Abstract
The adiponectin gene (ADIPOQ) plays an important role in energy homeostasis. In this study five separate regions (regions 1 to 5) of ovine ADIPOQ were analysed using PCR-SSCP. Four different PCR-SSCP patterns (A1-D1, A2-D2) were detected in region-1 and region-2, respectively, with seven and six SNPs being revealed. In region-3, three different patterns (A3-C3) and three SNPs were observed. Two patterns (A4-B4, A5-B5) and two and one SNPs were observed in region-4 and region-5, respectively. In total, nineteen SNPs were detected, with five of them in the coding region and two (c.46T/C and c.515G/A) putatively resulting in amino acid changes (p.Tyr16His and p.Lys172Arg). In region-1, -2 and -3 of 316 sheep from eight New Zealand breeds, variants A1, A2 and A3 were the most common, although variant frequencies differed in the eight breeds. Across region-1 and region-3, nine haplotypes were identified and haplotypes A1-A3, A1-C3, B1-A3 and B1-C3 were most common. These results indicate that the ADIPOQ gene is polymorphic and suggest that further analysis is required to see if the variation in the gene is associated with animal production traits.
Keywords: adiponectin, ADIPOQ, variation, haplotype, PCR-SSCP, sheep
1. Introduction
As a member of the adipocytokine family, adiponectin (ADIPOQ) is secreted mainly by the white adipose tissue, but it can also be produced by other tissues such as brown adipose tissue, bone marrow and skeletal muscle [1,2,3]. It has been demonstrated that ADIPOQ plays an important role in the adipokine signalling pathway and thus it can regulate energy homeostasis, glucose metabolism and lipid metabolism [4]. In humans and mice, the expression of ADIPOQ in adipose tissue is negatively correlated with obesity [5].
The human adiponectin gene (ADIPOQ) was first identified in 1995 and is located on human chromosome 3q27 [6,7]. It spans about 17 kb and contains three exons and two introns. Variation in the human gene has been reported to be associated with obesity, type 2 diabetes susceptibility, cancer risk, serum adiponectin levels and Chronic Obstructive Pulmonary Disease [8,9,10,11]. In livestock species, polymorphisms in ADIPOQ have been associated with various traits including chest circumference in goats [12], fat deposition, carcass traits and reproduction traits in pigs [13,14], and meat marbling, ribeye muscle area and carcass fat thickness in cattle [3]. Despite these findings the various functions of ADIPOQ remain poorly understood, but the reports published to date suggest that ADIPOQ may be an important gene for key animal production traits.
In this study, the objective was to search for polymorphism in different regions (spanning the promoter region to the 3'-UTR region) of ADIPOQ in different breeds of sheep, in anticipation of a much larger study to ascertain whether the variation has an effect on ovine growth, carcass and other production traits.
2. Materials and Methods
All research involving animals was carried out in accordance with the Animal Welfare Act 1999 (New Zealand Government) and the collection of sheep blood drops by nicking sheep ears is covered by Section 7.5 Animal Identification, of the Animal Welfare (Sheep and Beef Cattle) Code of Welfare 2010; a code of welfare issued under the Animal Welfare Act 1999 (New Zealand Government).
2.1. Sheep Investigated and DNA Collection
One hundred unrelated sheep, selected from a variety of common breeds in New Zealand (NZ) including Merinos (n = 18), Suffolks (n = 21), Texels (n = 22), Dorset Downs (n = 19) and NZ Romneys (n = 20), were used initially to screen for variation in ADIPOQ by methods that are described below. Having identified that variation existed in these sheep, an additional 216 unrelated sheep from a variety of breeds were added to the analysis to enable more accurate determination of individual variant frequencies.
Samples of blood were collected directly onto FTA cards (Whatman BioScience, Middlesex, UK) and DNA for analysis was purified from the dried blood spots, using a procedure described by Zhou et al. [15].
2.2. PCR Amplification and SSCP Analysis
Five different regions of ovine ADIPOQ located at or near the coding sequences were chosen for analysis. These were region-1 (containing part of the promoter, entire exon 1 and part of intron 1), region-2 (containing entire exon 2 and flanking sequences from introns 1 and 2), region-3 (containing part of intron 2 and part of exon 3), region-4 (containing part of exon 3) and region-5 (containing part of exon 3). Five sets of PCR primers were designed based on the ovine genome sequence (NC_019458.1) for amplification of these regions and their precise coordinates are described in Table 1. These primers were synthesised by Integrated DNA Technologies (Coralville, IA, USA).
