Abstract
More than a billion people lack access to safe drinking water that is free from pathogenic bacteria and toxic metals. The World Health Organization estimates several million people, mostly children, die every year due to the lack of good quality water. Driven by this need, we report the development of PGLa antimicrobial peptide and glutathione conjugated carbon nanotube (CNT) bridged three-dimensional (3D) porous graphene oxide membrane, which can be used for highly efficient disinfection of Escherichia coli O157:H7 bacteria and removal of As(III), As(V), and Pb(II) from water. Reported results demonstrate that versatile membrane has the capability to capture and completely disinfect pathogenic pathogenic E. coli O157:H7 bacteria from water. Experimentally observed disinfection data indicate that the PGLa attached membrane can dramatically enhance the possibility of destroying pathogenic E. coli bacteria via synergistic mechanism. Reported results show that glutathione attached CNT-bridged 3D graphene oxide membrane can be used to remove As(III), As(V), and Pb(II) from water sample at 10 ppm level. Our data demonstrated that PGLa and glutathione attached membrane has the capability for high efficient removal of E. coli O157:H7 bacteria, As(III), As(V), and Pb(II) simultaneously from Mississippi River water.
Keywords: 3D porous graphene oxide membrane, CNT-bridged graphene oxide, E. coli O157:H7 bacteria removal, bacteria disinfection, separation of As(III), As(V) and Pb(II) from water
Graphical abstract
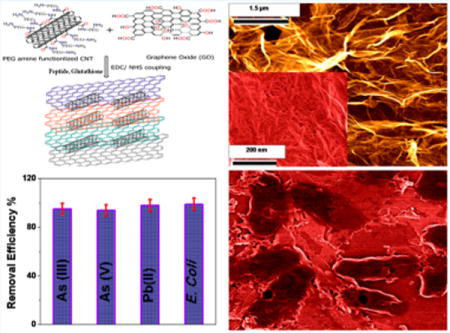
1. INTRODUCTION
Even in the 21st century, several billion people lack good quality drinking water that is free from pathogenic bacteria and toxic metals.1–4 According to the World Health Organization (WHO),3 water contamination by fecal indicator pathogenic bacteria such as Escherichia coli O157:H7 kills more than 10 million children/year under the age of five. Current technology to remove pathogenic bacteria from water is mainly chemical treatment.1,2,5,6 Due to the chemical resistance, several E. coli O157:H7 strains have become resistant to common disinfection methods, and thus, a need has arisen for new technologies that can be used for highly effective removal and disinfection of pathogens from water.1,2,5,6 On the other hand, due to the rapidly growing world population, industrialization, intensification of agricultural activities, and urbanization, society faces a major health risk due to the presence of various toxic heavy metals such as As and Pb in natural waters beyond acceptable limits.7–13
According to the World Health Organization (WHO), 180 million people from more than 70 countries have been exposed to toxic levels of arsenic from drinking water.4 It is well documented that long-time exposure of arsenic can cause skin, liver, lung, and bladder cancers, which have killed more than 40,000 people in Bangladesh in past few years.4,8 As a result, WHO has established arsenic tolerance level of maximum of 10 μg As/L (10 ppb) in drinking water.4 Other groundwater contaminant which is highly toxic to human health is Pb(II), which can affect the immune system.13–15 It is well documented that Pb(II) builds up in the body over the years and damages kidneys, liver, blood, and brain cells.15 Despite the availability of modern technology such as chemical precipitation, adsorption, membrane filtration, or photocatalytic degradation,7,8 more than 500 million people are drinking groundwater with As(III), As(V), and Pb(II) concentrations well above the toxicity level. Driven by this need, we report herein the development of peptide conjugated carbon nanotube (CNT) bridged hybrid graphene oxide based three-dimensional (3D) membrane for highly efficient of disinfection of E. coli O157:H7 bacteria, as shown in Scheme 1. We have also reported that same membrane can be used for the removal of As(III), As(V), and Pb(II) from water. The novelty of the reported graphene oxide material based technology is based on separation and removal of biological and chemical toxins selectively and simultaneously.
Scheme 1. Schematic Representation of the Disinfection of E. coli O157:H7 Pathogens and Separation of Toxic Metals Using PGLa and Glutathione-Conjugated CNT-Bridged Porous Graphene Oxide Membranea.
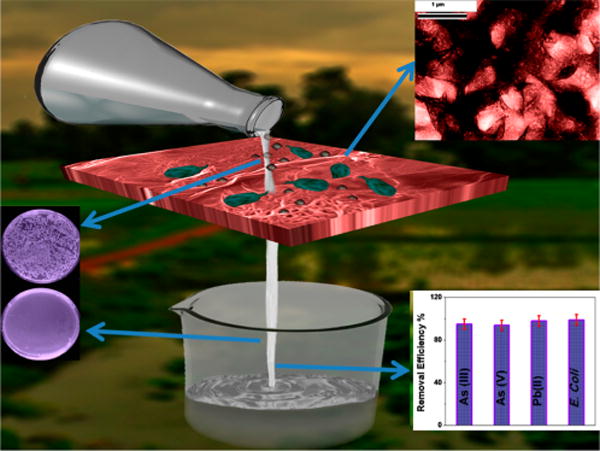
a(Inset, left) TEM picture of E. coli pathogens being captured by 3D membrane. (Inset, top right) Bacteria colony counting results show that no E. coli O157:H7 is present in the water once it has been passed through the membrane. (Inset, bottom right) Removal efficiency data shows the membrane cab be used to separate biological and chemical toxin simultaneously.
In the past few years, we and other groups have demonstrated that, due to possible large scale production at low cost, high specific surface area, low mass density, and good flexibility, graphene-based membranes with interconnected porous structures possess novel physical and chemical properties.16–25 Engineered graphene-material-based membranes have demonstrated significant potential for broad use in a variety of water treatment applications.26–33 Because two-dimensional (2D) graphene oxide (GO) has plenty of hydrophobic basal plane and hydrophilic oxygen-containing groups on the surface,34–39 we have developed three-dimensional (3D) graphene oxide based membrane via covalent functionalized using one-dimensional (1D) carbon nanotobe (CNT).
In the last two decades, due to unique properties, 1D CNTs have become advanced materials for possible future applications.40–47 In our synthesis, we used 1D CNTs to physically separate 2D graphene oxide sheets from accumulation and to increase the porous size of the membrane. To capture and kill pathogens, we have developed PGLa antimicrobial peptide conjugated membrane with a pore size of around 400 nm, which not only can capture E. coli O157:H7 pathogens from water but also has the capability to kill E. coli O157:H7 on contact. PGLa is an α-helical peptide of 21 amino acids (GMASKAGAIAGKIAKVALKAL-NH2), which is known to destroy bacteria by interacting with their lipid membranes.48–51 Similarly, for the selective removal of Pb(II) and As(III) from water sample, we have modified the membrane with glutathione. Glutathione (γ-L-glutamyl-L-cysteinyl-glycine) is a tripeptide which exists in the cells and protects cellular membranes from the toxic effects of heavy metals by forming complex with Pb(II), As(III), As(V) etc.10–15 Our experimental data show that PGLa and glutathione peptide conjugated porous membrane can be used as a versatile membrane to remove pathogens and toxic metals and to kill E. coli O157:H7 simultaneously.
