Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) remains a highly chemoresistant tumor entity for which no reliable molecular targets exist to predict or influence the success of chemotherapy. Recently, we identified a panel of microRNAs associated with induced gemcitabine chemoresistance in human PDAC cell lines. This clinical study evaluates these microRNAs and associated molecular markers as prognostic markers of outcome in 98 PDAC patients Union Internationale Contre le Cancer (UICC) stage II undergoing curative surgery with adjuvant gemcitabine chemotherapy. The primary end points of this study are recurrence-free survival and overall survival.
Results
Poor response to chemotherapy was significantly correlated to overexpression of microRNA-21 (p = 0.029), microRNA-99a (p = 0.037), microRNA-100 (p = 0.028), and microRNA-210 (p = 0.021) in tissue samples of PDAC patients UICC stage II. Upregulation of these microRNAs was associated with a significantly shorter overall survival and recurrence-free survival (p < 0.05). Overexpression of phosphatase and tensin homolog (PTEN) (p = 0.039) and low expression of multidrug resistance (MDR)-1 (p = 0.043) and breast cancer resistance protein (BCRP)-1 (p = 0.038) were significantly correlated to improved response to adjuvant chemotherapy. Adjuvant gemcitabine treatment (p < 0.0001) and low tumor grading (p = 0.047) were correlated to better outcome. MicroRNA-100, microRNA-21, and its targets PTEN and MDR-1 were independent factors of survival in multivariate analysis.
Conclusions
Multivariate survival analyses identified microRNA-21 and microRNA-100 as unfavorable prognostic factors in resected and adjuvant treated PDAC UICC stage II patients.
Keywords: Pancreatic ductal adenocarcinoma, Chemoresistance, microRNA
Background
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most deadly cancers in western countries with a median survival below 6 months and a dismal 5-year overall survival rate of less than 5 % [1]. Despite developments in novel diagnostic and surgical techniques over the past decades, only morbidity and postoperative mortality were improved, without significant impact on survival. Lack of early symptoms combined with rapid disease progression result in low resectability rates of about 15 % of PDAC patients with 5-year survival rates below 20 % [2].
Adjuvant first-line chemotherapy and palliative treatment by gemcitabine have slightly improved the clinical outcome. However, nearly half of the treated patients do not benefit from gemcitabine-based therapies [3, 4]. Chemotherapy response markers are needed to identify PDAC patients who may benefit from adjuvant gemcitabine chemotherapy.
PDAC chemoresistance is acquired through multiple molecular pathways and genetic alterations affecting cell cycle, apoptosis, and intracellular drug accumulation. Especially, members of the ATP binding cassette (ABC) superfamily including multidrug resistance (MDR)-1 P-glycoprotein, multidrug resistance protein (MRP)-1, and breast cancer resistance protein (BCRP)-1 have been shown to mediate resistance against several anticancer drugs by enhanced drug efflux across cellular plasma membranes [5]. Deoxycytidine kinase (DCK), a key activator of gemcitabine, and its regulator Hu antigen R (HuR) have been identified as powerful independent prognostic factors for PDAC patients undergoing adjuvant gemcitabine therapy [6–8]. Further, mutations and epigenetic downregulation of phosphatase and tensin homolog (PTEN), a tumor suppressor and the most important negative regulator of Akt/PI3K signaling, are regarded as crucial for PDAC development and chemoresistance [9].
Although a number of molecular players have been connected to chemoresistance, their clinical impact remains controversial [10].
We previously identified a panel of microRNAs associated with induced gemcitabine chemoresistance in human PDAC cell lines: Two PDAC cell lines were treated by repeated pulsatile gemcitabine treatment to induce acquired chemoresistance. Global microRNA profiling identified a panel of 16 significantly and concordantly dysregulated microRNAs in both chemoresistant PDAC cell clones [11].
In this clinical study, our aim was to validate these microRNA candidates as prognostic markers of chemotherapy response to gemcitabine in patients with resected PDAC stage II according to the Union Internationale Contre le Cancer (UICC).
Results
Clinicopathologic factors and chemotherapy response as survival markers
A total of 91 PDAC patients UICC stage II with curative pancreatic head (82.4 %) or left resection (17.6 %) and 13 patients with benign, noninflammatory pathology were analyzed (Table 1). Seven of initially 98 PDAC patients with an in-hospital-mortality of ≤30 days were excluded. The median age at diagnosis was 66 years, and the male to female ratio was approximately 1.2:1. The distribution by stage was 31.9 % UICC stage IIa and 68.2 % Stage IIb. The majority of patients (65; 71.4 %) had died by the time of final analysis with a median follow-up time of 30.5 months for the 26 patients still alive. The median survival time for all patients was 22 months.
Table 1.
Clinicopathologic characteristics of PDAC stage II patients
| patients n | Median overall survival (months) | 95 % CI | p | |
|---|---|---|---|---|
| Total | 91 | 21 | 16–26 | |
| Age (years) | ||||
| ≤65 | 41 | 27 | 21–33 | 0.1754 |
| >65 | 50 | 19 | 16–22 | |
| Gender | ||||
| Female | 41 | 26 | 18–34 | 0.8796 |
| Male | 50 | 20 | 15–25 | |
| UICC stage | ||||
| IIa | 29 | 27 | 13–40 | 0.3347 |
| IIb | 62 | 20 | 16–24 | |
| Grading | ||||
| G1 and G2 | 55 | 26 | 1–31 | 0.0474 |
| G3 | 36 | 15 | 8–22 | |
| Perineural invasion | ||||
| Pn0 | 32 | 19 | 9–29 | 0.7835 |
| Pn1 | 59 | 21 | 16–26 | |
| Vene invasion | ||||
| V0 | 74 | 23 | 16–30 | 0.5583 |
| V1 | 17 | 21 | 17–25 | |
| Lymphatic invasion | ||||
| L0 | 50 | 24 | 18–29 | 0.9821 |
| L1 | 41 | 21 | 15–26 | |
| Resection margin | ||||
| R0 | 77 | 23 | 15–31 | 0.3653 |
| R1 | 24 | 18 | 16–26 | |
| Type of surgery | ||||
| Pancreatic head resection | 75 | 32 | 17–25 | 0.2475 |
| Traverso-Longmire | 66 | |||
| Whipple-Kausch | 9 | |||
| Pancreatic left resection | 16 | 21 | 14–49 | |
| Adjuvant gemcitabine | ||||
| Yes | 64 | 31 | 24–37 | 2.9922E-8 |
| No | 27 | 12 | 9–14 | |
Indication of median overall survival in months and 95 % confidence interval (CI). p < 0.05 indicates significance
Univariate analyses indicated that tumor grading (p = 0.0474) correlated with worse overall survival. Patients who underwent adjuvant gemcitabine chemotherapy had significantly improved overall survival rates (p = 2.9922E-8) (Table 1). Patients with adjuvant gemcitabine treatment and good response (determined by median split) showed improved median overall and recurrence-free survival vs. bad response (33 months, 95 % confidence interval (CI) 26–49 vs. 19 months, 95 % CI 15–13, p = 1.3628E-09 and 32 months, 95 % CI 25–46 vs. 11 months, 95 % CI 8–12, p = 2.8043E-15) and vs. no adjuvant treatment (12 months, 95 % CI 8–17, p = 9.0791E-13 and 7 months, 95 % CI 5–9, p = 1.565E-15). Improved overall survival was even shown in patients with poor chemotherapy response vs. no adjuvant treatment (p = 0.0093) (Fig. 1). Subgroup analysis of response to chemotherapy groups (group II vs. III) revealed no significant differences in the distribution of clinicopathologic factors (Table 2).
