... |
| T. separata | ........................... - - .. | .......................... | ......................... | ......................C........ |
| T. capreoli | ........................... - - .. | .......................... | ......................... | ............................... |
| T. bicornis | ........................... - - .. | .....C.................... | ......................... | ............................... |
| T. taurotragi | ........................... - - .. | .......................... | ......................... | ............................... |
| T. mutans | ........................... - - .C | .C.......................C | .....C................... | ............................... |
| Theileria sp. OT3 | ........................... - - .. | .......................... | ......................... | ............................... |
| Theileria sp. NG | ........................... - - .. | .......................... | ......................... | ............................... |
| Theileria sp. YW | ........................... - - .. | .......................... | ......................... | ............................... |
| Theileria sp. B15 | ........................... - - .C | .C......................CC | .....C................... | .............................C. |
| B. vulpes | ......................A.... - - .. | .........................C | .....CT.C................ | ......G......................CT |
| B. hongkongensis | ......................A.... - - AA | .....................TG.C. | .....CTCA........G....... | ....................T....T...GT |
| B. divergens | ......................A.... - - AA | .....................TG.CC | .....CTCA........A....... | .........................T...GT |
| B. bovis | ...............C........... - - .A | .CTC.........C...GG..CG.CC | TC...CTCG..C.....C.C..... | ........................C.A-.GT |
| B. bigemina | ......................A.... - - .. | .....................TG..G | .C.A.CTCA........C....... | ....G...............T.....A..CT |
| B. gibsoni | ......................A.... - - AA | .....................TG.CG | ...ATCTCA........A....... | .A...........................GT |
| B. microti | ......................A.... - - .. | .........................C | .....CT.C................ | ......G......................CT |
| B. felis | ......................A.... - - .. | ......................G.CC | .....CT.C................ | ......G......................CT |
| B. canis | ......................A.... - - .A | .....................TG.C. | .....CTCA........G....... | .........................TA..GT |
| C. felis | .......................A... - - .. | ..................C..A.... | ..G.TCT....G............. | ............................... |
| H. americanum | ......................AA...AA.. | .CT................A.A.... | ....ACT..TT.A............ | ............................T.A |
| T. gondii | ...................C..T..AAAT.. | T...A..G...........A.....C | .....C...T.......A....... | ....................G.......... |
Primers and probes are shown at the head of the table. Dots indicate nucleotides identical to primers and probes, and dashes denote absence of the nucleotide. The upstream primer is used as the demonstrated sequences without gaps while the two probes and downstream primer are used as antisense oligonucleotides. The designed oligonucleotides show minimum mismatching with Theileria spp. The 6-FAM label is directly attached to the 3-terminal nucleotide of the fluorescein probe, and the LCRed-640 fluorescein label is added via a linker to the 5′-end of the LCRed-640 probe. The 18S rRNA sequences for the available recognized Theileria spp. on GenBank and other closely related protozoan species were obtained from GenBank: T. orientalis (HM538222), T. buffeli (HQ840967), T. annulata (KF429799), T. sergenti (EU083804), T. luwenshuni (JX469527), T. velifera (AF097993), T. ovis (AY508458), T. parva (L02366), T. uilenbergi (JF719835), T. equi (AY534882), T. lestoquardi (JQ917458), T. separata (AY260175), T. capreoli (AY726011), T. bicornis (AF499604), T. taurotragi (L19082), T. mutans (FJ213585), Babesia vulpes (JX454779), Theileria sp. OT3 (KF470868), Theileria sp. NG-2013a (KF597076), Theileria sp. YW-2014 (AB981984), Theileria sp. B15a (JN572700); B. hongkongensis (JQ867356), B. divergens (AJ439713), B. bovis (JQ723013), B. bigemia (JQ723014), B. gibsoni (EU583386), B. microti (AB219802), B. felis (AF244912), B. canis (HM590440), Hepatozoon americanum (AF176836), Cytauxzoon felis (AY679105), and Toxoplasma gondii (L37415).