2. EXPERIMENTAL SECTION
2.1. Materials
All the chemicals including PGLa, glutathione, single wall carbon nanotube, graphite, poly(ethylene glycol) (PEG), KMnO4, nitric acid, ethylene glycol, arsenic, and lead salts were purchased from Fisher Scientific and Sigma-Aldrich. E. coli O157:H7 bacteria and growth media to grow bacteria were purchased from the American Type Culture Collection (ATCC, Rockville, MD).
2.2. Synthesis of Water-Soluble 2D Graphene Oxide
We have synthesized water-soluble 2D graphene oxide using modified Hummers method, as we have reported recently.21,25 For this purpose, we have performed graphite exfoliation by strong oxidizing agents, as shown in shown in Scheme 2A.16–25
Scheme 2.
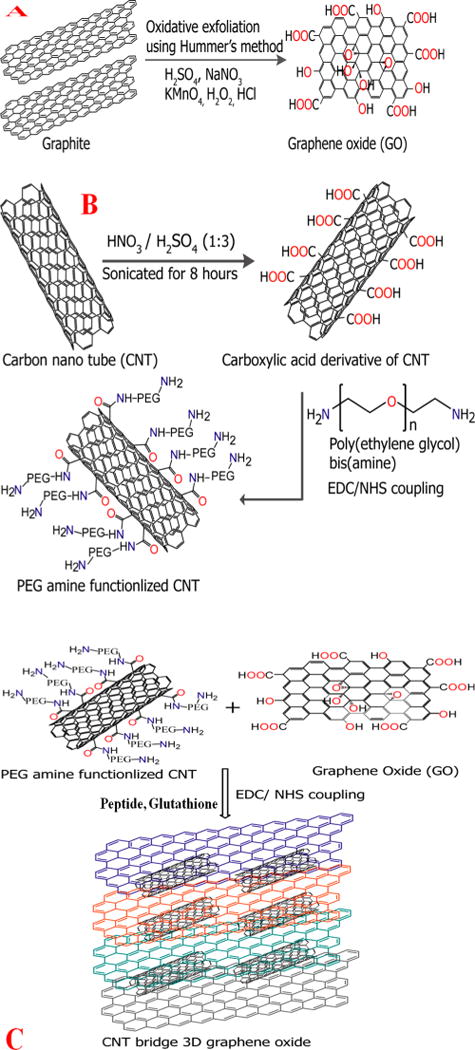
Schematic Representation Showing (A) the Synthesis Procedure for Graphene Oxide from Graphite, (B) Our Synthesis Procedure to Develop PEG-Amine-Functionalized CNT, and (C) the Synthesis Procedure for the CNT-Bridged 3D Graphene Oxide Membrane
In brief, in the first step, 1.8 g of graphite flakes was treated with 1.8 g of NaNO3 in 80 mL of H2SO4 and 5g of KMnO4 for 30 min without changing the temperature. After the recation, black paste was obtained. In the next step, we filtered and redispersed the obtained graphene oxide in 150 mL of water and sonicated the mixture for several hours for exfoliation. At the end, water-soluble 2D graphene oxide was obtained. We used ultrahigh resolution field emission scanning electron microscopy (FE-SEM HITACHI) coupled with a BF/DF Duo-STEM detector and energy-dispersive X-ray spectroscopy (EDX; Bruker) to characterize 2D graphene oxide. As shown in Figure 1A, EDX mapping shows the presence of C and O in water-soluble graphene oxide.
Figure 1.
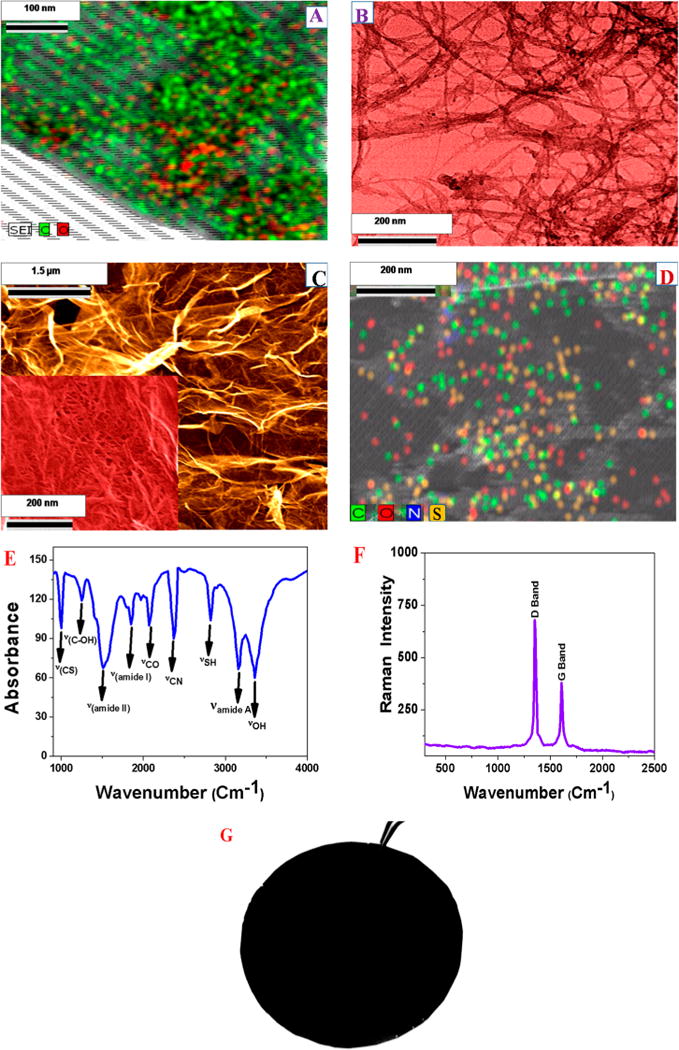
(A) EDX mapping shows the presence of C and O in water-soluble graphene oxide. (B) TEM image of freshly prepared PEG amine functionalized SWCNT. (C) SEM image of PGLa and glutathione-conjugated CNT-bridged graphene oxide membrane shows the 3D structure with pore sizes of 300–500 nm. (D) EDX mapping shows the presence of C, N, S, and O in PGLa and glutathione-conjugated CNT-bridged graphene oxide 3D network membranes. (E) FTIR spectrum shows the existence of amide A, I, and II bands, which indicates the presence of peptides on the 3D membrane. Similarly, the presence of −SH and −CS bands clearly indicate the presence of glutathione on the 3D membrane. The stretches of −CO, −OH, −CN, and –C–OH groups from graphene oxide and CNT can also be seen on the FTIR spectra. (F) Raman spectrum from freshly prepared hybrid membrane clearly indicates the presence of D and G bands in PGLa and glutathione-conjugated CNT-bridged graphene oxide 3D network membranes. (G) Photograph shows freshly prepared PGLa and glutathione-conjugated CNT-bridged porous hybrid graphene oxide membrane.