Table 1.
The primer sequences and PCR-SSCP conditions used for analysis of ovine ADIPOQ.
| Region Amplified | Primer Binding Region a | Primer Sequence (5'–3') | PCR Annealing Temperature | Amplified Size | SSCP Condition |
|---|---|---|---|---|---|
| Region-1 | 198617873_198617893 | F: TTCCTGCTTCTGATCTTGACC | 58 °C | 388 bp | 300 V, 14%, 17 °C |
| 198618239_198618260 | R: CAGCCTAGAAATTGAATCAGTC | ||||
| Region-2 | 198627771_198627789 | F: ACAGCGTGGATCTGGGTTC | 62 °C | 390 bp | 200 V, 14%, 32.5 °C |
| 198628140_198628159 | R: CACAATTCACTTTCGGCTGC | ||||
| Region-3 | 198628889_198628909 | F: GGTCTTCTTGTTCTCTAGGTC | 58 °C | 391 bp | 200 V, 12%, 20 °C |
| 198629260_198629279 | R: TGGTCCACGTTCTGGTTCTG | ||||
| Region-4 | 198629237_198629256 | F: TGCTCTTCACCCACGACCAG | 58 °C | 373 bp | 200 V, 14%, 26 °C |
| 198629583_198629605 | R: GTCCTGCGAACATAGTATATC | ||||
| Region-5 | 198629532_198629553 | F: GGATTCTGAACATCATTCATTC | 58 °C | 455 bp | 390 V, 14%, 5 °C |
| 198629967_198629986 | R: CAGATGAGTTGGTGGGAGAC |
a The primer binding position is given relative to the ovine genome sequence (NC_019458.1).
Amplifications were performed in a 15 μL reaction containing the purified genomic DNA on one punch of the FTA card, 0.25 μM of each primer, 150 μM of each dNTP (Bioline, London, UK), 2.5 mM of Mg2+, 0.5 U of Taq DNA polymerase (Qiagen, Hilden, Germany) and 1 × reaction buffer supplied with the enzyme. The thermal profile for the five regions amplified consisted of 2 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at the annealing temperatures shown in Table 1 and 30 s at 72 °C, with a final extension of 5 min at 72 °C. Amplification was carried out in S1000 thermal cyclers (Bio-Rad, Hercules, CA, USA).
The amplicons produced were visualized by electrophoresis in 1% agarose (Quantum Scientific, Queensland, Australia) gels, using 1 × TBE buffer (89 mM Tris, 89 mM Boric acid, 2 mM Na2EDTA), containing 200 ng/mL of ethidium bromide. They were then subjected to SSCP analysis. A 0.7 μL aliquot of each amplicon was mixed with 7 μL of loading dye (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene-cyanol). After denaturation at 95 °C for 5 min, the samples were cooled rapidly on wet ice and loaded onto 16 cm × 18 cm, 12 or 14% acrylamide:bisacrylamide (37.5:1) (Bio-Rad) gels. Electrophoresis was performed using Protean II xi cells (Bio-Rad) for 19 h in 0.5 × TBE using the conditions described in Table 1. The gels were silver-stained by the method of Byun et al. [16].
Amplicons that were identified as potentially homozygous by PCR-SSCP analysis were directly sequenced at the Lincoln University DNA Sequencing Facility. Those DNA sequences only found in a heterozygous form were analysed using an approach that has been described previously [17]. Briefly, a band corresponding to the allele was excised as a gel slice from the polyacrylamide gel, macerated, and then used as a template for re-amplification with the original primers. This second amplicon was then sequenced.
2.3. Sequence Analyses
The frequency of occurrence of sequence variants in the different regions was calculated using POPGEN version 3.2 (Molecular Biology and Biotechnology Centre, University of Alberta, Canada). Sequence alignments, translations and comparisons were carried out using DNAMAN version 5.2.10 (Lynnon BioSoft, Vaudreuil, Canada). The BLAST algorithm was used to search the NCBI GenBank databases (http://blast.ncbi.nlm.nih.gov/) for homologous sequences.