Fig. 1.
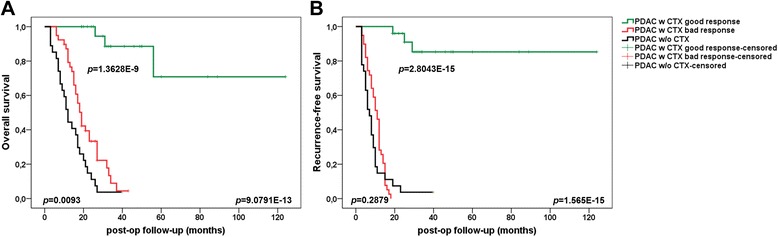
Prognostic impact of adjuvant gemcitabine treatment in PDAC UICC stage II patients. Improved overall (a) and recurrence-free survival (b) in patients with adjuvant gemcitabine treatment and good (n = 25) vs. bad (n = 39) response (p = 1.3628E-09; p = 2.8043E-15) and vs. no adjuvant gemcitabine treatment (n = 27) (p = 9.0791E-13; p = 1.565E-15). Improved overall survival in patients with bad chemotherapy response vs. no adjuvant treatment (p = 0.0093)
Table 2.
Clinicopathologic characteristics of PDAC patients UICC stage II with different adjuvant chemotherapy response
| Total | PDAC II w/o CTX | PDAC II w CTX | P | P | ||
|---|---|---|---|---|---|---|
| Good response | Bad response | |||||
| (n = 91) | I (n = 27) | II (n = 25) | III (n = 39) | II vs. III | ||
| Age (years) | ||||||
| ≤65 | 41 | 10 | 12 | 19 | 0.5092 | 1 |
| median (range) | 63 (54–65) | 56 (41–64) | 53 (31–65) | |||
| >65 | 50 | 17 | 13 | 20 | ||
| median (range) | 72 (66–84) | 73 (66–84) | 71 (67–79) | |||
| Gender | ||||||
| Female | 41 | 14 | 11 | 16 | 0.6801 | 1 |
| Male | 50 | 13 | 14 | 23 | ||
| UICC stage | ||||||
| IIa | 29 | 7 | 9 | 13 | 0.6469 | 1 |
| IIb | 62 | 20 | 16 | 23 | ||
| Grading | ||||||
| G1 | 5 | 0 | 3 | 2 | 0.0231 | 0.5008 |
| G2 | 50 | 10 | 16 | 24 | ||
| G3 | 36 | 17 | 6 | 13 | ||
| Perineural invasion | ||||||
| Pn0 | 32 | 10 | 10 | 12 | 0.7305 | 0.5904 |
| Pn1 | 59 | 17 | 15 | 27 | ||
| Vene invasion | ||||||
| V0 | 74 | 23 | 22 | 29 | 0.3256 | 0.2204 |
| V1 | 17 | 4 | 3 | 10 | ||
| Lymphatic invasion | ||||||
| L0 | 50 | 17 | 15 | 18 | 0.3367 | 0.3149 |
| L1 | 41 | 10 | 10 | 21 | ||
| Resection margin | ||||||
| R0 | 67 | 20 | 21 | 26 | 0.3071 | 0.1553 |
| R1 | 24 | 7 | 4 | 13 | ||
| Type of surgery | ||||||
| Pancreatic head resection | 75 | 26 | 17 | 32 | 0.0276 | 0.2347 |
| Traverso-Longmire | 66 | 22 | 15 | 29 | ||
| Whipple-Kausch | 9 | 4 | 2 | 3 | ||
| Pancreatic left resection | 16 | 1 | 8 | 7 | ||
| Adjuvant gemcitabine | ||||||
| Yes | 64 | 0 | 25 | 39 | 0.0000 | 1 |
| No | 27 | 27 | 0 | 0 | ||
| Tumor recurrence | ||||||
| ≤18 months | 63 | 24 | 0 | 39 | 0.0000 | 0.0001 |
| >18 months | 5 | 2 | 3 | 0 | ||
| Tumor related death | ||||||
| ≤18 months | 39 | 20 | 0 | 19 | 0.0234 | 0.0856 |
| >18 months | 22 | 6 | 3 | 13 | ||
p < 0.05 indicates significance
MicroRNAs as markers of chemotherapy response and survival
Macrodissected PDAC tissue of patients with poor (n = 39) and good chemotherapy response (n = 25) was evaluated by quantitative real-time PCR (qRT‐PCR) for the expression of in vitro chemoresistance-specific microRNAs (Fig. 2, Table 3). MicroRNA-21 (p = 4.3817E-07), microRNA-31 (p = 0.0003), microRNA-99a (p = 0.0406), microRNA-100 (p = 4.0492E-08), microRNA-125b (p = 0.0001), microRNA-138 (p = 0.0301), microRNA-147b (p = 0.0028), and microRNA-210 expression (p = 0.0044) were significantly upregulated in PDAC stage II vs. benign controls. Further, microRNA-148 was significantly downregulated in PDAC tissues (p = 3.9814E-09).
Fig. 2.
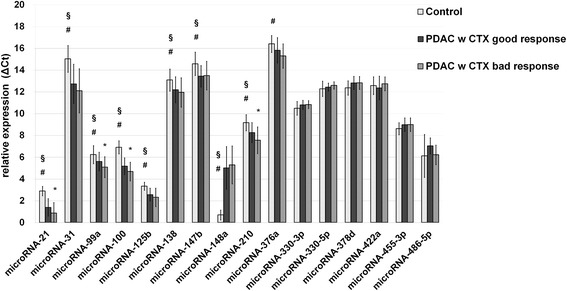
ΔCT expression level (means with standard deviation) of in vitro chemoresistant-specific microRNAs in benign and cancerous human pancreatic specimen. ΔCt levels are inversely proportional to the amount of target microRNA in the sample. Asterisk indicates to a significant difference of p < 0.05 between PDAC patients with good and poor chemotherapy (w CTX) response. Number sign and section sign indicate to a significant difference of p < 0.05 between benign noninflammatory pancreas controls and PDAC with good or bad CTX response, respectively
Table 3.