2.3. Synthesis of Water-Soluble PEG Amine Functionalized SWCNT
For the development of water-soluble PEG amine functionalized single wall carbon nnaotube (SWCNT), in the initial site, we have performed the oxidative treatments of SWCNT using 3:1 ratio of concentrated sulfuric acid and nitric acid,40–48 as shown in Scheme 2B. During this process, SWCNT tips were chemically functionalized by carboxy group and water-soluble carboxy functionalized SWCNT was obtained. In the next step, we have functionalized bis amine PEG (H2N-PEG-NH2) with water-soluble SWCNTs by treating the – CO2H of SWCNT with aimne group of PEG in the presence of EDC cross-linking agent under inert medium. For this development, we have used the coupling chemistry between –CO2H group of SWCNT and -NH2 group of H2N-PEG-NH2 to develop amine PEG coupled water-soluble SWCNT, a shown in Scheme 2B. At the end, we have used JEOL 2010-F microscope (TEM) for the characterization of water-soluble PEG amine functionalized SWCNT.
2.4. Synthesis of PGLa and Glutathione-Conjugated CNT-Bridged 3D Graphene Oxide Membrane
Oxygen-containing functional groups in water-soluble 2D graphene oxide and amine group in PEG-coupled water-soluble SWCNTs are serve as anchoring points for developing CNT-bridged 3D graphene oxide membrane. For the selective disinfection of E. coli bacteria and separation of As(III) and Pb(II), we have developed PGLa and glutathione-conjugated porous membrane. To accomplish this, we used coupling chemistry between –CO2H group of 2D graphene oxide and –NH2 group of glutathine, PGLa peptide and PEG coupled water-soluble SWCNTs via amide linkages as shown in Scheme 2C. For this purpose, 20 mL of 2D graphene oxide, 20 mL of amine PEG-coupled SWCNT, 100 mg of glutathione, and 80 mg of PGLa peptides were mixed together and sonicated for 10 min. After sonication, samples were kept on oil bath at about 60 °C under a hood for 80 min. At the end, we have obtained PGLa and glutathione-conjugated CNT-bridged 3D graphene oxide semisolid foam, which was used to develop 8 × 8 cm membrane using spin-casting. The membrane structure is shown in Figure 1G. Then, we characterized 3D membrane using scanning electron microscope (SEM), EDX, Fourier transform infrared spectroscopy (FTIR), and Raman spectroscopy. Figure 1C,D display high-resolution SEM image with energy-dispersive X-ray spectroscopy (EDX) mapping of the PGLa peptide and glutathione-conjugated CNT-bridged membranes microstructure, which show an interconnected 3D network with a pore size of 300–500 nm. The inset high-resolution picture in Figure 1C clearly shows the presence of CNT in 3D membrane. The EDX mapping, as reported in Figure 1D, clearly shows the presence of C, N, S, and O in the 3D hybrid graphene oxide network, which indicates the presence CNT, GO, glutathione and peptides. Next, we have used Thermo Nicolet Nexus 870 FTIR spectrometer equipped with three beam splitters to find out the chemical composition of PGLa peptide and glutathione attached CNT bridges 3D graphene oxide. As shown in the Figure 1E, the FTIR spectrum of the PGLa peptide and glutathione-conjugated CNT-bridged membranes show a very strong band at ∼3325 cm−1, which can be attributed as peptide Amide A band. In the reported IR spectra, we have observed an amide I band at ∼1750 cm−1 which is mainly due to the stretching vibrations of the C=O bond of the amide. We have also observed an amide II band at ∼1550 cm−1, which is mainly due to the in-plane NH bending vibration from peptide. Similarly, an amide III band ∼1260 cm−1 is mainly due to the peptides. Reported IR spectra at Figure 1E also show a strong band at ∼3600 cm−1, which is mainly due to the –OH stretching vibration. Similarly, IR peaks were observed at ∼1722 cm−1 for the carbonyl (–C=O) stretch of carboxylic acid from graphene oxide and CNT. Figure 1F shows the Raman spectra from CNT-bridged graphene oxide membrane which clearly shows the strong D-band ∼1345 cm−1 and a G-band ∼1625 cm−1.30–40 Experimentally observed Raman spectra show strong D band which indicates that the surface modification extent for graphene oxide and CNT is high.
2.5. Membrane Characterization
Using nitrogen adsorption analysis via the Brunauer–Emmett–Teller (BET) method, we found that the specific surface area for the membranes was 663 m2 g−1, and the pore volume was 0.560 cm3 g−1. From BET analysis, we measured the pore size distribution, which shows an average pore diameter of 400 nm. We measured the water flux by collecting the permeate water through the membrane using an electronic balance and calculated water flux of our membrane is 186.3 L m−2 h−1 bar−1.
2.6. E. coli O157:H7 Bacteria Sample Preparation
E. coli O157:H7 were purchased from the ATCC and then cultured using ATCC protocol as instructed. In brief, E. coli O157:H7 was rehydrated on 8 to 10 mL of Bacto tryptic soy broth (BD) and incubated at 37 °C for 24 h. In the next step, a single colony of E. coli O157:H7 from tryptic agar plate was inoculated into 15 mL of tryptic soy broth for 12 h. We diluted the stock solution of bacteria several times to vary the concentration of E. coli O157:H7 from 102–106 CFU (colony forming unit)/mL.
2.7. Fluorescence Imaging of E. coli O157:H7 Bacteria
For fluorescence imaging of E. coli O157:H7, we used Cy3 attached PGLa peptide. Imaging experiment was performed using Olympus IX71 inverted confocal fluorescence microscope. For imaging purpose, we used 580 nm light as an excitation source. We used SPOT Insight digital camera for fluorescence collection. Olympus DP capture software has been used for data processing.
2.8. Determination of the Percentage of live E. coli O157:H7 bacteria
After removal by 3D membrane, we transferred E. coli O157:H7 bacteria to colony-countable plates and incubated for 24 h at 37 °C. After that, the colony number for each plate was counted with a colony counter (Bantex, Model 920 A).