2.4. Haplotype Determination
Samples were genotyped at region-1 and region-3 of ovine ADIPOQ. Haplotypes spanning these two regions of the gene were constructed using those samples that were homozygous in either region. For those samples that were heterozygous in both regions, haplotypes could not be determined, as these two regions span over 11 kb and haplotype-specific PCR amplification [18] would be difficult using the DNA polymerase enzyme employed in this study.
3. Results
3.1. Identification of Sequence Variants of Ovine ADIPOQ
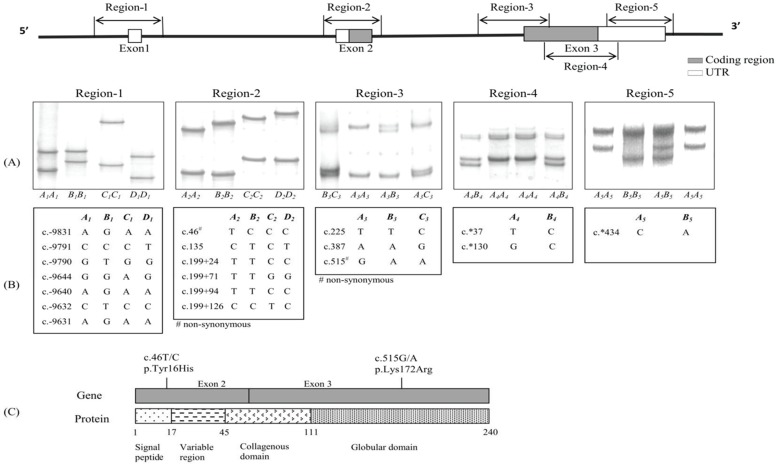
Of the 100 samples from the initial five breeds investigated, four SSCP patterns were obtained from the region-1 and region-2 amplicons, three SSCP patterns from the region-3 amplicons, and two SSCP patterns from the region-4 and region-5 amplicons (Figure 1A).
Figure 1.
Variation identified in ovine ADIPOQ. Unique PCR-SSCP patterns (A) representing different DNA sequences (B) were detected in region-1 (1); region-2 (2); region-3 (3); region-4 (4); and region-5 (5). Two non-synonymous SNPs that would result in amino acid changes in the signal peptide and the globular domain were identified (C). The coordinates of the SNPs are annotated below the patterns based on the ovine whole genome sequence (NC_019458.1) and the numbering of positions follows the guidelines presented at http://www.hgvs.org/mutnomen/.
After sequencing, these patterns were confirmed as novel variant sequences of ovine ADIPOQ and the sequences were deposited into GenBank with accession numbers as follows: A1-D1: KP903754-KP903757; A2-D2: KP903758-KP903761; A3-C3: KP903762-KP903764; A4-B4: KR9011984-KR011985; and A5-B5: KP903765-KP903766. There were seven, six, three, two and one SNPs identified in region-1, region-2, region-3, region-4 and region-5, respectively (Figure 1B). It is notable that the c.46 T/C substitution in exon 2 putatively results in a non-conservative amino acid change (p.Tyr16His) in the signal peptide, and the c.515A/G substitution in exon 3 putatively results in a conservative amino acid change (p.Lys172Arg) in the globular domain (Figure 1C).
3.2. Initial Screen for Variation in ADIPOQ
In the initial 100 samples investigated, polymorphism in region-4 and region-5 was low. There were only two genotypes observed in region-4: A4/A4 (91%) and A4/B4 (9%) with individual variant frequencies of 95.5% for A4 and 4.5% for B4; and two genotypes in region-5: A5/A5 (89%) and A5/B5 (11%) with individual variant frequencies of 94.5% for A5 and 5.5% for B5. Variants in these two regions were accordingly not investigated in additional sheep.
3.3. Frequencies of the Sequence Variants in Different Breeds
In the 316 sheep of various breeds, A1 and B1 (region-1) were the most common variants and observed in all breeds, with an overall frequency of 55.7% and 42.1%, respectively (Table 2). B1 was the most common variant with frequency of 47.3% in Corriedale sheep and A1 was the second most common variant with a frequency of 39.0%. C1 was only observed in Romney sheep, while D1 was observed in Merino, Romney, Corriedale and Perendale sheep, with the lowest overall average frequency of 1.1%.
Table 2.