MicroRNA expression of PDAC patients with good and poor chemotherapy (w CTX) response
| Control vs. PDAC w CTX good response | Control vs. PDAC w CTX bad response | PDAC w CTX good response vs. PDAC w CTX bad response | |
|---|---|---|---|
| microRNA-21 | 2.89 vs. 1.39 (+51.6 %) p = 4.3817E-07 |
2.89 vs. 0.86 (+70.3 %) p = 3.3364E-09 |
1.39 vs. 0.86 (+38.7 %) p = 0.0295 |
| microRNA-31* | 15.02 vs. 12.74 (+15.2 %) p = 0.0003 |
15.02 vs. 12.10 (+19.4 %) p = 1.3354E-05 |
12.74 vs. 12.10 (+5.0 %) p = 0.2232 |
| microRNA-99a | 6.23 vs. 5.61 (+10.0 %) p = 0.0406 |
6.23 vs. 5.09 (+18.4 %) p = 0.0004 |
5.61 vs. 5.09 (+9.3 %) p = 0.0370 |
| microRNA-100 | 6.91 vs. 5.18 (+25.0 %) p = 4.0492E-08 |
6.91 vs. 4.69 (+32.2 %) p = 2.9936E-11 |
5.18 vs. 4.69 (+9.5 %) p = 0.0275 |
| microRNA-125b | 3.34 vs. 2.57 (+22.9 %) p = 0.0001 |
3.34 vs. 2.31 (+30.8 %) p = 0.0001 |
2.57 vs. 2.31 (+10.2 %) p = 0.1968 |
| microRNA-138 | 13.08 vs. 12.19 (+6.8 %) p = 0.0301 |
13.08 vs. 11.95 (+8.7 %) p = 0.0082 |
12.19 vs. 11.95 (+2.0 %) p = 0.4705 |
| microRNA-147b | 14.57 vs. 13.44 (+7.8 %) p = 0.0028 |
14.57 vs. 13.49 (+7.4 %) p = 0.0105 |
13.44 vs. 13.49 (−0.4 %) p = 0.8662 |
| microRNA-148a | 0.69 vs. 5.03 (−622.1 %) p = 3.9814E-09 |
0.69 vs. 5.29 (−660.5 %) p = 2.1259E-12 |
5.03 vs. 5.29 (−5.3 %) p = 0.5821 |
| microRNA-210 | 9.17 vs. 8.26 (+9.9 %) p = 0.0044 |
9.17 vs. 7.56 (+17.6 %) p = 4.9353E-05 |
8.26 vs. 7.56 (+8.5 %) p = 0.0208 |
| microRNA-376a | 16.39 vs. 15.82 (+3.5 %) p = 0.1332 |
16.39 vs. 15.28 (+6.8 %) p = 0.0026 |
15.82 vs. 15.28 (+3.4 %) p = 0.0849 |
Benign, noninflammatory pancreatic specimens of patients with pancreatic cystadenoma or papillary adenoma served as controls. All data are given as median ΔCt level expression differences in percent. p < 0.05 indicates significance
Expression of in vitro dysregulated microRNA-376a, microRNA-330-3p, microRNA-330-5p, microRNA-378d, microRNA-422a, microRNA-455-3p, and microRNA-486-5p did not differ between PDAC and control.
After normalization to benign noninflammatory controls (n = 13) by the ΔΔCt method, poor adjuvant gemcitabine mono-chemotherapy response was significantly related to overexpression of microRNA-21 (p = 0.0366), microRNA-99a (p = 0.0163), microRNA-100 (p = 0.0157), and microRNA-210 (p = 0.0252) (Fig. 3).
Fig. 3.
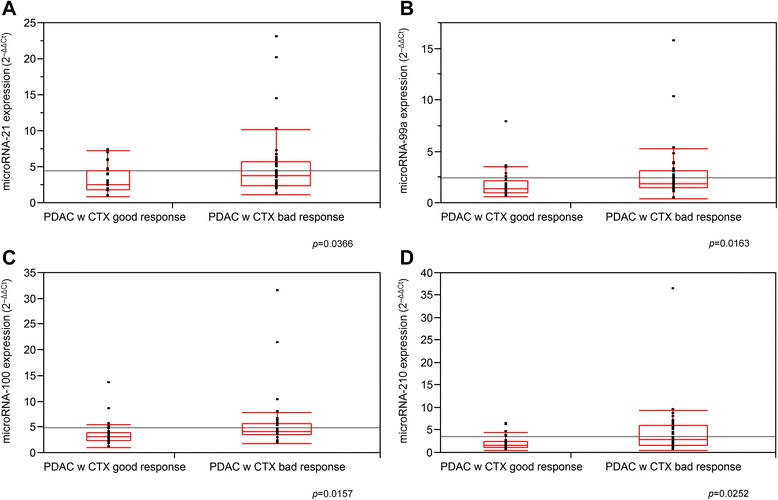
The 2-ΔΔCt expression level of microRNA-21 (a), microRNA-99a (b), microRNA-100 (c), and microRNA-210 (d) in PDAC UICC stage II with good and bad response. p < 0.05 indicates significance
Kaplan-Meier survival analysis revealed significantly improved overall survival and recurrence-free survival rates in PDAC patients with low expression of microRNA-21 (cutoff 4.7; p = 0.0181; p = 0.0149), microRNA-99a (cutoff 2.5; p = 0.0325; p = 0.1711), microRNA-100 (cutoff 5.0; p = 0.0004; p = 0.0111), microRNA-125b (cutoff 1.6; p = 0.0491; p = 0.0373), and microRNA-210 (cutoff 4.6; p = 0.0161; p = 0.0116) in the adjuvant setting (Fig. 4). Univariate Cox regression overall and recurrence-free survival analyses identified microRNA-21 (p = 0.0231; p = 0.0211), microRNA-99a (p = 0.0393; p = 0.1864), microRNA-100 (p = 0.0013; p = 0.0163), microRNA-125b (p = 0.0578; p = 0.0472), and microRNA-210 (p = 0.0211; p = 0.0168) as unfavorable prognostic factors in resected and adjuvant-treated PDAC UICC stage II patients (Table 5). These microRNAs did not correlate significantly with standard clinicopathologic parameters or immunohistological markers (data not shown).
Fig. 4.
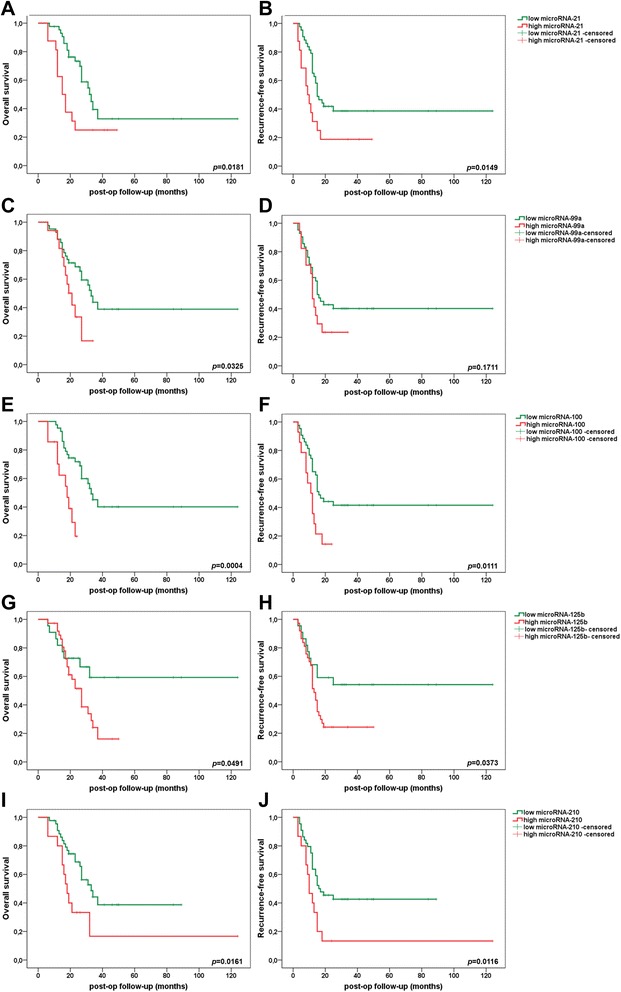
Prognostic impact of microRNA-21 (a, b), microRNA-99a (c, d), microRNA-100 (e, f), microRNA-125b (g, h), and microRNA-210 (i, j) on overall survival (right column) and recurrence-free survival (left column) in PDAC UICC stage II patients. p < 0.05 indicates significance
Table 5.