3. RESULTS AND DISCUSSION
To understand whether PGLa-conjugated CNT-bridged 3D graphene oxide membranes can be used for the removal of E. coli bacteria from infected water, we spiked 3.2 × 106 colony-forming units (CFU)/mL of E. coli with 100 mL of distilled water. After 110 min of gentle shaking, we used our 3D membrane to filter the E. coli infected water sample. In the next step, to find the removal efficiency of E. coli bacteria, we performed reverse transcription polymerase chain reaction (RT-PCR) technique,52,53 as shown in Figure 2A. We also used colony plating technique on LB agar in water sample as shown in Figure 2D,E. Reported experimental data using colony plating technique and RT-PCR clearly show that about 100% E. coli were removed by the PGLa conjugated CNT-bridged 3D graphene oxide membrane.
Figure 2.
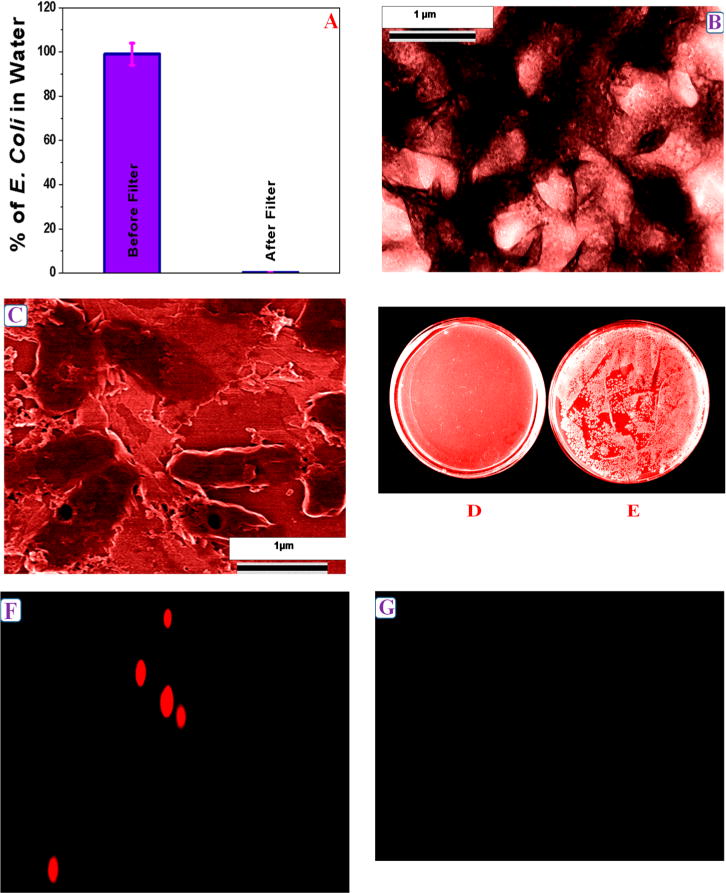
(A) Plot shows E. coli O157:H7 removal efficiency using PGLa conjugated CNT-bridged 3D graphene oxide membrane. Reverse transcription polymerase chain reaction (RTPCR) was used to quantify the amount of E. coli O157:H7 present. (B) TEM image shows the capture of E. coli O157:H7 by CNT-bridged hybrid graphene oxide membrane. (C) SEM image demonstrating the capture of E. coli O157:H7 by PGLa-conjugated CNT-bridged 3D graphene oxide based membrane. Colonies of E. coli O157:H7 showing the amount of live E. coli O157:H7 bacteria (D) after filtration by our membrane and (E) before filtration. (F) Fluorescence image shows the presence of E. coli O157:H7 bacteria on membrane after E. coli O157:H7 infected water was filtered by membrane. (G) Fluorescence image shows the absence of E. coli O157:H7 bacteria in water after separation by membrane.
This very high efficient removal of E. coli bacteria using the CNT-bridged membrane is due to the fact that the size of E. coli bacteria is about 1.5–2 μm, whereas the pore size of the developed CNT-bridged membrane is about 400 nm. Due to the above fact, only water can pass through the porous membrane and E. coli bacteria will not able to go through the PGLa attached membrane. To characterize E. coli bacteria captured by the membrane, we also used high-resolution TEM, SEM, and fluorescence imaging techniques, as shown in Figures 2B,C,F,G. Reported TEM and SEM images clearly indicate that E. coli bacteria are captured on the surface of the membrane. To understand better, we also performed fluorescence imaging as shown in Figures 2F,G. For this purpose, we used Cy3-modified PGLa peptide which was attached with the CNT-bridged 3D membrane. The fluorescence images, as reported in Figures 2F,G, also confirmed the RT-PCR and colony plating technique results, which clearly indicate that after filtration, E. coli bacteria were captured by the membrane and as a result, we have not observed any fluorescence from water sample.
Because E. coli O157:H7 is pathogenic and known to be transmitted through contaminated water or by contact, it is very important that the membrane should be able to disinfect E. coli O157:H7 bacteria after separation, so that after filtration, the membrane should not be harmful for society. As a result, to understand whether the captured E. coli O157:H7 by PGLa conjugated CNT-bridged 3D graphene oxide membranes are alive or dead, we washed the E. coli O157:H7 bacteria captured membrane thoroughly using 150 mL of water. After that, we estimated the amount of live E. coli O157:H7 bacteria using RT-PCR and colony plating technique. As reported in Figure 3A,B, almost 100% of E. coli O157:H7 bacteria were killed when we used PGLa conjugated CNT-bridged 3D graphene oxide membranes.
Figure 3.
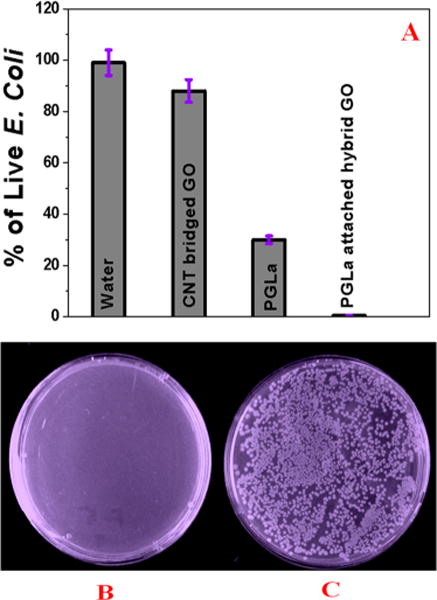
(A) RTPCR data show E. coli O157:H7 killing efficiency using CNT-bridged 3D GO without PGLa, with only PGLa, and with PGLa conjugated CNT-bridged 3D graphene oxide membrane. Colonies of E. coli O157:H7 bacteria demonstrating the amount of live bacteria after filtration by membrane in the presence of (B) PGLa-conjugated CNT-bridged 3D graphene oxide membrane and (C) CNT-bridged 3D graphene oxide based membrane without PGLa.