2. Materials and Methods
2.1. Samples Collection
Jugular venipuncture was used to collect blood in EDTA from convenience samples of apparently healthy livestock (cattle, goats, sheep, donkeys, and horses) on five Caribbean islands [22]. This study was reviewed and approved by the Institutional Animal Care and Use Committee of the Ross University School of Veterinary Medicine (RUSVM), St. Kitts. Owners of animals gave permission for the blood samples to be collected.
2.2. DNA Extraction
The DNA was extracted from aliquots (200 μL) of the whole blood samples with the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's instructions. The DNAs were eluted into 200 μL Buffer AE and couriered to Yangzhou University College of Veterinary Medicine, China, at room temperature where they were frozen at −80°C until PCRs were performed.
2.3. PCRs for Theileria Detection and Species Determination
All the PCRs were performed on a Roche Light-Cycler 480-II platform with the HMBS gene as an endogenous control [23]. Samples found positive in the pan-Theileria FRET-qPCR were tested in a conventional PCR with primers targeting a highly polymorphic 584–610-nucleotide region of the 18S rRNA gene of Theileria spp. (Table 2, Figure 1) [21]. The amplicons from positive conventional PCRs were sequenced directly with forward and reverse primers to determine the Theileria spp. present (BGI, Shanghai, China) [21] as has been done with 18S rRNA sequences in a number of previous studies [11, 14, 17, 21, 24].
Table 2.
Primers and probes used in this study.
| PCR | Primer/probe | Nucleotides sequence | Amplicon |
|---|---|---|---|
| FRET-qPCR | Forward Reverse 6-FAM LCRed-640 |
5′-TAGTGACAAGAAATAACAATACGGGGCTT-3′ 5′-CAGCAGAAATTCAACTACGAGCTTTTTAACT-3′ 5′-CCAATTGATACTCTGGAAGAGGTTT-(6-FAM)-3′ 5′-(LCRed640)-AATTCCCATCATTCCAATTACAAGAC-Phosphate-3′ |
178 bp |
|
| |||
| Conventional PCR | Upstream Downstream |
5′-CCTGAGAAACGGCTACCACATCT-3′ 5′-GGACTACGACGGTATCTGATCG-3′ |
T. orientalis 593 bp; T. buffeli 591 bp; T. annulata 591 bp; T. sergenti 591 bp; T. luwenshuni 594 bp; T. velifera 592 bp; T. ovis 595 bp; T. parva 592 bp; T. uilenbergi 592 bp; T. equi 596 bp; T. cervi 595 bp; T. lestoquardi 591 bp; T. separata 593 bp; T. capreoli 599 bp; T. bicornis 610 bp; T. taurotragi 587 bp; T. mutans 585 bp; B. vulpes 609 bp; Theileria sp. OT3 600 bp; Theileria sp. NG 597 bp; Theileria sp. YW 593 bp; Theileria sp. B15 584 bp |
Figure 1.
Alignment of the sequences in the polymorphic region of the 18S RNA gene of Theileria spp. and related organisms that was targeted in the standard PCR used in the study. Nucleotides that are identical for all species are highlighted in blue while those that vary between species and can be used for differentiation are not highlighted.
3. Results and Discussion
The sensitive and specific pan-Theileria FRET-qPCR [21] we used in our study demonstrated that a substantial proportion of livestock (8.6%; 65/752) on the five Caribbean islands we studied were infected with Theileria (Table 3). Each island had positive animals and each livestock species we studied was found to be infected with Theileria spp. Sequencing of representative samples of positive 18S rRNA PCR amplicons we obtained (n = 43) showed that there was one recognized Theileria spp. (T. equi) present in the Caribbean, along with five less well characterized Theileria sp. (Tables 3 and 4, Figure 2). The average copy number of 18S rRNA per mL whole blood was relatively low at 116.6 ± 440.8, indicating that the animals we studied were chronically infected.
Table 3.