Univariate Cox regression overall and recurrence-free survival analysis
| Variable | Subset | Overall survival | Recurrence-free survival | ||
|---|---|---|---|---|---|
| Hazard ratio (95 % CI) | p value | Hazard ratio (95 % CI) | p value | ||
| PTEN | low/high | 3.095 (1.5502–6.1805) | 0.0014 | 2.7944 (1.4826–5.2669) | 0.0015 |
| DCK | low/high | 0.5536 (0.2772–1.1057) | 0.0939 | 0.5178 (0.2813–0.9532) | 0.0345 |
| HuR | low/high | 0.6799 (0.3448–1.3409) | 0.2656 | 0.6569 (0.3559–1.2125) | 0.1789 |
| MDR-1 | low/high | 2.2213 (1.1291–4.3698) | 0.0208 | 2.5516 (1.3799–4.7179) | 0.0028 |
| MRP-1 | low/high | 2.0104 (0.9778–4.1331) | 0.0576 | 2.5762 (1.3334–4.9773) | 0.0049 |
| BCRP-1 | low/high | 2.7438 (1.3117–5.7395) | 0.0074 | 3.9656 (1.9563–8.0385) | 0.0001 |
| VEGF-1 | low/high | 1.5977 (0.6059–4.2123) | 0.3435 | 1.3889 (0.6157–3.1331) | 0.4286 |
| N | N0/N1 | 1.2670 (0.6269–2.5605) | 0.5096 | 1.16965 (0.6213–2.2018) | 0.6273 |
| G | G3/G1-2 | 0.9045 (0.4425–1.8491) | 0.7833 | 0.7767 (0.4036–1.4946) | 0.4492 |
| L | L0/L1 | 1.2793 (0.6478–2.5264) | 0.4781 | 1.1996 (0.6541–2.1998) | 0.5565 |
| V | V0/V1 | 1.4841 (0.6944–3.1716) | 0.3083 | 1.7039 (0.8545–3.3975) | 0.1301 |
| Pn | Pn0/Pn1 | 1.3512 (0.6629–2.7538) | 0.4073 | 1.2389 (0.6426–2.3889) | 0.5224 |
| R | 0/1 | 1.3996 (0.6839–2.8644) | 0.3576 | 1.8921 (0.9923–3.6081) | 0.0528 |
| gender | f/m | 0.9404 (0.4784–1.8487) | 0.8586 | 1.0039 (0.5444–1.8513) | 0.9899 |
| age | >65/≤65 | 0.7334 (0.3747–1.4354) | 0.3655 | 0.7598 (0.4142–1.3938) | 0.3749 |
| operation | DP/PD | 1.1089 (0.5034–2.4431) | 0.7974 | 1.2319 (0.5889–2.5774) | 0.5796 |
| CTX response | good/bad | 23.3621 (5.5057–99.1321) | 0.00002 | 159.7685 (11.1902–2281.0912) | 0.0002 |
| microRNA-21 | low/high | (2.3127) (1.1218–4.7675) | 0.0231 | 2.2006 (1.1256–4.3022) | 0.0211 |
| microRNA-31* | low/high | 1.4769 (0.7365–2.9614) | 0.2719 | 1.7738 (0.9362–3.3606) | 0.0788 |
| microRNA-99 | low/high | 2.2291 (1.0399–4.7781) | 0.0393 | 1.5775 (0.8023–3.1019) | 0.1864 |
| microRNA-100 | low/high | 4.0097 (1.7189–9.3536) | 0.0013 | 2.3653 (1.1714–4.7762) | 0.0163 |
| microRNA-125b | low/high | 2.1879 (0.9745–4.9127) | 0.0578 | 2.0935 (1.0093–43426) | 0.0472 |
| microRNA-138 | low/high | 1.4770 (0.6583–3.3141) | 0.3442 | 1.3268 (0.6556–2.6854) | 0.4318 |
| microRNA-147b | low/high | 0.8217 (0.4094–1.6489) | 0.5805 | 0.7387 (0.3891–1.4022) | 0.3543 |
| microRNA-148a | low/high | 0.9135 (0.4533–1.8411) | 0.8003 | 1.1714 (0.6249–2.1957) | 0.6217 |
| microRNA-210 | low/high | 2.4021 (1.1408–5.0578) | 0.0211 | 2.2881 (1.1611–4.5091) | 0.0168 |
| microRNA-376a | low/high | 1.9456 (0.9518–3.9771) | 0.0681 | 1.7029 (0.8906–3.2563) | 0.1075 |
Indication of hazard ratio and 95 % confidence interval (CI). p < 0.05 indicates significance
Classic immunohistological PDAC chemoresistance und survival markers
Corresponding formalin-fixed paraffin-embedded (FFPE) sections of our study groups with 91 patients with PDAC UICC stage II and 13 patients with pancreatic cystadenoma or papillary adenoma (total n = 104) were evaluated immunohistochemically by tissue microarray for the expression of the tumor suppressor PTEN, the gemcitabine metabolism regulators DCK and HuR, and the ATP transporters MDR-1, MRP-1, and BCRP-1, as well as the tumor angiogenesis marker vascular epithelial growth factor (VEGF)-1. MDR-1 (p = 0.0426) and BCRP-1 (p = 0.0378) overexpression as well as PTEN (p = 0.0394) underexpression correlated significantly with poor chemotherapy response in PDAC patients. VEGF-1 was significantly downregulated in PDAC tissue vs. benign controls (p < 0.0001) (Fig. 5, Table 4).
Fig. 5.
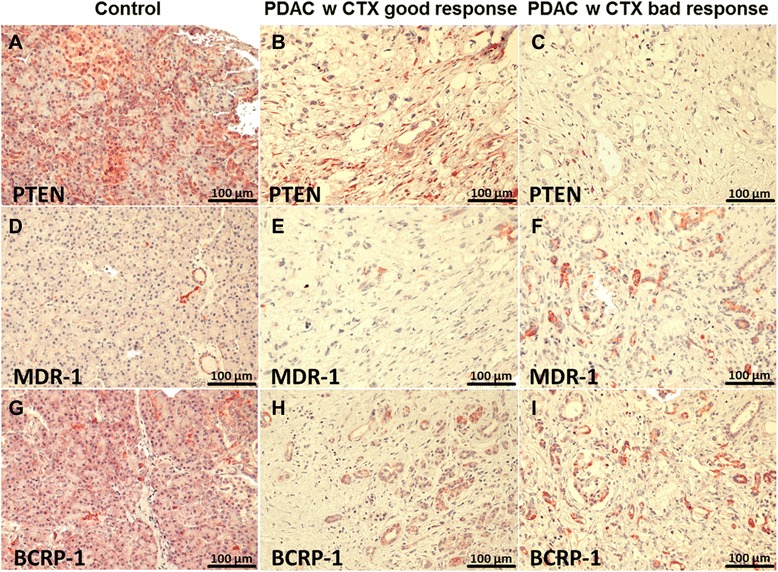
Immunohistochemical staining of PTEN, MDR-1, und BCRP-1 in benigne pancreas (a, d, g), PDAC UICC stage II with good (b, e, h) and poor chemotherapy response (c, f, i)
Table 4.