Frequencies of ovine ADIPOQ variants in various sheep breeds.
| Breed | n | Region-1 | Region-2 | Region-3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | B1 | C1 | D1 | A2 | B2 | C2 | D2 | A3 | B3 | C3 | |||||
| Merino | 68 | 50.0 | 48.5 | 0.0 | 1.5 | 55.2 | 5.2 | 9.6 | 30.2 | 66.9 | 11.8 | 21.3 | |||
| Romney | 71 | 52.8 | 41.6 | 4.9 | 0.7 | 76.8 | 0.0 | 4.9 | 18.3 | 49.3 | 1.4 | 49.3 | |||
| Suffolk | 42 | 53.6 | 46.4 | 0.0 | 0.0 | 75.0 | 1.2 | 0.0 | 23.8 | 35.7 | 0.0 | 64.3 | |||
| Texel | 22 | 56.8 | 43.2 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 65.9 | 6.8 | 27.3 | |||
| Corriedale | 41 | 39.0 | 57.3 | 0.0 | 3.7 | 75.6 | 2.4 | 6.1 | 15.9 | 28.1 | 3.7 | 68.3 | |||
| Perendale | 14 | 57.1 | 39.3 | 0.0 | 3.6 | 89.3 | 0.0 | 0.0 | 10.7 | 42.9 | 0.0 | 57.1 | |||
| Dorper | 39 | 76.9 | 23.1 | 0.0 | 0.0 | 56.4 | 1.3 | 38.5 | 3.9 | 53.9 | 5.1 | 41.0 | |||
| Dorset Down | 19 | 81.6 | 18.4 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 34.2 | 0.0 | 65.8 | |||
| Overall | 316 | 55.7 | 42.1 | 1.1 | 1.1 | 72.78 | 1.7 | 8.7 | 16.8 | 49.1 | 4.4 | 46.5 | |||
In region-2, A2 was observed in all of the breeds studied and was the most common variant with an overall frequency of 72.8% (Table 2). Only A2 was observed in the Texel and Dorset Down sheep and B2 was not observed in the Texel, Dorset Down, Romney and Perendale sheep. B2 was the least common variant with an overall frequency of 1.7%. D2 was the second most common variant with an overall frequency of 16.8% and was observed in all of the breeds except Texel and Dorset Down. C2 was observed in all of the breeds, except the Suffolk, Perendale, Texel and Dorset Down sheep.
In region-3, A3 was the most common variant and observed in all of the breeds, with an overall frequency of 49.1% (Table 2). C3 was also found in all breeds with an overall frequency of 46.52%, while B3 was the least common variant with an overall frequency of 4.4%. B3 was not observed in Suffolk, Perendale and Dorset Down sheep.
3.4. Haplotypes Spanning Region-1 to Region-3
Nine haplotypes that spanned region-1 to region-3 of ovine ADIPOQ were identified in the 316 sheep (Table 3). Of these haplotypes, A1-C3, A1-A3, B1-A3 and B1-C3 were the most common with an overall frequency of 30.1%, 26.4%, 23.2% and 14.9%, respectively. The other five haplotypes were rare, each occurring at frequency of less than 5%.
Table 3.
ADIPOQ haplotypes and frequencies for the region spanning region-1 to region-3.
| Haplotype | Frequency | Number (n) |
|---|---|---|
| A1-C3 | 30.1% | 139 |
| A1-A3 | 26.4% | 123 |
| B1-A3 | 23.2% | 108 |
| B1-C3 | 14.9% | 69 |
| B1-B3 | 3.3% | 15 |
| C1-A3 | 1.1% | 5 |
| A1-B3 | 0.7% | 3 |
| C1-B3 | 0.2% | 1 |
| C1-C3 | 0.2% | 1 |
4. Discussion
This is the first study to report sequence variation in ovine ADIPOQ. Five different regions were investigated in eight commercial sheep breeds from NZ, including meat, wool and dual-purpose breeds. Nineteen SNPs were found suggesting that the ovine gene is quite variable. Extensive heterogeneity has also been described in humans [8,11], pigs [14], cattle [3] and goats [12].