Next, to find out whether PGLa peptide conjugation is necessary to destroy E. coli O157:H7 bacteria, we also performed the same experiment with CNT-bridged 3D graphene oxide membrane in the absence of PGLa antimicrobial peptide attachment. Figure 3A,C clearly shows that most of the E. coli O157:H7 bacteria were alive when CNT-bridged 3D graphene oxide membrane was used. The observed very high killing by PGLa peptide attached CNT-bridged membrane can be due to the several facts: It is reported34–38 that PGLa has the capability to penetrate the outer membrane of bacteria. Due to the insertion of the PGLa into the inner E. coli O157:H7 bacteria membrane, lateral expansion of the lipid bilayer occurs. Also, PGLa peptide binding can destabilize the inner membrane of E. coli O157:H7 bacteria. Both mechanisms, as discussed above, can be responsible for the E. coli O157:H7 bacteria cell death. Another possibility is that 3D graphene oxide can kill E. coli O157:H7 bacteria by mechanical wrapping,33,36,37 as shown in the SEM and TEM images in Figure 2B,C. This mechanical wrapping, as discussed above, may cause induced membrane stress on the E. coli O157:H7 membrane by disrupting and damaging cell membranes. The final output can be cell lysis, as reported before.39–41 Next, to understand how much percentage of E. coli O157:H7 bacteria were killed due to the mechanical wrapping, we performed the same experiment by using only CNT-bridged 3D graphene oxide without PGLa peptide. On the other hand, to find out how much E. coli O157:H7 bacteria is killed by PGLa peptide, we performed an E. coli O157:H7 bacteria killing experiment by adding PGLa directly to the water solution. Figure 3A indicates that PGLa is able to kill roughly 48% of E. coli O157:H7 bacteria in the absence of CNT-bridged 3D graphene oxide, whereas CNT-bridged 3D graphene can kill only 11% E. coli O157:H7 bacteria. Our experimental data, as reported in Figure 3A, clearly indicate that PGLa attached CNT-bridged 3D graphene oxide membrane exhibit synergistic killing effect, where almost 100% of E. coli O157:H7 bacteria were killed. The observed synergistic mechanism can be due to the fact that E. coli O157:H7 bacteria are trapped by 3D porus graphene oxide, which causes membrane stress. This condition allows PGLa to bind easily with E. coli O157:H7 bacteria and helps to penetrate the outer membrane of bacteria. As a result, E. coli O157:H7 bacteria are killed due to the disrupted and damaged cell membranes. So, our reported experimental data clearly show that the multimodal mechanism by PGLa attached CNT-bridged graphene oxide membrane can dramatically enhance the possibility of destroying E. coli O157:H7 bacteria due to the synergistic killing mechanism effect.
Next, to understand whether glutathione-conjugated CNT-bridged 3D graphene oxide membranes can be used for the separation and removal of the As(III) and Pb(II) from water sample, we performed filtration of 100 mL water sample containing 10 ppm As(III), 10 ppm As(V), and 10 ppm Pb(II) using our 3D membrane. We have used inductively coupled plasma mass spectrometry (ICP-MS) for finding the As(III), As(V), and Pb(II) removal amount.
As shown in Figure 4A–C, our data clearly show that more than 96% As(II), 92% As(V), and 98% Pb(II) have been captured by the membrane. This high removal efficiency is due to As (III) and Pb(II) having very high affinity for glutathione, where both metal ions can bind with glutathione via –SH. And for that reason, glutathione is used to protect cellular membranes from the toxic effects of heavy metals. It is also known that glutathione has the capability to reduce As(V) to As(III) and then make a complex with As(III). As a result, a glutathione-attached 3D membrane can remove As(V), As(III), and Pb(II) via the formation of a chemical complex. Another possible mechanism for high removal of toxic metal ions is due to the adsorption by 3D porous graphene oxide. Because the 3D membrane contains an open pore network, which facilitates fast diffusion of As(III), As(V), and Pb(II) inside 3D network, it allows CNT-bridged graphene oxide membrane to be a good adsorbent with high adsorption capacity for the removal of heavy metal ions. Reported heavy metal removal efficiency for multiple toxic metals together is around 20% higher than previously reported efficiency for individual toxic metal.35,38
Figure 4.
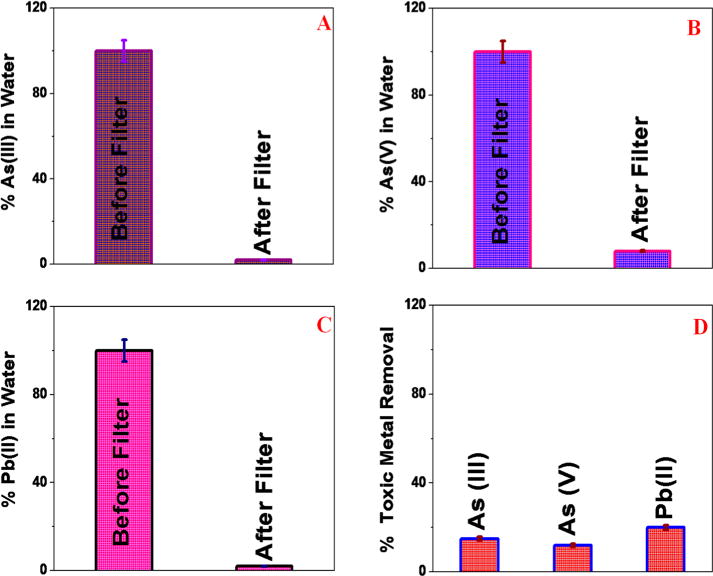
(A) As(III) removal efficiency using glutathione-conjugated CNT-bridged 3D graphene oxide membrane. ICP-MS was used to quantify the amount of As(III) present. (B) As(V) removal efficiency using glutathione-conjugated CNT-bridged 3D graphene oxide membrane. ICP-MS was used to quantify the amount of As(V) present. (C) Pb(II) removal efficiency using glutathione-conjugated CNT-bridged 3D graphene oxide membrane. ICP-MS was used to quantify the amount of Pb(II) present. (D) Percentage of removal efficiency for different toxic metals using CNT-bridged 3D graphene oxide membrane without glutathione. Reported data clearly show that the presence of gluthione is very important for high efficiency As(III), As(V), and Pb(II) removal.
To understand the contribution of each possible mechanism for the very high toxic metals separation efficiency using 3D graphene oxide membrane, we developed a CNT-bridged 3D membrane without glutathione. As shown in Figure 4D, our experimental data clearly show that only 12% of As(III), 9% of As(V), and 18% of Pb(II) can be removed via adsorption mechanism using CNT-bridged 3D graphene oxide membrane. On the other hand, more than 95% of As(III) and Pb(II) removal is possible when glutathione is present. Our reported experimental data clearly show that the presence of glutathione is necessary for very highly efficient removal of toxic metals like As(III), As(V), and Pb(II).