Immunohistochemical staining density of different study groups
| Antibody | Control (I) | PDAC w CTX | PDAC w/o CTX | p | |
|---|---|---|---|---|---|
| Good response (II) | Bad response (III) | ||||
| PTEN | |||||
| +4 | 3 | 1 | 0 | 0 | I vs. II: 0.0901 |
| +3 | 6 | 7 | 6 | 0 | I vs. III: 0.0003 |
| +2 | 4 | 14 | 16 | 12 | II vs. III: 0.0394 |
| +1 | 0 | 3 | 17 | 15 | |
| DCK | |||||
| +4 | 7 | 10 | 6 | 1 | I vs. II: 0.7549 |
| +3 | 4 | 8 | 13 | 16 | I vs. III: 0.0287 |
| +2 | 2 | 6 | 15 | 9 | II vs. III: 0.1147 |
| +1 | 0 | 1 | 5 | 1 | |
| HuR | |||||
| +4 | 0 | 0 | 0 | 0 | I vs. II: 0.0687 |
| +3 | 5 | 2 | 3 | 1 | I vs. III: 0.0137 |
| +2 | 5 | 13 | 13 | 8 | II vs. III: 0.3054 |
| +1 | 3 | 10 | 23 | 18 | |
| MDR-1 | |||||
| +4 | 0 | 0 | 0 | 0 | I vs. II: 0.7427 |
| +3 | 0 | 1 | 7 | 1 | I vs. III: 0.1068 |
| +2 | 2 | 3 | 11 | 5 | II vs. III: 0.0426 |
| +1 | 11 | 21 | 21 | 21 | |
| MRP-1 | |||||
| +4 | 9 | 1 | 4 | 1 | I vs. II: 0.00001 |
| +3 | 4 | 3 | 12 | 3 | I vs. III: 0.0001 |
| +2 | 0 | 6 | 11 | 10 | II vs. III: 0.1005 |
| +1 | 0 | 15 | 12 | 13 | |
| BCRP-1 | |||||
| +4 | 2 | 0 | 0 | 0 | I vs. II: 0.00003 |
| +3 | 8 | 1 | 7 | 2 | I vs. III: 0.0005 |
| +2 | 3 | 12 | 24 | 19 | II vs. III: 0.0378 |
| +1 | 0 | 12 | 8 | 6 | |
| VEGF-1 | |||||
| +4 | 10 | 0 | 1 | 0 | I vs. II: <0.0001 |
| +3 | 3 | 4 | 6 | 2 | I vs. III: <0.0001 |
| +2 | 0 | 14 | 23 | 12 | II vs. III: 0.8456 |
| +1 | 0 | 7 | 9 | 13 | |
p < 0.05 indicates significance
Kaplan-Meier survival analysis revealed significantly improved overall survival and recurrence-free survival rates in PDAC patients with high expression of PTEN (p = 0.0007 each) and low expression of MDR-1 (p = 0.0163; p = 0.0015) and BCRP-1 (p = 0.0047; p = 0.00003) in the adjuvant setting (Fig. 6). Univariate Cox regression overall and recurrence-free survival analyses identified PTEN (p = 0.0014; p = 0.0015) and DCK (p = 0.0939; p = 0.0345) as favorable prognostic factors as well as MDR-1 (p = 0.0208; p = 0.0028), BCRP-1 (p = 0.0074; p = 0.0001), and MRP-1 (p = 0.0576; p = 0.0049) as unfavorable prognostic factors in resected and adjuvant-treated PDAC UICC stage II patients (Table 5).
Fig. 6.
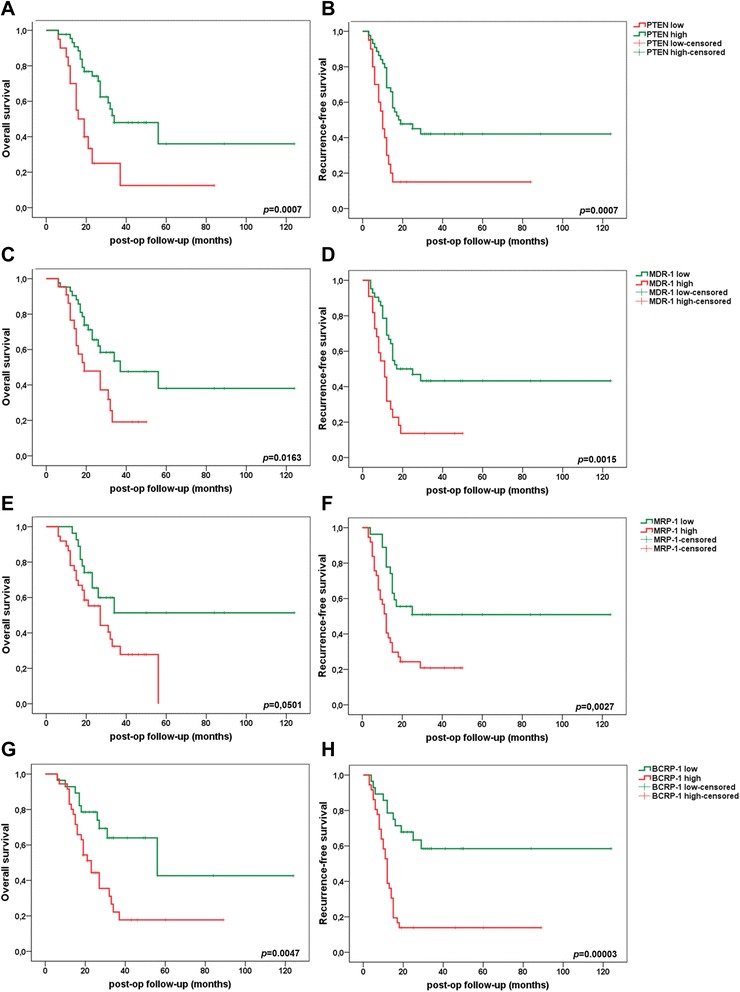
Prognostic impact of PTEN (a, b), MDR-1 (c, d), MRP-1 (e, f), and BCRP-1 (g, h) on overall survival (right column) and recurrence-free survival (left column) in PDAC UICC stage II patients. p < 0.05 indicates significance
Identification of independent prognostic factors by multivariate analysis
Multivariate Cox regression overall and recurrence-free survival analyses identified microRNA-21 (p = 0.0064; p = 0.0338), microRNA-100 (p = 0.0045; p = 0.1683), and MDR-1 (p = 0.0003; p = 0.0021) as unfavorable prognostic factors while PTEN (p = 0.1008; p = 0.0211) was a favorable prognostic factor in resected and adjuvantly treated PDAC UICC stage II patients (Table 6). As combinations of microRNAs are likely to provide more accurate prognostic information, the most accurate subset of variables (microRNA-21, -31*, -99, -100, -125b, -138, -147b, -148a, -210, and -376a) was identified using multivariate survival analysis with a forward stepwise variable selection procedure based on the likelihood ratio. Co-expression of microRNA-21 and microRNA-100 as remaining independent prognostic factors results in a hazard ratio of 2.620 (95 % CI 1,251-5,487) and 3.543 (95 % CI 1.504-8.345), respectively.
Table 6.
Multivariate Cox regression overall and recurrence-free survival analysis
| Variable | Subset | Overall survival | Recurrence-free survival | ||
|---|---|---|---|---|---|
| Hazard ratio (95 % CI) | p value | Hazard ratio (95 % CI) | p value | ||
| PTEN | low/high | 0.6076 (0.3351–1.1017) | 0.1008 | 0.5431 (0.3232–0.9125) | 0.0211 |
| MDR-1 | low/high | 3.0434 (1.6602–5.5790) | 0.0003 | 2.1309 (1.3164–3.4494) | 0.0021 |
| BCRP-1 | low/high | 0.7729 (0.3924–1.5225) | 0.4565 | 1.3209 (0.7463–2.3378) | 0.3394 |
| microRNA-21 | low/high | 3.0649 (1.3709–6.8522) | 0.0064 | 2.2521 (1.0640–4.7666) | 0.0338 |
| microRNA-100 | low/high | 4.8450 (1.6306–14.3961) | 0.0045 | 1.8714 (0.7673–4.5643) | 0.1683 |
| microRNA-210 | low/high | 1.2116 (0.4849–3.0270) | 0.6812 | 1.7623 (0.7634–4.0681) | 0.1843 |
Indication of hazard ratio and 95 % confidence interval (CI). p < 0.05 indicates significance
Discussion
As pancreatic cancer surgery and perioperative treatment have significantly improved over the past decades, today, resistance to chemotherapy and chemotoxicity remain as important aspects of treatment failure in resectable PDAC patients.