Of the nineteen SNPs detected in this study, only two were non-synonymous. One (c.46T/C) was located in the exon 2 coding region and the other (c.515G/A) was located in the exon 3 coding region. These would potentially result in the amino acid changes p.Tyr16His and p.Lys172Arg, respectively. The substitution p.Tyr16His is in the signal peptide and in the vicinity of the boundary between the signal peptide and variable region, while p.Lys172Arg is in a globular domain of ADIPOQ (see Figure 1C). p.Tyr16His may change the structure and thus function of the single peptide, leading to either a change in how the peptide is targeted post-translationally, or a change in the rate or nature of signal peptide processing. It is notable that these two amino acid substitutions occur at amino acid positions that are conserved in many mammalian species including humans, pigs, goats, rabbits, horses and cattle, with Tyr16 and Lys172 being the common residues in these other species.
Two substitutions (rs2241766 G/T in exon 2 and rs17366743 C/T in exon 3) in humans have been reported by Yang et al. [19] in the vicinity of ovine c.46T/C and c.515G/A. In humans, these substitutions have been reported to be associated with variation in adiponectin levels in some populations [20]. This would suggest that c.46T/C and c.515G/A are worthy of further investigation in other breeds of sheep including investigation of whether these SNPs are associated with variation in carcass fat traits.
We detected seven SNPs in the ovine ADIPOQ promoter, including c.-9831A/G, c.-9791C/T, c.-9790G/T, c.-9644G/A, c.-9640A/G, c.-9632C/T and c.-9631A/G. The effect of these substitutions is difficult to ascertain in the absence of functional studies of the ovine ADIPOQ promoter region. It is notable that in the human ADIPOQ promoter, multiple physiologically important binding sites have been reported including sites for Sp1, SREBP, AP1 and C/EBP [21]. In humans a proximal substitution (c.-11377C/G) has been reported and this substitution is located in a SP1 binding site [22]. The G nucleotide substitution at c.-11377C/G results in a change in SP1 bonding and causes a reduction in ADIPOQ activity [22]. Three other substitutions in the promoter (c.-19166T/G, c.-11426A/G and c.-11391G/A) have also been detected in humans and reported to be associated with type 2 diabetes and obesity [23]. While these SNPs are even further removed from the ovine SNPs described here, it supports the conclusion that additional investigation of promoter region variation is warranted.
In pigs, ADIPOQ has been mapped to chromosome 13, and is located near a quantitative trait locus (QTL) for backfat thickness [24] and variation in the average length of the fatty acid chains in the longissimus dorsi muscle [25]. Porcine nucleotide substitutions (c.-90C/A, c.-67G/A and c.-829C/T) have been reported and c.-67G/A and c. -892C/T segregate as two haplotypes (G-C and A-T) that have been associated with carcass and meat quality traits [26].
In cattle, ADIPOQ has been mapped to chromosome 1 and is a positional candidate gene for the QTL that affects meat marbling, meat quality grade, ribeye muscle area and weaning weight [27,28]. The SNP c.-176A/G (g.1454) in the bovine ADIPOQ promoter has been associated with longissimus dorsi muscle area and backfat thickness, and c.-199C/T (g.1431) and c.-34G/A (g.1596) in the bovine ADIPOQ promoter have been associated with fat thickness and ribeye muscle area [3,29].
This evidence from pigs and cattle supports the contention that the variation described here in ovine ADIPOQ may affect key sheep traits, including potentially fat deposition, fat composition and meat quality, and possibly also growth traits.
Four of the ovine substitutions c.199+24T/C, c.199+71T/G, c.199+94T/C and c.199+126T/C were found in intron 2. They would be unlikely to affect the structure or function of ADIPOQ directly unless they are linked to other variation in coding regions of the gene, or regions that may affect the processing of the primary transcript including its stability or longevity. It is also known that non-coding RNAs transcribed from intronic regions can affect transcription efficiency by affecting regulatory elements, such as enhancers, silencers or other DNA structures [30,31]. In human ADIPOQ, intron 2 variation (rs17366743: c.214+276A/C) has been linked with promoter polymorphisms in the gene, including c.-11426A/G, c.-11377C/G and c.-11391G/A, and is associated with ADIPOQ levels in diabetes and obesity [20]. In pig, variation in the non-coding regions of ADIPOQ has been associated with production traits, with SNP c.178A/G (g.1735) in porcine intron 2 having a significant effect on shoulder fat thickness [13].