Prevalence of Theileria spp. in livestock from five Caribbean islands.
| Bovine | Goat | Sheep | Donkey | Horse | Total | Theileria spp. | |
|---|---|---|---|---|---|---|---|
| Dominica | 3/77 (3.9%) |
0/70 (0.0%) |
1/15 (6.7%) |
N/A∗ | N/A | 4/162 (2.5%) | T. equi, Theileria sp. NG-2013a, B. vulpes, or closely related organism |
|
| |||||||
| Grenada | N/A | 2/31 (6.5%) |
N/A | N/A | N/A | 2/31 (6.5%) | Theileria sp. B15a |
|
| |||||||
| Montserrat | 0/12 (0.0%) |
8/19 (42.1%) |
24/62 (38.7%) |
N/A | N/A | 32/93 (34.4%) | Theileria sp. OT3, B. vulpes, or closely related organism |
|
| |||||||
| Nevis | 19/43 (44.2%) |
2/114 (1.8%) |
0/41 (0.0%) |
N/A | N/A | 21/198 (10.6%) |
T. equi, Theileria sp. NG-2013a |
|
| |||||||
| St. Kitts | 0/193 (0.0%) |
0/4 (0.0%) |
0/26 (0.0%) |
5/25 (20.0%) |
1/20 (5.0%) |
6/268 (2.2%) |
T. equi, Theileria sp. YW-2014 |
|
| |||||||
| Total | 22/325 (6.8%) |
12/238 (5.0%) |
25/144 (17.4%) |
5/25 (20.0%) |
1/20 (5.0%) |
65/752 (8.6%) |
|
|
| |||||||
| Theileria spp. |
T. equi, Theileria sp. NG-2013a |
T. equi, Theileria sp. OT3, Theileria sp. B15a, B. vulpes, or closely related organism |
T. equi, Theileria sp. OT3, B. vulpes, or closely related organism |
T. equi, Theileria sp. YW-2014 |
T. equi | ||
∗No specimen was available.
Table 4.
The Theileria spp. identified in livestock from five Caribbean islands and their similarity with reported organisms on GenBank.
| Sequences identified in this study | Highly similar sequences in GenBank | Mismatch | |||
|---|---|---|---|---|---|
| Theileria spp. | Number | Source | GenBank # | Source | |
| T. equi | 14 | 8 cattle, 1 goat from Nevis; 1 cow from Dominica; 3 donkeys, 1 sheep from St. Kitts |
KF559357 | Horse from China | 0/550 |
|
| |||||
| Theileria sp. OT3 | 11 | 8 sheep, 3 goats from Montserrat | KF470868 | Sheep from China | 0/555 |
|
| |||||
| Theileria sp. NG-2013a | 7 | 6 cattle from Nevis; 1 cow from Dominica |
KF597076 | Waterbuck from Kenya | 0/552 |
|
| |||||
| Theileria sp. YW-2014 | 1 | 1 donkey from St. Kitts | AB981984 | Sika deer from Japan | 0/549 |
|
| |||||
| Theileria sp. B15a | 1 | 1 goat from Grenada | JN572700 | Buffalo from South Africa | 0/539 |
|
| |||||
|
B. vulpes or closely related organism |
9 | 4 sheep, 4 goats from Montserrat; 1 goat from Dominica |
JX454779 | Dog from France | 0/178 |
Figure 2.
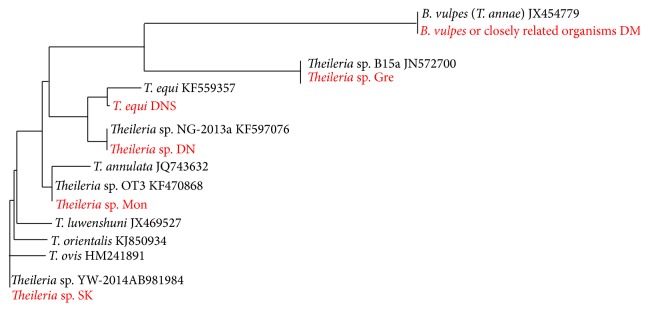
Phylogenetic tree of sequences identified in this study and closely related Theileria spp. Sequences from GenBank have gene accession numbers in black font; those from this study are in red font. B. vulpes or closely related organism DM indicates the organism detected on Dominica and Montserrat; Theileria sp. Gre indicates the Theileria sp. from Grenada; T. equi DNS indicates the sequence of T. equi from Dominica, Nevis, and St. Kitts; Theileria sp. Mon indicates the Theileria sp. detected from Montserrat; and Theileria sp. SK is the sequence of Theileria sp. from St. Kitts.