The high mortality of locally advanced and metastatic PDAC could be reduced by increasing susceptibility to adjuvant gemcitabine chemotherapy. Currently, no tissue biomarkers are available to guide therapeutic strategies or predict prognosis and chemotherapy response in PDAC patients.
Several prognostic clinical factors of survival have been identified in PDAC patients [12, 13]. In accordance with the literature, low tumor grading and adjuvant gemcitabine chemotherapy were correlated to improved survival in PDAC UICC stage II patients in this study. However, with the exception of adjuvant gemcitabine treatment, none of the investigated clinicopathological parameters were independent prognostic markers in the adjuvant setting. Instead, we identified microRNA-100 and microRNA-21 with its targets PTEN and MDR-1 as independent prognostic survival and chemotherapy response markers in PDAC UICC stage II patients.
The oncogenic role of microRNA-21 in PDAC patients and its association with poor survival and induction of chemoresistance to gemcitabine is already well known [9, 14]. Increased microRNA-21 expression has been shown to promote chemoresistance through repression of the tumor suppressor PTEN and decreased expression of programmed cell death 4 (PDCD4). Interestingly, decrease in PDCD4 levels in turn increases the expression of apoptosis inhibitors and the multidrug resistance protein MDR-1 [15–17]. In this study, high expression of PTEN and low expression of MDR-1 was accompanied by significantly improved overall and recurrence-free survival as well as chemotherapy response. However, the clinical significance of MDR-1 expression in human PDAC is controversial. Some research groups failed to demonstrate MDR-1 expression in PDAC tissues, whereas other studies showed a common and high MDR-1 expression potentially contributing to the chemoresistant biology of PDAC [5, 18–21]. VEGF has been reported as a tumor angiogenesis marker in PDAC, positively correlating with microRNA-21 expression [9, 22]. However, the significance of VEGF in tumor neoangiogenesis and progression in PDAC is still unclear. Literature on the prognostic value of VEGF in PDAC shows conflicting findings, and phase II–III clinical trials did not reveal any benefit from combined gemcitabine anti-VEGF PDAC treatment [23]. Our data could not confirm a prognostic role of VEGF in PDAC.
We validated several microRNAs in clinical tumor samples that we had previously identified to be dysregulated in vitro [11]. The identified chemoresistance-associated microRNAs 99a, 100, 125b, and 210 are already known as potential oncogenes in PDAC. In smaller clinical studies, microRNA-210 has been reported to be overexpressed in PDAC patients and associated with a worse outcome [24]. microRNA-99a and microRNA-100, two members of the microRNA-99 family, were found to be overexpressed in PDAC tissue compared with normal pancreatic tissue and chronic pancreatitis [25]. Overexpression of microRNA-125b was demonstrated in prostate cancer, glioblastoma, and drug resistance in pediatric leukemia and breast cancer [26–29]. Interestingly, co-overexpression of miR-125b, miR-99a, and miR-100 was reported in different cancers suggests possible co-regulation of these miRs in chemoresistant PDAC as well [30, 31].
In addition to microRNA-21, microRNA-100 remained as a further independant prognostic survival and chemotherapy response marker in multivariate analysis. Further multi-center studies with a larger number of PDAC patients are required to reevaluate the impact of microRNA-99a, microRNA-125b, and microRNA-210 as independant survival predictors.
Conclusions
We have identified a panel of microRNAs that could serve as survival and chemotherapy response markers in PDAC patients UICC stage II. In particular, microRNA-21 and microRNA-100 show promise as molecular markers and key regulators for targeted therapies in chemorefractive PDAC patients.
Methods
Patients and samples
A tissue bank and follow-up database are maintained prospectively by the Department of General and Visceral Surgery and the Comprehensive Cancer Center Muenster, University Hospital Muenster, Muenster, Germany. From these, 98 PDAC UICC stage II tumor specimens having undergone resection between 2003 and 2012 were retrieved. Benign, noninflammatory pancreatic specimens of 13 patients with pancreatic cystadenoma or papillary adenoma served as controls. Tissue samples were immediately fixed in 10 % buffered formalin and then processed into a paraffin-embedded block and stored at room temperature. Sections from each of the 111 specimens were examined by a pathologist and graded histologically. All cancerous specimens showed vital tumor tissue.
Ethical approval for postoperative tissue collection was obtained (Ethics committee, University Muenster, Az: 1IXHai v. 19.9.2001), and all patients provided informed written consent. All patients underwent radical resection and were assigned to duodenopancreatectomy or left pancreatic resection. Patients that received immunosuppression, neoadjuvant chemo-, or radiotherapy were excluded to avoid potential influences on microRNA expression. Perioperative clinical data, histopathological information, and follow-up data were collected for all patients using the clinical cancer registry. A poor chemotherapy response was defined as a tumor recurrence or a tumor-related death of ≤18 months (cutoff) postoperatively. The cutoff of 18 months was selected due to the median survival of 13–18 months of PDAC patients with curative treatment intent (median split method) [32]. The primary end points of this study were recurrence-free survival and overall survival.
Tumor macrodissection and RNA purification
For each histological sample, a representative hematoxylin and eosin (H&E)-stained section was reviewed by a trained and experienced pathologist. Tissue samples included in this study had >60 % viable cells and <20 % necrosis. Tumor and stromal tissue areas were selectively distinguished with a permanent marker by the pathologist to guide macrodissection. Macrodissection of samples with adenoma was carried out to create benign controls using sample material distant from adenoma. Twenty-micrometer sections from each FFPE tissue sample were cut with a microtome and processed immediately. After melting the paraffin block at 65 °C for 1 h to liberate the tissue from the surrounding paraffin, separation of the tumor tissue from the stromal area with a scalpel was carried out. Cut sections were placed in 1.5 ml tubes for RNA extraction.
Total RNA enriched in microRNAs fraction was purified through robotic workstation (QIAcube, Qiagen) by using miRNeasy FFPE Kit isolation system following manufacturer’s protocols (Qiagen). RNA concentration and purity were assessed by Agilent 2100 Bioanalyzer and RNA 6000 Nano/Pico LabChip (Agilent Tech., Boeblingen, Germany). Samples with RIN >7 were considered intact and used for analysis. RNA was stored at −80 °C.
Selection of microRNAs
Sixteen significantly deregulated microRNAs in two human PDAC cell lines (MIA-PaCa-2 and PANC-1) with acquired gemcitabine chemoresistance had been previously identified. Our prior in vitro study with Affymetrix GeneChip miRNA microarray (Affymetrix UK Ltd., High Wycombe, UK) analysis of 1733 human microRNAs and validation by qRT-PCR showed microRNA-21, microRNA-99a, microRNA-100, microRNA-125b, microRNA-138, microRNA-147b, microRNA-148a, microRNA-210, microRNA-376a, and microRNA-455-3p to be significantly upregulated, whereas microRNA-31-star, microRNA-330-3p, microRNA-330-5p, microRNA-378d, microRNA-422a, and microRNA-486-5p were significantly downregulated. Bioinformatic analysis and Western blot validation suggested involvement of these microRNAs in pathways controlling cell death and cycle [11].
MicroRNA expression data were normalized to expression levels of the four housekeeping genes RNU1A, RNU6-2, SNORD68, and SNORD96A selected from a total of ten tested housekeeping genes.