We detected three SNPs in the ovine ADIPOQ 3'-UTR (c.*37T/C, c.*130G/C and c.*434C/A). In humans, at least ten SNPs in the 3'-UTR of ADIPOQ have been described, but most of them are rare. Human SNPs rs1063537 T/C, rs1063539 C/G and rs6773957 A/G are more common and have an association with type 2 diabetes and obesity in a Chinese population [20,32]. In goats, two nucleotide substitutions (c.*88T/A and c.*210A/G) have been described in the 3'-UTR. The relationships between these two SNPs and body weight at different ages, has been analysed, but no associations were found [33].
In this work, different frequencies were found for ovine ADIPOQ variants in different breeds. The different origins of these breeds may be reflected in the variant frequencies found in ADIPOQ. The majority of NZ sheep breeds are derived from European breeding stock. For example, the difference in variant frequencies in Romney and Perendale breeds in this study was small. This is probably because the Perendale breed was developed from a Romney-Cheviot cross. However, although the Corriedale sheep was developed by crossing Lincoln rams with Merino ewes, the variant frequencies in the Corriedale and Merino sheep are different. This may be a consequence of the foundation sheep for the Corriedale breed having different variant frequencies than the base Merino population.
Haplotypes can span a large region of the gene and help increase the Polymorphism Information Content (PIC). In this context, A1 and B1 were the most common variants (55.70% and 42.09%) in region-1; A2 was the most common (72.78%) in region-2; and A3 and C3 were the most common variants (49.05% and 46.52%) in region-3. This prompted us to undertake further analysis of region-1 and region-3. Nine haplotypes were observed spanning the two regions. Only A1-A3, A1-C3, B1-A3 and B1-C3 were common. Due to the promoter (region-1) and intron 2-exon 3 region (region-3) of ADIPOQ playing an important role in regulating ADIPOQ function in humans, these regions and haplotypes may be worthy of further investigation in sheep. More haplotypes may also be found when more of the gene is analysed in larger numbers of sheep.
In summary, the work presented in this study suggests that ovine ADIPOQ has high levels of polymorphism in sheep, and it could be speculated that even more new variants will be found when more samples and different breeds are analysed. Given the apparently important role of ADIPOQ in metabolism, we can also speculate that some of the genetic variation described in this study may be associated with sheep carcass traits. However, because only a small number of sheep were investigated here a much larger association study will be required.
5. Conclusions
This study used PCR-SSCP to screen for variation in ovine ADIPOQ and identified a total of 19 SNPs. The majority of these were found in non-coding regions. There were nine haplotypes of ADIPOQ identified and the frequencies of these haplotypes varied in different breeds of sheep. The effect of this variation in ovine ADIPOQ on sheep production traits warrants investigation.
Acknowledgments
This study was financially supported by the International S & T Cooperation Program of China (Grant No. 2011DFG33310), the Funds for Creative Research Groups of Gansu Province (Grant No. 1210RJIA005), the International S & T Cooperation Program of Gansu Province (Grant No. 1304WCGA178) and Lincoln University Gene-Marker Laboratory.
Author Contributions
Qing-Ming An, Hui-Tong Zhou, Yu-Zhu Luo and Jon G. H. Hickford conceived and designed the project. Qing-Ming An and Hui-Tong Zhou performed the experiments. Qing-Ming An and Hui-Tong Zhou analyzed the data. Qing-Ming An, Hui-Tong Zhou, Jiang Hu, Yu-Zhu Luo and Jon G. H. Hickford wrote the manuscript. All authors reviewed and commented on the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
References
- 1.Yokota T., Meka C.S.R., Medina K.L., Igarashi H., Comp P.C., Takahashi M., Nishida M., Oritani K., Miyagawa J.-I., Funahashi T., et al. Paracrine regulation of fat cell formation in bone marrow cultures via adiponectin and prostaglandins. J. Clin. Invest. 2002;109:1303–1310. doi: 10.1172/JCI0214506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadowaki T., Yamauchi T. Adiponectin and adiponectin receptors. Endocr. Rev. 2005;26:439–451. doi: 10.1210/er.2005-0005. [DOI] [PubMed] [Google Scholar]