Next, to find out whether our membrane can be used for the separation of pathogens and toxic metals from the environmental sample, we used Mississippi River water spiked with E. coli, arsenic, and lead. Because Mississippi River water contains different metal ions, by using river water, we are able to test if our membrane can be used for targeted metal ion separation in the presence of other metal ions. For this purpose, we collected water samples from Mississippi River and spiked 3 × 106 colony-forming units (CFU)/mL of E. coli and 10 ppm As(III), 10 ppm As(V), and 10 ppm Pb(II). After that, we performed filtration of 100 mL spiked Mississippi River water sample using our PGLa and glutathione conjugated CNT-bridged 3D membrane. At the end, we have used inductively coupled plasma mass spectrometry (ICP-MS) for finding the As(III), As(V) and Pb(II) removal amount. We have also used RT-PCR to find out the removal efficiency for E. coli O157:H7 bacteria. As shown in Figure 5, our data clearly show that PGLa and glutathione conjugated CNT-bridged 3D graphene oxide membrane can be used for the removal of more than 99% of E. coli O157:H7 bacteria, 98% of As(III), 94% of As(V), and 98% of Pb(II) simultaneously from infected Mississippi River water.
Figure 5.
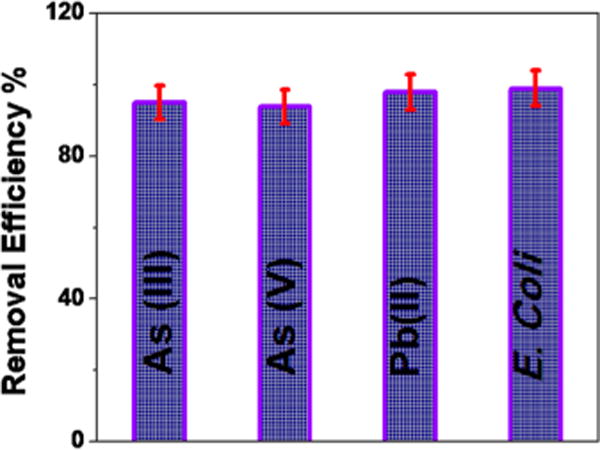
Plot showing percentage of removal efficiency of different toxic metals and E. coli bacteria by PGLa and glutathione attached CNT-bridged 3D graphene oxide membrane from Mississippi River water.
4. CONCLUSIONS
In conclusion, we have reported the development of PGLa and glutathione conjugated CNT-bridged 3D graphene oxide membrane for disinfection of pathogenic E. coli O157:H7 bacteria and removal of As(III), As(V), and Pb(II) with very high efficiency. We demonstrated that PGLa antimicrobial peptide conjugated CNT-bridged porous 3D graphene oxide membrane has the capability to capture and completely disinfect pathogenic pathogenic E. coli O157:H7 bacteria from water. Our reported data indicate that because the pore size of the CNT-bridged membrane (∼400 nm) is much smaller than pathogenic E. coli O157:H7 bacteria (∼1.5 μm), only water can pass through the porous membrane, whereas E. coli O157:H7 bacteria were captured by the membrane, which had been confirmed by SEM, TEM, and fluorescence images. Using RT-PCR and colony counting data, we have shown that almost 100% of pathogenic E. coli O157:H7 bacteria can be captured from the water sample using PGLa peptide conjugated membrane. Our reported disinfection data indicate that the PGLa-attached CNT-bridged graphene oxide membrane can dramatically enhance the possibility of destroying pathogenic E. coli O157:H7 bacteria via synergistic effect where CNT-bridged 3D graphene oxide helps to trap E. coli and allow PGLa to bind easily with E. coli O157:H7 bacteria. This helps PGLa to penetrate the outer membrane of bacteria and kill E. coli O157:H7 bacteria.
Our experimental results show that glutathione-attached CNT-bridged 3D graphene oxide membrane can be used to remove As(III), As(V), and Pb(II) from water samples at the 10 ppm level. We have shown that the most efficient way to remove toxic metals is to use a glutathione-attached membrane, where glutathione binds with As(III), As(V), and Pb(II) and removes them from water. Our data demonstrate that PGLa and glutathione-attached CNT-bridged 3D graphene oxide membrane has the capability for highly efficient and simultaneous removal of E. coli O157:H7 bacteria, As(III), As(V), and Pb(II) from Mississippi River water. Though we are in a relatively early stage of development, our experimental data reported here open up a new possibility for effective removal of pathogenic bacteria and toxic metals from environmental samples. Before the membrane can be used for society, vigorous research needs to be conducted to find a cost-effective process for large-scale development and methods to improve the long-term performance.
Acknowledgments
P.C.R. thanks NSF-PREM (grant no. DMR-1205194) and NIH RCMI (grant no. G12MD0007581) for their generous funding. F.P. thanks NIH RCMI (grant no. G12 MD007591) for his fellowship.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Drinking Water Chlorination: A Review of Disinfection Practices and Issues. http://www.waterandhealth.org/drinkingwater/wp.html, date of access 03/12/2015.
- 2.Billions affected daily by water and sanitation crisis. http://water.org/water-crisis/one-billion-affected/, date of access 03/12/2015.
- 3.Health through safe drinking water and basic sanitation. http://www.who.int/water_sanitation_health/mdg1/en/, date of access 03/12/2015.
- 4.Health Topics: Arsenic. http://www.who.int/topics/arsenic/en/, date of access 03/12/2015.