Quantification of microRNAs
qRT-PCR was performed using the miScript PCR system (Qiagen). Total RNA samples (1 μg) were reverse transcribed to complementary DNA (cDNA) using miScript II Reverse Transcription Kit (Qiagen). For each sample, 2 μl of the generated cDNA was mixed with 10 μl 2× QuantiTect SYBR, 2 μl 10× miScript Universal Primer, 2 μl gene specific 10× miScript Primer Assay (for five different miRNAs), and 1 μl nuclease free water. All samples were analyzed in triplicate using Bio-Rad CFX384TM RT PCR cycler (Bio-Rad Laboratories, Munich, Germany) with miScript SYBR Green PCR Kit (Qiagen). No template and minus RT controls were run in parallel. The cycling program involved an initial incubation step of 15 min at 95 °C to activate HotStarTaq DNA Polymerase, followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s and elongation at 70 °C for 30 s. Quantitative miR analysis was performed using CFX Manager Software v2.1 (Bio-Rad Laboratories). Expression of the selected microRNAs was analyzed quantitatively relative to the four housekeeping genes (hg) by the 2–ΔΔCT method using the equation: relative quantity R = 2–ΔΔCt, where ΔΔCt = (Ct microRNA − Ct hg) cancer − (Ct microRNA − CtT hg) benign and Ct is the cycle threshold [33].
Tissue array immunohistochemistry
For the tissue microarray (TMA), corresponding H&E-stained sections from each FFPE block were used to define and to mark representative tumor areas by the pathologist, two to four random, representative 2-mm tissue cores were punched from the designated area using a biopsy needle and inserted in a grid pattern into a recipient paraffin block using a tissue arrayer [34]. The final TMA contained 44 tissue cores processed in duplicate. Four-micrometer-thick sections were cut from each completed TMA array block and transferred to silanized glass slides. Sections from these arrays were then stained with the panel of antibodies: phosphatase and tensin homolog (PTEN) (monoclonal antibody, 1:200, clone NCH-38, Dako, Hamburg, Germany), deoxycytidine kinase (DCK) (monoclonal antibody, 1:5000, clone V9, Dako, Hamburg, Germany), HuR, MRP-1, MDR-1, BCRP-1, VEGF-1, and lymphatic endothelial marker D2-40. These antibodies were chosen based on their use as markers of tumor suppression, gemcitabine transport, and metabolism [5, 6, 8, 9, 35–37].
All immunohistochemical TMA studies were performed by the avidin-biotin-peroxidase complex method of Hsu et al. [38] For immunostaining, the Dako Autostainer Plus S3400 and the Dako Real Detection System (Alkaline Phosphatase/RED, Rabbit/Mouse, K5005) were used according to the manufacturer’s instructions (Dako, Hamburg, Germany). Briefly, after dewaxing and epitope recovery for 35 min at 98 °C, endogenous peroxidase activity was quenched by Dako Real Peroxidase-Blocking Solution for 10 min at room temperature. Afterwards, slides were incubated with primary antibodies and diluted in Dako Real antibody diluent, for 25 min. This step was followed by incubation with Dako Real Link, biotinylated secondary antibodies (20 min), incubation with Dako Real Streptavidin Alkaline Phosphatase (20 min), and visualization by RED chromogen (2 × 8 min) using the Dako autostaining system with appropriate controls for each run. Finally, slides were counterstained with Mayer’s hematoxylin and dehydrated in graded alcohols. Positive controls were included in all experiments using the same experimental conditions. A negative control was prepared by the same staining procedure without primary antibodies.
Immunohistochemical staining was evaluated semi-quantitatively, according to the percentage of cells showing specific immunoreactivity. Two independent investigators evaluated, in a blinded manner, antibody staining using light microscopy (Eclipse E1000M and NIS-Elements D3.1 imaging software, Nikon, Tokyo, Japan) and a scoring system with a scale ranging from 0 to 4+ [5]. Disagreements were resolved by joint review. For each case, the core with the highest percentage of tumor cell staining was used for analysis. Grade 0 was defined as complete absence of staining. Grade 1+ was defined as a low positive staining up to 25 %. Grade 2+ was defined as a moderate positive staining density of ≥25 but <50 %. A positive high staining density ≥50 % but <75 % of cells was defined as grade 3+. Finally, a positive staining density ≥75 % was assessed as grade 4+. Staining intensity was scored as weak, moderate, or strong.
Statistical analysis
Statistical analysis was performed with the SPSS® Statistics Version 22 (IBM Corp. Armonk, NY) for Windows®. Data were expressed as medians of mean normalized expression with standard deviation (SD). Statistical significance was determined by using the two-tailed Student’s ttest to compare two data sets.
Box-and-whisker plots demonstrating the median (middle quartile) were used to show normalized microRNA expression data (2-ΔΔCt) in the different therapy response groups. The middle half of a data set falls within the interquartile range represented by the width of the box. The whiskers indicate variability outside the upper and lower quartiles. The grand mean representing the overall mean of all data combined is displayed as a horizontal line across the entire graph. Comparison between gene expression and categorical variables was conducted using the nonparametric Mann-Whitney U test or the Kruskal-Wallis test to test more than two groups. To test the correlation between the clinicopathological data and the expression of the microRNAs, we used the Fisher two-tailed exact test and whenever appropriate the χ2 test. All of the variables were dichotomized. For analysis of follow-up data, life table curves were calculated using the Kaplan-Meier method. Log-rank test was used for comparison of relapse-free survival rates and survival rates between the groups with high and low microRNA or TAM marker expression in PDAC tissue compared to benign controls. For the identification of the best cutoff value for a supposed prognostic variable the log-rank test was performed for all observed values within the entire data set according to LeBlanc et al. [39] The primary end points were disease-specific survival or relapse-free survival, as measured from the date of surgery to the time of the last follow-up or cancer-related death or tumor relapse, respectively. Data of patients who were still alive and without evidence of tumor relapse at the end of the study were censored. A Cox proportional-hazards regression model was used to estimate hazard ratios and 95 % confidence intervals (CIs) and to perform multivariate survival analysis using a forward stepwise variable selection procedure based on the likelihood ratio. Variables with significant p values in the univariate analysis were included in the multivariate analysis. p value <0.05 was considered to be statistically significant. Housekeeping gene selection was performed by geNorm software (Biogazelle NV, Zwijnaarde, Belgium), a popular algorithm to determine the most stable reference (housekeeping) genes from a set of tested candidate reference genes (n = 10). The gene expression stability measure (M) for a reference gene is calculated by geNorm as the average pairwise variation for that gene with all other tested reference genes. Housekeeping genes with an average expression stability M ≤ 0.5 were considered to have a high target reference stability [40]. By this way, we identified four housekeeping genes for accurate microRNA qRT-PCR data normalization.
Acknowledgements
We thank the patients and clinicians contributing to this study. The authors thank particularly Ms Frauke Spiecker, Ms Petra Fischer, Ms Cordula Westermann, and Ms Inka Buchroth for their support and expert technical assistance.
Abbreviations
- ABC
ATP binding cassette
- BCRP
breast cancer resistance protein
- DCK
deoxycytidine kinase
- FFPE
formalin-fixed paraffin-embedded
- H&E
hematoxylin and eosin
- HuR
Hu antigen R
- MDR
multidrug resistance
- MRP
multidrug resistance protein
- PDAC
pancreatic ductal adenocarcinoma
- PTEN
phosphatase and tensin homolog
- TMA
tissue microarray
- UICC
Union Internationale Contre le Cancer
- VEGF
vascular epithelial growth factor
Footnotes
Competing interests
The authors declare that they have no competing interests or funding to disclose.