- 3.Morsci N.S., Schnabel R.D., Taylor J.F. Association analysis of adiponectin and somatostatin polymorphisms on BTA1 with growth and carcass traits in Angus cattle. Anim. Genet. 2006;37:554–562. doi: 10.1111/j.1365-2052.2006.01528.x. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y., Zhang Y.H., Jiang H., Xiao S.Q., Wang S., Ma Q., Sun G.J., Li F.J., Deng Q., Dai L.S., et al. Detection of differentially expressed genes in the longissimus dorsi of Northeastern Indigenous and Large White pigs. Genet. Mol. Res. 2011;10:779–791. doi: 10.4238/vol10-2gmr1170. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi M., Arita Y., Yamagata K., Matsukawa Y., Okutomi K., Horie M., Shimomura I., Hotta K., Kuriyama H., Kihara S., et al. Genomic structure and mutations in adipose-specific gene, adiponectin. Int. J. Obes. Relat. Metab. Disord. 2000;24:861–868. doi: 10.1038/sj.ijo.0801244. [DOI] [PubMed] [Google Scholar]
- 6.Scherer P.E., Williams S., Fogliano M., Baldini G., Lodish H.F. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 7.Hsueh W.-C., Jean P.L., St., Mitchell B.D., Pollin T.I., Knowler W.C., Ehm M.G., Bell C.J., Sakul H., Wagner M.J., Burns D.K., et al. Genome-wide and fine-mapping linkage studies of type 2 diabetes and glucose traits in the Old Order Amish: Evidence for a new diabetes locus on chromosome 14q11 and confirmation of a locus on chromosome 1q21-q24. Diabetes. 2003;52:550–557. doi: 10.2337/diabetes.52.2.550. [DOI] [PubMed] [Google Scholar]
- 8.Chu H., Wang M., Zhong D., Shi D., Ma L., Tong N., Zhang Z. ADIPOQ polymorphisms are associated with type 2 diabetes mellitus: A meta‐analysis study. Diabetes Metab. Res. Rev. 2013;29:532–545. doi: 10.1002/dmrr.2424. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y., Zhang F., Ding R., Skrip L., Wang Y., Lei H., Hu D. ADIPOQ gene polymorphisms and cancer risk: A meta-analysis. Cytokine. 2013;61:565–571. doi: 10.1016/j.cyto.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Ramya K., Ayyappa K.A., Ghosh S., Mohan V., Radha V. Genetic association of ADIPOQ gene variants with type 2 diabetes, obesity and serum adiponectin levels in south Indian population. Gene. 2013;532:253–262. doi: 10.1016/j.gene.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y., Jiang H., Kuang J., Hou X., Feng Y., Su Z. Genetic variations in ADIPOQ gene are associated with chronic obstructive pulmonary disease. PLoS ONE. 2012;7:e50848. doi: 10.1371/journal.pone.0050848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X., Du Y., Zhang C., Shi X., Chen D., Sun J., Jin Q., Lan X., Chen H. Polymorphism in a microsatellite of the acrp30 gene and its association with growth traits in goats. Biochem. Genet. 2011;49:533–539. doi: 10.1007/s10528-011-9428-6. [DOI] [PubMed] [Google Scholar]
- 13.Dai L.H., Xiong Y.Z., Deng C.Y., Jiang S.W., Zuo B., Zheng R., Li F.E., Lei M.G. Association of the AG polymorphism in porcine adiponectin gene with fat deposition and carcass traits. Asian-Australas. J. Anim. Sci. 2006;19:779–783. doi: 10.5713/ajas.2006.779. [DOI] [Google Scholar]
- 14.Houde A.A., Murphy B.D., Mathieu O., Bordignon V., Palin M.F. Characterization of swine adiponectin and adiponectin receptor polymorphisms and their association with reproductive traits. Anim. Genet. 2008;39:249–257. doi: 10.1111/j.1365-2052.2008.01714.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H., Hickford J.G.H., Fang Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006;354:159–161. doi: 10.1016/j.ab.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Byun S.O., Fang Q., Zhou H., Hickford J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009;385:174–175. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Gong H., Zhou H., Hickford J.G.H. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011;38:31–35. doi: 10.1007/s11033-010-0074-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H., Li S., Liu X., Wang J., Luo Y., Hickford J.G.H. Haplotyping using a combination of polymerase chain reaction-single-strand conformational polymorphism analysis and haplotype-specific PCR amplification. Anal. Biochem. 