- 5.Pal S, Joardar J, Song J. Removal of E. coli from Water Using Surface-Modified Activated Carbon Filter Media and Its Performance over an Extended Use. Environ Sci Technol. 2006;40:6091–6097. doi: 10.1021/es060708z. [DOI] [PubMed] [Google Scholar]
- 6.Deng D, Zhang N, Mustapha A, Xu D, Wuliji T, Farley M, Yang J, Hua B, Liu F, Zheng G. Differentiating Enteric Escherichia coli from Environmental Bacteria Through the Putative Glucosyltransferase Gene. Water Res. 2014;61:224–231. doi: 10.1016/j.watres.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Fu FL, Wang QJ. Removal of Heavy Metal Ions From Wastewaters: A review. J Environ Manage. 2011;92:407–418. doi: 10.1016/j.jenvman.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborti D, Rahman MM, Das B, Murrill M, Dey S, Mukherjee SC, Dhar RK, Biswas BK, Chowdhury UK, Roy S. Status of Groundwater Arsenic Contamination in Bangladesh: A 14-year Study Report. Water Res. 2010;44:5789–5802. doi: 10.1016/j.watres.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 9.Scott N, Hatlelid KM, MacKenzie NE, Carter DE. Reactions of Arsenic(III) and Arsenic(V) Species with Glutathione. Chem Res Toxicol. 1993;6(10):102–106. doi: 10.1021/tx00031a016. [DOI] [PubMed] [Google Scholar]
- 10.Spuches AM, Kruszyna HG, Rich AM, Wilcox DE. Thermodynamics of the As(III)–Thiol Interaction: Arsenite and Monomethylarsenite Complexes with Glutathione, Dihydrolipoic Acid, and other Thiol Ligands. Inorg Chem. 2005;44:2964–2972. doi: 10.1021/ic048694q. [DOI] [PubMed] [Google Scholar]
- 11.Shen SW, Li XF, Cullen WR, Weinfeld M, Le XC. Arsenic Binding to Proteins. Chem Rev. 2013;113:7769–7792. doi: 10.1021/cr300015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri JR, Arbneshi T, Khan SA, Neely A, Candice P, Varisli BM, Washington MS, Robinson B, Banerjee S, Singh AK, Senapati D, Ray PC. Use of Gold Nanoparticles in a Simple Colorimetric and Ultrasensitive Dynamic Light Scattering Assay: Selective Detection of Arsenic in Groundwater. Angew Chem, Int Ed. 2009;48:9668–9671. doi: 10.1002/anie.200903958. [DOI] [PubMed] [Google Scholar]
- 13.Mah V, Jalilehvand F. Lead(II) Complex Formation with Glutathione. Inorg Chem. 2012;51:6285–6298. doi: 10.1021/ic300496t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai F, Wang CA, Wang TT, Li L, Su ZM. Colorimetric Detection of Pb2+ Using Glutathione Functionalized Gold Nanoparticles. ACS Appl Mater Interfaces. 2010;2:1466–1470. doi: 10.1021/am100107k. [DOI] [PubMed] [Google Scholar]
- 15.Beqa L, Singh AK, Khan SA, Senapati D, Arumugam SR, Ray PC. Gold Nanoparticle-Based Simple Colorimetric and Ultrasensitive Dynamic Light Scattering Assay for the Selective Detection of Pb(II) from Paints, Plastics, and Water Samples. ACS Appl Mater Interfaces. 2011;3:668–673. doi: 10.1021/am101118h. [DOI] [PubMed] [Google Scholar]
- 16.Geim AK, Novoselov KS. The Rise of Graphene. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Wang X, Zhang L, Lee S, Dai H. Chemically Derived, Ultrasmooth Graphene Nanoribbon Semiconductors. Science. 2008;319:1229–1232. doi: 10.1126/science.1150878. [DOI] [PubMed] [Google Scholar]
- 18.Gao W, Alemany LB, Ci L, Ajayan PM. New Insights into the Structure and Reduction of Graphite Oxide. Nat Chem. 2009;1:403–408. doi: 10.1038/nchem.281. [DOI] [PubMed] [Google Scholar]
- 19.Sun P, Zheng F, Zhu M, Song Z, Wang K, Zhong M, Wu D, Little RB, Xu Z, Zhu H. Selective Trans-membrane Transport of Alkali and Alkaline Earth Cations Through Graphene Oxide Membranes Based on Cation-π Interactions. ACS Nano. 2014;8:850–859. doi: 10.1021/nn4055682. [DOI] [PubMed] [Google Scholar]
- 20.Mi B. Graphene Oxide Membranes for Ionic and Molecular Sieving. Science. 2014;343:740–742. doi: 10.1126/science.1250247. [DOI] [PubMed] [Google Scholar]
- 21.Viraka Nellore BP, Kanchanapally R, Pramanik A, Sinha SS, Chavva SR, Hamme A, Ray PC. Aptamer-Conjugated Graphene Oxide Membranes for Highly Efficient Capture and Accurate Identification of Multiple Types of Circulating Tumor Cells. Bioconjugate Chem. 2015;26:235–242. doi: 10.1021/bc500503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun P, Zheng F, Zhu M, Song Z, Wang K, Zhong M, Wu D, Little RB, Xu Z, Zhu H. Selective Trans-Membrane Transport of Alkali and Alkaline Earth Cations through Graphene Oxide Membranes Based on Cation-π Interactions. ACS Nano. 2014;8:850–859. doi: 10.1021/nn4055682. [DOI] [PubMed] [Google Scholar]
- 23.Nair NN, Wu HA, Jayaram PN, Grigorieva IV, Geim AK. Unimpeded Permeation of Water Through Helium-Leak-Tight Graphene-Based Membraness. Science. 2012;335:442–444. doi: 10.1126/science.1211694. [DOI] [PubMed] [Google Scholar]
- 24.Han Y, Xu Z, Gao C. Ultrathin Graphene Nanofiltration Membrane for Water Purification. Adv Funct Mater. 2013;23:3693–3700. [Google Scholar]
- 25.Fan Z, Yust B, Nellore BOV, Sinha SS, Kanchanapally R, Crouch RA, Pramanik A, Reddy SC, Sardar D, Ray PC. Accurate Identification and Selective Removal of Rotavirus Using a Plasmonic–Magnetic 3D Graphene Oxide Architecture. J Phys Chem Lett. 2014;5:3216–3221. doi: 10.1021/jz501402b. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Song Z, Zhang X, Huang Y, Li S, Mao Y, Ploehn HJ, Bao Y, Yu M. Ultrathin, Molecular-Sieving Graphene Oxide Membraness for Selective Hydrogen Separation. Science. 2013;342:95–98. doi: 10.1126/science.1236686. [DOI] [PubMed] [Google Scholar]
- 27.Joshi RK, Carbone P, Wang FC, Kravets VG, Su Y, Grigorieva IV, Wu HA, Geim AK, Nair RR. Precise and Ultrafast Molecular Sieving through Graphene Oxide Membraness. Science. 2014;343:752–754. doi: 10.1126/science.1245711. [DOI] [PubMed] [Google Scholar]
- 28.Kim HW, Yoon HW, Yoon SM, Yoo BM, Ahn BK, Cho YH, Shin HJ, Yang H, Paik U, Kwon S. Selective Gas Transport Through Few-Layered Graphene and Graphene Oxide Membraness. Science. 2013;342:91–95. doi: 10.1126/science.1236098. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh T, Biswas C, Oh J, Arabale G, Hwang T, Luong ND, Jin M, Lee YH, Nam JD. Solution-Processed Graphite Membrane from Reassembled Graphene Oxide. Chem Mater. 2012;24:594–599. [Google Scholar]