Theileria equi and the Theileria sp. YW-2014 were the species we identified in equids. Theileria sp. YW-2014 has been described in a Sika deer (Cervus nippon) from Japan but there is little sequence data on the organism with only a 552 bp sequence of the 18S rRNA gene reported in GenBank (AB981984). On the other hand, T. equi is a well-recognized cause of equine piroplasmosis [25], an important disease of horses which has been recognized and studied in the Caribbean [14–16]. The organism has also been found in dogs in Spain [26], South Africa [27], and Nigeria [28] with some having clinical signs that responded to appropriate treatment [29]. Our finding that T. equi also occurs in domestic ruminants further expands the host range of the organism. The significance, extent, and consequences of infections with T. equi in domestic ruminants require further investigation.
Numerous species of Amblyomma, Dermacentor, Hyalomma, Ixodes, and Rhipicephalus are confirmed or suspected vectors of T. equi [29]. Of these, only A. cajennense [30], R. microplus [30], R. sanguineus [31], and R. turanicus (unpublished observation) occur in the Caribbean. There is conflicting data that Amblyomma cajennense is a competent vector of T. equi [32] but the tick is localized to Jamaica, Trinidad, and Cuba in the Caribbean [30] and it appears, then, not to be the vector of T. equi we found on Nevis, St. Kitts, and Dominica. Dermacentor nitens, the tropical horse tick, is very common in the Caribbean and the tropical Americas [33]. Although there is no data on the competence of D. nitens as a vector of T. equi and there is contradictory epidemiological evidence [32], PCR positive D. nitens have been found [32] and Asgarali et al. [16] have suggested that this tick is a vector in Trinidad. They also suggested that R. microplus [16], which is very common on cattle throughout the Caribbean, might also be a vector. While there is some evidence that R. microplus is a competent vector [34] and our PCR identified T. equi in cattle on two islands, it seems unlikely that R. microplus is an important natural vector as it is a one host species and transovarial transmission has not been demonstrated [32]. Although R. sanguineus and R. turanicus have been implicated as vectors of T. equi, more recent studies have failed to confirm their role [29]. Further studies are needed to establish the epidemiology of T. equi and its vectors in the Caribbean and the neighboring Americas [32].
Theileria sp. OT3 was first described in sheep, deer, and chamois in Spain [35–37] and later in sheep in Italy [38], China [39], and Turkey [40]. Its pathogenicity and vectors have yet to be determined. Ours is the first report of the organism in the Caribbean and also the first report of the Theileria sp. OT3 in goats which further demonstrates that the organism has a wide distribution and host range. Although we used only convenience samples which were not representative of the islands, it is of note that Theileria sp. OT3 was the most prevalent species we detected in ruminants. Similar high prevalences of infections have also been reported in the other countries where the organism has been described. The sequences of our Theileria sp. OT3 were identical to one another and to that of an isolate from China (KF470868) [39]. A phylogenetic relationship tree established for the Chinese isolate showed that the Theileria sp. OT3 forms a separated cluster and that the organism is closely related to T. uilenbergi, T. luwenshuni, and T. ovis. Although the organism has only been found in apparently healthy animals, studies on its pathogenicity seem indicated as, with its high prevalence and wide distribution, it might be causing substantial economic losses for livestock farmers.