2014;466:59–64. doi: 10.1016/j.ab.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Yang H., Ye E., Si G., Chen L., Cai L., Ye C., Zhang C., Lu X. Adiponectin gene polymorphism rs2241766 T/G is associated with response to pioglitazone treatment in type 2 diabetic patients from Southern China. PLoS ONE. 2014;9:e112480. doi: 10.1371/journal.pone.0112480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H.F. Biomarkers of adiponectin: Plasma protein variation and genomic DNA polymorphisms. Biomarker Insights. 2009;4:123–133. doi: 10.4137/bmi.s3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barth N., Langmann T., Schölmerich J., Schmitz G., Schäffler A. Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: Role of SP1/SP3 and TNF-α as regulatory pathways. Diabetologia. 2002;45:1425–1433. doi: 10.1007/s00125-002-0895-5. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D., Ma J., Brismar K., Efendic S., Gu H.F. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J. Diabetes Complications. 2009;23:265–272. doi: 10.1016/j.jdiacomp.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Chung H.F., Long K.Z., Hsu C.-C., al Mamun A., Chiu Y.-F., Tu H.-P., Chen P.-S., Jhang H.-R., Hwang S.-J., Huang M.-C. Adiponectin gene (ADIPOQ) polymorphisms correlate with the progression of nephropathy in Taiwanese male patients with type 2 diabetes. Diabetes Res. Clin. Pract. 2014;105:261–270. doi: 10.1016/j.diabres.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Liu G., Jennen D.G.J., Tholen E., Juengst H., Kleinwächter T., Hölker M., Tesfaye D., Ün G., Schreinemachers H.-J., Murani E., et al. A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc‐Pietrain resource population. Anim. Genet. 2007;38:241–252. doi: 10.1111/j.1365-2052.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 25.Guo T., Ren J., Yang K., Ma J., Zhang Z., Huang L. Quantitative trait loci for fatty acid composition in longissimus dorsi and abdominal fat: Results from a White Duroc × Erhualian intercross F2 population. Anim. Genet. 2009;40:185–191. doi: 10.1111/j.1365-2052.2008.01819.x. [DOI] [PubMed] [Google Scholar]
- 26.Cieslak J., Flisikowska T., Schnieke A., Kind A., Szydlowski M., Switonski M., Flisikowski K. Polymorphisms in the promoter region of the adiponectin (ADIPOQ) gene are presumably associated with transcription level and carcass traits in pigs. Anim. Genet. 2013;44:340–343. doi: 10.1111/j.1365-2052.2012.02397.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Yang M., Li C., Xu Y., Sun J., Lei C., Lan X., Zhang C., Chen H. Identification and genetic effect of a variable duplication in the promoter region of the cattle ADIPOQ gene. Anim. Genet. 2014;45:171–179. doi: 10.1111/age.12112. [DOI] [PubMed] [Google Scholar]
- 28.Kim J.B., Kim D.J., Lee J.K., Lee C.Y. Genetic relationship between carcass traits and carcass price of Korean cattle. Asian-Australas. J. Anim. Sci. 2010;23:848–854. doi: 10.5713/ajas.2010.90555. [DOI] [Google Scholar]
- 29.Shin S., Chung E. Novel SNPs in the bovine ADIPOQ and PPARGC1A genes are associated with carcass traits in Hanwoo (Korean cattle) Mol. Biol. Rep. 2013;40:4651–4660. doi: 10.1007/s11033-013-2560-0. [DOI] [PubMed] [Google Scholar]
- 30.Chorev M., Carmel L. The function of introns. Front. Genet. 2012 doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaunitz F., Heise K., Gebhardt R. A silencer element in the first intron of the glutamine synthetase gene represses induction by glucocorticoids. Mol. Endocrinol. 2004;18:63–69. doi: 10.1210/me.2003-0062. [DOI] [PubMed] [Google Scholar]
- 32.Jiang B., Liu Y., Liu Y., Fang F., Wang X., Li B. Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Mol. Biol. Rep. 2014;41:925–933. doi: 10.1007/s11033-013-2937-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan X.Y., Liu J.B., Zhu J.L., Liu T.G., Zhang L.Z., Zhang Y., Lei C.Z., Chen H. Identification of a novel mutation within the goat adiponectin gene and its effect on body weight in Chinese indigenous breeds. Biochem. Genet. 2012;50:94–102. doi: 10.1007/s10528-011-9474-0. [DOI] [PubMed] [Google Scholar]