- 30.Zhao G, Jiang L, He Y, Li J, Dong H, Wang X, Hu W. Sulfonated Graphene for Persistent Aromatic Pollutant Management. Adv Mater. 2011;23:3959–3963. doi: 10.1002/adma.201101007. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Tang Z-R, Fu X, Xu Y-J. TiO2–Graphene Nanocomposites for Gas-phase Photocatalytic Degradation of Volatile Aromatic Pollutant: Is TiO2–Graphene Truly Different from other TiO2–Carbon Composite Materials? ACS Nano. 2010;4:7303–7314. doi: 10.1021/nn1024219. [DOI] [PubMed] [Google Scholar]
- 32.Koenig SP, Wang L, Pellegrino J, Bunch JS. Selective Molecular Sieving through Porous Graphene. Nat Nanotechnol. 2012;7:728–732. doi: 10.1038/nnano.2012.162. [DOI] [PubMed] [Google Scholar]
- 33.Kanchanapally R, Viraka Nellore BP, Sinha SS, Pedraza F, Jones SJ, Pramanik A, Chavva SR, Tchounwou C, Shi Y, Vangara A, Sardar D, Ray PC. Antimicrobial Peptide-Conjugated Graphene Oxide Membrane for Efficient Removal and Effective Killing of Multiple Drug Resistant Bacteria. RSC Adv. 2015;5:18881–18887. doi: 10.1039/C5RA01321F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei N, Peng X, Xu Z. Understanding Water Permeation in Graphene Oxide Membranes. ACS Appl Mater Interfaces. 2014;6:5877–5883. doi: 10.1021/am500777b. [DOI] [PubMed] [Google Scholar]
- 35.Vadahanambi S, Lee SH, Kim WJ, Oh IK. Arsenic Removal from Contaminated Water Using Three-Dimensional Graphene-Carbon Nanotube-Iron Oxide Nanostructures. Environ Sci Technol. 2013;47:10510–10517. doi: 10.1021/es401389g. [DOI] [PubMed] [Google Scholar]
- 36.Tian TF, Shi XZ, Cheng L, Luo YC, Dong ZL, Gong H, Xu LG, Zhong ZT, Peng R, Liu Z. Graphene-Based Nanocomposite as an Effective, Multifunctional, and Recyclable Antibacterial Agent. ACS Appl Mater Interfaces. 2014;6:8542–8548. doi: 10.1021/am5022914. [DOI] [PubMed] [Google Scholar]
- 37.Hu W, Peng C, Luo W, Lv M, Li X, Li D, Huang Q, Fan C. Graphene-Based Antibacterial Paper. ACS Nano. 2010;4:4317–4323. doi: 10.1021/nn101097v. [DOI] [PubMed] [Google Scholar]
- 38.Madadrang CJ, Kim HY, Gao GH, Wang N, Zhu J, Feng H, Gorring M, Kasner ML, Hou SF. Adsorption Behavior of EDTA-Graphene Oxide for Pb(II) Removal. ACS Appl Mater Interfaces. 2012;4:1186–1193. doi: 10.1021/am201645g. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Nair RR, Pillai PB, Gupta SN, Lyengar MAR, Sood AK. Graphene Oxide-MnFe2O4 Magnetic Nanohybrids for Efficient Removal of Lead and Arsenic from Water. ACS Appl Mater Interfaces. 2014;6:17426–17436. doi: 10.1021/am504826q. [DOI] [PubMed] [Google Scholar]
- 40.De Volder MFL, Tawfick SH, Baughman RH, Hart AJ. Carbon Nanotubes: Present and Future Commercial Applications. Science. 2013;339:535–539. doi: 10.1126/science.1222453. [DOI] [PubMed] [Google Scholar]
- 41.Hu LB, Hecht DS, Grüner G. Carbon Nanotube Thin Films: Fabrication, Properties, and Applications. Chem Rev. 2010;110:5790–5844. doi: 10.1021/cr9002962. [DOI] [PubMed] [Google Scholar]
- 42.Schnorr JM, Swager TM. Emerging Applications of Carbon Nanotubes. Chem Mater. 2011;23:646–657. [Google Scholar]
- 43.Beqa L, Fan Z, Singh AK, Senapati D, Ray PC. Gold Nano-Popcorn Attached SWCNT Hybrid Nanomaterial for Targeted Diagnosis and Photothermal Therapy of Human Breast Cancer Cells. ACS Appl Mater Interfaces. 2011;3:3316–3324. doi: 10.1021/am2004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tung VC, Chen LM, Allen MJ, Wassei JK, Nelson K, Kaner RB, Yang Y. Low-Temperature Solution Processing of Graphene–Carbon Nanotube Hybrid Materials for High-Performance Transparent Conductors. Nano Lett. 2009;9:1949–1955. doi: 10.1021/nl9001525. [DOI] [PubMed] [Google Scholar]
- 45.Huang JH, Fang J-H, Liu C-C, Chu C-W. Effective Work Function Modulation of Graphene/Carbon Nanotube Composite Films As Transparent Cathodes for Organic Optoelectronics. ACS Nano. 2011;5:6262–627. doi: 10.1021/nn201253w. [DOI] [PubMed] [Google Scholar]
- 46.Cheng YW, Lu ST, Zhang HB, Varanasi V, Liu J. Synergistic Effects from Graphene and Carbon Nanotubes Enable Flexible and Robust Electrodes for High-Performance Supercapacitors. Nano Lett. 2012;12:4206–4211. doi: 10.1021/nl301804c. [DOI] [PubMed] [Google Scholar]
- 47.Chen P, Xiao TY, Qian YH, Li SS, Yu SH. A Nitrogen-Doped Graphene/Carbon Nanotube Nanocomposite with Synergistically Enhanced Electrochemical Activity. Adv Mater. 2013;25:3192–3196. doi: 10.1002/adma.201300515. [DOI] [PubMed] [Google Scholar]
- 48.Tu Y, Lv M, Xiu P, Huynh T, Zhang M, Castelli M, Liu Z, Huang Q, Fan C, Fang H. Destructive Extraction of Phospholipids from Escherichia coli Membranes by Graphene Nanosheets. Nat Nanotechnol. 2013;8:594–601. doi: 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- 49.Melo MN, Ferre R, Castanho MARB. Antimicrobial Peptides: Linking Partition, Activity and High Membrane-bound Concentrations. Nat Rev Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 50.da Silva A, Jr, Teschke O. Effects of the Antimicrobial Peptide PGLa on Live Escherichia coli. Biochim Biophys Acta, Mol Cell Res. 2003;1643:95–103. doi: 10.1016/j.bbamcr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Nishida M, Imura Y, Yamamoto M, Kobayashi S, Yano Y, Matsuzaki K. Interaction of a Magainin-PGLa Hybrid Peptide With Membranes: Insight Into the Mechanism of Synergism. Biochemistry. 2007;46:14284–14290. doi: 10.1021/bi701850m. [DOI] [PubMed] [Google Scholar]
- 52.Jothikumar N, Griffiths MW. Rapid Detection of Escherichia coli O157:H7 with Multiplex Real-Time PCR Assays. Appl Environ Microbiol. 2002;68:3169–3171. doi: 10.1128/AEM.68.6.3169-3171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ibekwe AM, Watt PM, Grieve CM, Sharma VK, Lyons SR. Multiplex Fluorogenic Real-Time PCR for Detection and Quantification of Escherichia coli O157:H7 in Dairy Wastewater Wetlands. Appl Environ Microbiol. 2002;68:4853–4862. doi: 10.1128/AEM.68.10.4853-4862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