Theileria mutans and T. velifera are African species that have been reported on Guadeloupe [12] and appear most likely to have been imported with cattle from West Africa in the 18th Century. This might also have been the case with the other African Theileria sp. we identified. Seven of the Theileria we found had an identical sequence to that of Theileria sp. NG-2013a (KF597076) described from waterbuck (Kobus defassa) in Kenya [41]. This organism has been found to cluster with T. equi but might represent a novel taxon. Its vectors and pathogenicity are unknown. One goat we studied had a Theileria spp. with a sequence identical to that of Theileria sp. B15a (JN572700) from an African buffalo (Syncerus caffer) in South Africa [42]. This organism is close to T. mutans which is widespread in Africa where it is transmitted by Amblyomma spp. and causes benign theileriosis. Previously T. mutans has been reported on Guadeloupe based on serological test results [12] and, since we found no confirmatory molecular evidence for the presence of T. mutans, it might be that the cross-reacting antibodies detected in Guadeloupe were against this closely related Theileria sp. B15.
The only non-Theileria sp., the pan-Theileria FRET-qPCR identified in our study, was one that appeared to be Babesia vulpes [43] or a closely related species. This organism was previously known as “T. annae” or the “Babesia microti-like organism” but was reclassified based on 18S rRNA and tubulin-beta gene sequence data [43]. The sequences of the short amplicons we obtained in the pan-Theileria FRET-qPCR were identical to those of B. vulpes (JX454779; KF773740) and had one mismatch with B. microti (AB219802, HQ629933, and LC005772). We were unable, however, to obtain longer amplicons with the standard PCR, probably because there were only very low copy numbers present in the positive animals. We were, then, unable to use this additional sequencing data to confirm that the organism we detected was B. vulpes or determine if it was a closely related species or strain. The pan-Theileria FRET-qPCR we used in our study was designed to detect seventeen recognized Theileria spp., which did not include “T. annae” or B. vulpes (Table 1) [21]. These seventeen recognized Theileria spp. differed from one another by only a maximum of 4 nucleotides in the regions of the primers and the probes used in the pan-Theileria FRET-qPCR. It is of note, then, that these primers and probes enabled the multiplication and detection of B. vulpes, or a closely related species, with 8 nucleotide differences and it therefore appears that the pan-Theileria FRET-qPCR might not be as genus specific as first thought. Further work is currently underway in our laboratory to more clearly characterize the B. vulpes or closely related organism found in the Caribbean.
4. Conclusions
Our study has confirmed the sensitivity of the pan-Theileria PCR in the rapid detection of a wide range of Theileria spp. but has also shown it might detect B. vulpes or closely related organisms. We found livestock infected with Theileria spp. on each of the five islands we studied. While we could not confirm previous reports of T. mutans and T. velifera in cattle, we found that one recognized species, T. equi, four poorly characterized Theileria spp., and B. vulpes or a closely related organism are present in the region. Further studies are indicated to more precisely determine the phylogenetic relationships of these organisms in the Caribbean with closely related organisms from other parts of the world. Also, the prevalences of infections on the different islands should be determined as well as the impact these poorly characterized organisms might have on livestock production, both in the Caribbean and around the world where they are found.
Acknowledgments
This project was supported by grants from the National Natural Science Foundation of China (no. 31472225) and the Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou, Jiangsu, China and Grant 2006 34135 6930 from the United States Department of Agriculture through its program for Tropical and Subtropical Agricultural Research (T-STAR) and by Ross University School of Veterinary Medicine.
Conflict of Interests
The authors declare that they have no competing interests.
Authors' Contribution
Chengming Wang, Patrick Kelly, and Jilei Zhang designed the study, analyzed the data, and wrote the paper. Jilei Zhang, Jing Li, and Chuanling Xu carried out the experiments. All authors read and approved the final paper.