Authors’ contributions
SAD designed the experiments, performed the statistical analysis, and prepared the manuscript. SAD, BA, GK, and WAM performed the immunohistological and molecular experiments. SAD, NS, GK, and JH contributed to the specimens and clinicopathologic data. Critical revision of the manuscript was performed by all participants. All authors read and approved the final manuscript.
Contributor Information
Sameer Abdallah Dhayat, Phone: +49 251 83 56326, Email: sameer.dhayat@ukmuenster.de.
Baha Abdeen, Email: bahaald_33@hotmail.com.
Gabriele Köhler, Email: gabriele.koehler@uni-muenster.de.
Norbert Senninger, Email: Senning@ukmuenster.de.
Jörg Haier, Email: Joerg.Haier@ukmuenster.de.
Wolf Arif Mardin, Email: WolfArif.Mardin@ukmuenster.de.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol. 2011;29(34):4548–54. doi: 10.1200/JCO.2011.36.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.O’Driscoll L, Walsh N, Larkin A, Ballot J, Ooi WS, Gullo G, et al. MDR1/P-glycoprotein and MRP-1 drug efflux pumps in pancreatic carcinoma. Anticancer Res. 2007;27(4B):2115–20. [PubMed] [Google Scholar]
- 6.Marechal R, Mackey JR, Lai R, Demetter P, Peeters M, Polus M, et al. Deoxycitidine kinase is associated with prolonged survival after adjuvant gemcitabine for resected pancreatic adenocarcinoma. Cancer. 2010;116(22):5200–6. doi: 10.1002/cncr.25303. [DOI] [PubMed] [Google Scholar]
- 7.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69(11):4567–72. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252(3):499–505. doi: 10.1097/SLA.0b013e3181f1fd44. [DOI] [PubMed] [Google Scholar]
- 9.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70(11):4528–38. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 10.Dhayat S, Mardin WA, Mees ST, Haier J. Epigenetic markers for chemosensitivity and chemoresistance in pancreatic cancer—a review. Int J Cancer. 2011;129(5):1031–41. doi: 10.1002/ijc.26078. [DOI] [PubMed] [Google Scholar]
- 11.Dhayat SA, Mardin WA, Seggewiß J, Ströse AJ, Matuszcak C, Hummel R, et al. MicroRNA profiling implies new markers of gemcitabine chemoresistance in mutant p53 pancreatic ductal adenocarcinoma. PLoS One. 2015;10(11):e0143755. doi: 10.1371/journal.pone.0143755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luttges J, Schemm S, Vogel I, Hedderich J, Kremer B, Kloppel G. The grade of pancreatic ductal carcinoma is an independent prognostic factor and is superior to the immunohistochemical assessment of proliferation. J Pathol. 2000;191(2):154–61. doi: 10.1002/(SICI)1096-9896(200006)191:2<154::AID-PATH603>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Lim JE, Chien MW, Earle CC. Prognostic factors following curative resection for pancreatic adenocarcinoma: a population-based, linked database analysis of 396 patients. Ann Surg. 2003;237(1):74–85. doi: 10.1097/00000658-200301000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5(5):e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284(39):26533–46. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen KE, Weiss GJ. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther. 2010;9(12):3126–36. doi: 10.1158/1535-7163.MCT-10-0397. [DOI] [PubMed] [Google Scholar]
- 17.Gisel A, Valvano M, El Idrissi IG, Nardulli P, Azzariti A, Carrieri A, et al. miRNAs for the detection of multidrug resistance: overview and perspectives. Molecules. 2014;19(5):5611–23. doi: 10.3390/molecules19055611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugawara I, Kataoka I, Morishita Y, Hamada H, Tsuruo T, Itoyama S, et al. Tissue distribution of P-glycoprotein encoded by a multidrug-resistant gene as revealed by a monoclonal antibody, MRK 16. Cancer Res. 1988;48(7):1926–9. [PubMed] [Google Scholar]
- 19.Benard J, Bourhis J, Riou G. Clinical significance of multiple drug resistance in human cancers. Anticancer Res. 1990;10(5A):1297–302. [PubMed] [Google Scholar]
- 20.Chen M, Xue X, Wang F, An Y, Tang D, Xu Y, et al. Expression and promoter methylation analysis of ATP-binding cassette genes in pancreatic cancer. Oncol Rep. 2012;27(1):265–9. doi: 10.3892/or.2011.1475. [DOI] [PubMed] [Google Scholar]
- 21.Suwa H, Ohshio G, Arao S, Imamura T, Yamaki K, Manabe T, et al. Immunohistochemical localization of P-glycoprotein and expression of the multidrug resistance-1 gene in human pancreatic cancer: relevance to indicator of better prognosis. Jpn J Cancer Res. 1996;87(6):641–9. doi: 10.1111/j.1349-7006.1996.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8(5):1067–74. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 23.O’Reilly EM. Pancreatic adenocarcinoma: new strategies for success. Gastrointest Cancer Res. 2009;3(2 Suppl):S11–5. [PMC free article] [PubMed] [Google Scholar]
- 24.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3(2):109–13. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. Jama. 2007;297(17):1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 26.Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, White RW. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2011;71(5):538–49. doi: 10.1002/pros.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu N, Lin X, Zhao X, Zheng L, Xiao L, Liu J, et al. MiR-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38MAPK-independent pathways. Br J Cancer. 2013;109(11):2853–63. doi: 10.1038/bjc.2013.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285(28):21496–507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmrich S, Rasche M, Schoning J, Reimer C, Keihani S, Maroz A, et al. miR-99a/100 ~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev. 2014;28(8):858–74. doi: 10.1101/gad.233791.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song T, Xia W, Shao N, Zhang X, Wang C, Wu Y, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11(4):905–11. [PubMed] [Google Scholar]
- 31.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 32.Wilkowski R, Wolf M, Heinemann V. Primary advanced unresectable pancreatic cancer. Recent Results Cancer Res. 2008;177:79–93. doi: 10.1007/978-3-540-71279-4_10. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10(7):657–62. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- 35.Nath S, Daneshvar K, Roy LD, Grover P, Kidiyoor A, Mosley L, et al. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2 doi: 10.1038/oncsis.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamada S, Satoh K, Hirota M, Kanno A, Umino J, Ito H, et al. The homeobox gene MSX2 determines chemosensitivity of pancreatic cancer cells via the regulation of transporter gene ABCG2. J Cell Physiol. 2012;227(2):729–38. doi: 10.1002/jcp.22781. [DOI] [PubMed] [Google Scholar]
- 37.Georgiadou D, Sergentanis TN, Sakellariou S, Filippakis GM, Zagouri F, Vlachodimitropoulos D, et al. VEGF and Id-1 in pancreatic adenocarcinoma: prognostic significance and impact on angiogenesis. Eur J Surg Oncol. 2014;40(10):1331–7. doi: 10.1016/j.ejso.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Hsu SM, Raine L, Fanger H. A comparative study of the peroxidase-antiperoxidase method and an avidin-biotin complex method for studying polypeptide hormones with radioimmunoassay antibodies. Am J Clin Pathol. 1981;75(5):734–8. doi: 10.1093/ajcp/75.5.734. [DOI] [PubMed] [Google Scholar]
- 39.LeBlanc M, Jacobson J, Crowley J. Partitioning and peeling for constructing prognostic groups. Stat Methods Med Res. 2002;11(3):247–74. doi: 10.1191/0962280202sm286ra. [DOI] [PubMed] [Google Scholar]
- 40.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]