References
- 1.Shaw M. K., Tilney L. G., Musoke A. J. The entry of Theileria parva sporozoites into bovine lymphocytes: evidence for MHC class I involvement. The Journal of Cell Biology. 1991;113(1):87–101. doi: 10.1083/jcb.113.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop R., Musoke A., Morzaria S., Gardner M., Nene V. Theileria: intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parasitology. 2004;129(supplement):S271–S283. doi: 10.1017/s0031182003004748. [DOI] [PubMed] [Google Scholar]
- 3.Florin-Christensen M., Schnittger L. Piroplasmids and ticks: a long-lasting intimate relationship. Frontiers in Bioscience. 2009;14(8):3064–3073. doi: 10.2735/3435. [DOI] [PubMed] [Google Scholar]
- 4.Glass E. J. The balance between protective immunity and pathogenesis in tropical theileriosis: what we need to know to design effective vaccines for the future. Research in Veterinary Science. 2001;70(1):71–75. doi: 10.1053/rvsc.2000.0428. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed J. S., Glass E. J., Salih D. A., Seitzer U. Innate immunity to tropical theileriosis. Innate Immunity. 2008;14(1):5–12. doi: 10.1177/1753425907087258. [DOI] [PubMed] [Google Scholar]
- 6.Sivakumar T., Hayashida K., Sugimoto C., Yokoyama N. Evolution and genetic diversity of Theileria . Infection, Genetics and Evolution. 2014;27:250–263. doi: 10.1016/j.meegid.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Rjeibi M. R., Darghouth M. A., Rekik M., Amor B., Sassi L., Gharbi M. First molecular identification and genetic characterization of Theileria lestoquardi in sheep of the Maghreb region. Transboundary and Emerging Diseases. 2014 doi: 10.1111/tbed.12271. [DOI] [PubMed] [Google Scholar]
- 8.Bawm S., Shimizu K., Hirota J.-I., et al. Molecular prevalence and genetic diversity of bovine Theileria orientalis in Myanmar. Parasitology International. 2014;63(4):640–645. doi: 10.1016/j.parint.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Belotindos L. P., Lazaro J. V., Villanueva M. A., Mingala C. N. Molecular detection and characterization of Theileria species in the Philippines. Acta Parasitologica. 2014;59(3):448–453. doi: 10.2478/s11686-014-0256-9. [DOI] [PubMed] [Google Scholar]
- 10.Hussain M. H., Saqib M., Raza F., et al. Seroprevalence of Babesia caballi and Theileria equi in five draught equine populated metropolises of Punjab, Pakistan. Veterinary Parasitology. 2014;202(3-4):248–256. doi: 10.1016/j.vetpar.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Chen Z., Liu Z., et al. Molecular identification of Theileria parasites of northwestern Chinese Cervidae. Parasites and Vectors. 2014;7, article 225 doi: 10.1186/1756-3305-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uilenberg G., Camus E., Barré N. Existence of Theileria mutans and Theileria velifera (Sporozoa, Theileriidae) in Guadeloupe (French West Indies) Revue d'Elevage et de Médecine Vétérinaire des Pays Tropicaux. 1983;36(3):261–264. [PubMed] [Google Scholar]
- 13.Alonso M., Camus E., Rodriguez Diego J., Bertaudière L., Tatareau J. C., Liabeuf J. M. Current status of bovine haemoparasitic diseases in Martinique (French West Indies) Revue d'Elevage et de Medecine Veterinaire des Pays Tropicaux. 1992;45(1):9–14. [PubMed] [Google Scholar]
- 14.Rampersad J., Cesar E., Campbell M. D., Samlal M., Ammons D. A field evaluation of PCR for the routine detection of Babesia equi in horses. Veterinary Parasitology. 2003;114(2):81–87. doi: 10.1016/s0304-4017(03)00129-8. [DOI] [PubMed] [Google Scholar]
- 15.Georges K. C., Ezeokoli C. D., Sparagano O., et al. A case of transplacental transmission of Theileria equi in a foal in Trinidad. Veterinary Parasitology. 2011;175(3-4):363–366. doi: 10.1016/j.vetpar.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Asgarali Z., Coombs D. K., Mohammed F., Campbell M. D., Caesar E. A serological study of Babesia caballi and Theileria equi in Thoroughbreds in Trinidad. Veterinary Parasitology. 2007;144(1-2):167–171. doi: 10.1016/j.vetpar.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Katzer F., McKellar S., Kirvar E., Shiels B. Phylogenetic analysis of Theileria and Babesia equi in relation to the establishment of parasite populations within novel host species and the development of diagnostic tests. Molecular and Biochemical Parasitology. 1998;95(1):33–44. doi: 10.1016/s0166-6851(98)00085-1. [DOI] [PubMed] [Google Scholar]
- 18.Gubbels J. M., de Vos A. P., van der Weide M., et al. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. Journal of Clinical Microbiology. 1999;37(6):1782–1789. doi: 10.1128/jcm.37.6.1782-1789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnittger L., Yin H., Qi B., et al. Simultaneous detection and differentiation of Theileria and Babesia parasites infecting small ruminants by reverse line blotting. Parasitology Research. 2004;92(3):189–196. doi: 10.1007/s00436-003-0980-9. [DOI] [PubMed] [Google Scholar]
- 20.Mans B. J., Pienaar R., Latif A. A. A review of Theileria diagnostics and epidemiology. International Journal for Parasitology: Parasites and Wildlife. 2015;4(1):104–118. doi: 10.1016/j.ijppaw.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y., Mao Y., Kelly P., et al. A pan-Theileria FRET-qPCR survey for Theileria spp. in ruminants from nine provinces of China. Parasites and Vectors. 2014;7, article 413 doi: 10.1186/1756-3305-7-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly P., Lucas H., Beati L., Yowell C., Mahan S., Dame J. Rickettsia africae in Amblyomma variegatum and domestic ruminants on eight Caribbean islands. Journal of Parasitology. 2010;96(6):1086–1088. doi: 10.1645/ge-2552.1. [DOI] [PubMed] [Google Scholar]
- 23.Wei L., Kelly P., Zhang J., et al. Use of a universal hydroxymethylbilane synthase (HMBS)-based PCR as an endogenous internal control and to enable typing of mammalian DNAs. Applied Microbiology and Biotechnology. 2014;98(12):5579–5587. doi: 10.1007/s00253-014-5659-x. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins E., Kock R., McKeever D., et al. Prevalence of Theileria equi and Babesia caballi as well as the identification of associated ticks in sympatric grevy’s zebras (Equus grevyi) and donkeys (Equus africanus asinus) in northern Kenya. Journal of Wildlife Diseases. 2015;51(1):137–147. doi: 10.7589/2013-11-316. [DOI] [PubMed] [Google Scholar]
- 25.Wise L. N., Kappmeyer L. S., Mealey R. H., Knowles D. P. Review of equine piroplasmosis. Journal of Veterinary Internal Medicine. 2013;27(6):1334–1346. doi: 10.1111/jvim.12168. [DOI] [PubMed] [Google Scholar]
- 26.Criado-Fornelio A., Martinez-Marcos A., Buling-Saraña A., Barba-Carretero J. C. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe. Part II. Phylogenetic analysis and evolutionary history. Veterinary Parasitology. 2003;114(3):173–194. doi: 10.1016/s0304-4017(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 27.Rosa C. T., Pazzi P., Nagel S., et al. Theileriosis in six dogs in South Africa and its potential clinical significance. Journal of the South African Veterinary Association. 2014;85(1, article 1114) doi: 10.4102/jsava.v85i1.1114. [DOI] [PubMed] [Google Scholar]
- 28.Adamu M., Troskie M., Oshadu D. O., Malatji D. P., Penzhorn B. L., Matjila P. T. Occurrence of tick-transmitted pathogens in dogs in Jos, Plateau State, Nigeria. Parasites and Vectors. 2014;7(1, article 119) doi: 10.1186/1756-3305-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scoles G. A., Ueti M. W. Vector ecology of equine piroplasmosis. Annual Review of Entomology. 2015;60:561–580. doi: 10.1146/annurev-ento-010814-021110. [DOI] [PubMed] [Google Scholar]
- 30.Camus E., Barre N. Vector situation of tick-borne diseases in the Caribbean islands. Veterinary Parasitology. 1995;57(1–3):167–176. doi: 10.1016/0304-4017(94)03118-g. [DOI] [PubMed] [Google Scholar]
- 31.Kelly P. J., Xu C., Lucas H., et al. Ehrlichiosis, babesiosis, anaplasmosis and hepatozoonosis in dogs from St. Kitts, West Indies. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0053450.e53450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peckle M., Pires M. S., Dos Santos T. M., et al. Molecular epidemiology of Theileria equi in horses and their association with possible tick vectors in the state of Rio de Janeiro, Brazil. Parasitology Research. 2013;112(5):2017–2025. doi: 10.1007/s00436-013-3360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garris G. I., Scotland K. Ticks on livestock in St. Lucia. Veterinary Parasitology. 1985;18(4):367–373. doi: 10.1016/0304-4017(85)90071-8. [DOI] [PubMed] [Google Scholar]
- 34.Stiller D., Goff W. L., Johnson L. W., Knowles D. P. Dermacentor variabilis and Boophilus microplus (Acari: Ixodidae): experimental vectors of Babesia equi to equids. Journal of Medical Entomology. 2002;39(4):667–670. doi: 10.1603/0022-2585-39.4.667. [DOI] [PubMed] [Google Scholar]
- 35.Nagore D., García-Sanmartín J., García-Pérez A. L., Juste R. A., Hurtado A. Identification, genetic diversity and prevalence of Theileria and Babesia species in a sheep population from Northern Spain. International Journal for Parasitology. 2004;34(9):1059–1067. doi: 10.1016/j.ijpara.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 36.García-Sanmartín J., Aurtenetxe O., Barral M., et al. Molecular detection and characterization of piroplasms infecting cervids and chamois in Northern Spain. Parasitology. 2007;134(Part 3):391–398. doi: 10.1017/s0031182006001569. [DOI] [PubMed] [Google Scholar]
- 37.Ros-García A., Barandika J. F., García-Pérez A. L., Juste R. A., Hurtado A. Assessment of exposure to piroplasms in sheep grazing in communal mountain pastures by using a multiplex DNA bead-based suspension array. Parasites and Vectors. 2013;6(1, article no. 277) doi: 10.1186/1756-3305-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giangaspero A., Marangi M., Papini R., Paoletti B., Wijnveld M., Jongejan F. Theileria sp. OT3 and other tick-borne pathogens in sheep and ticks in Italy: molecular characterization and phylogeny. Ticks and Tick-borne Diseases. 2015;6(1):75–83. doi: 10.1016/j.ttbdis.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Tian Z., Liu G., Yin H., et al. First report on the occurrence of Theileria sp. OT3 in China. Parasitology International. 2014;63(2):403–407. doi: 10.1016/j.parint.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Aydin M. F., Aktas M., Dumanli N. Molecular identification of Theileria and Babesia in sheep and goats in the Black Sea Region in Turkey. Parasitology Research. 2013;112(8):2817–2824. doi: 10.1007/s00436-013-3452-x. [DOI] [PubMed] [Google Scholar]
- 41.Githaka N., Konnai S., Bishop R., et al. Identification and sequence characterization of novel Theileria genotypes from the waterbuck (Kobus defassa) in a Theileria parva-endemic area in Kenya. Veterinary Parasitology. 2014;202(3-4):180–193. doi: 10.1016/j.vetpar.2014.02.056. [DOI] [PubMed] [Google Scholar]
- 42.Chaisi M. E., Collins N. E., Potgieter F. T., Oosthuizen M. C. Sequence variation identified in the 18S rRNA gene of Theileria mutans and Theileria velifera from the African buffalo (Syncerus caffer) Veterinary Parasitology. 2013;191(1-2):132–137. doi: 10.1016/j.vetpar.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Baneth G., Florin-Christensen M., Cardoso L., Schnittger L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasites & Vectors. 2015;8(1, article 207) doi: 10.1186/s13071-015-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]