Abstract
Despite the importance of signaling lipids, many questions remain about their function because we have few tools to chart lipid gradients in vivo. Here we describe a sphingosine 1-phosphate (S1P) reporter mouse, and use this mouse to define S1P distribution in the spleen. Surprisingly, the presence of blood does not predict the concentration of signaling-available S1P. Large areas of the red pulp are S1P-low, while S1P can be sensed by cells inside the white pulp near the marginal sinus. Lipid phosphate phosphatase 3 maintains low S1P concentrations in the spleen, and enables efficient marginal zone B cell shuttling. The exquisitely tight regulation of S1P availability may explain how a single lipid can simultaneously orchestrate many immune cell movements.
Introduction
Signaling lipids direct many steps of immune cell migration and activation7. For example, eicosanoids are among the first signals directing innate immune cells to a site of injury1. The efficiency of T cell entry into lymph nodes is increased by lysophosphatidic acid, which regulates both T and endothelial cell motility2,3. Within lymphoid organs, thromboxane A2 modulates T cell interactions with dendritic cells, and 7α-25 hydroxycholesterol recruits activated B cells to the rim of the follicles and positions dendritic cells in splenic bridging channels4–6. Lymphocyte exit from lymphoid organs into circulation is guided by sphingosine 1-phosphate (S1P)7. Yet despite the clear importance of signaling lipids, many questions remain about their function, in part because we have few tools to map their location in vivo.
A series of elegant studies has shed light on the distribution of protein chemokines by gene targeting fluorescent reporters into the chemokine-coding locus8–10. But lipids are not encoded genetically, and the abundance of signaling-available lipids is determined by the balance of the activity of synthetic enzymes, degrading enzymes and transporters. For example, S1P is made intracellularly by two sphingosine kinases, degraded intracellularly by two phosphatases and S1P lyase, exported out of the cell by the transporter SPNS2 and at least one additional unidentified transporter, and degraded extracellularly by three phosphatases11. Because of the complexity of the pathway, expression of any one of these enzymes is not predictive. Mass spectrometry has been widely used to quantify lipids, but measurements in tissues cannot distinguish extracellular lipid from intracellular lipid and lipid in the inner leaflet of the plasma membrane. Intracellular lipid can substantially confound interpretation, because many lipids act both extracellularly as ligands for cell-surface receptors and intracellularly as metabolic intermediates and protein co-factors. For example, extracellular S1P guides immune cell migration by signaling through G protein-coupled receptors. At the same time, intracellular phosphorylation of sphingosine to S1P and subsequent cleavage by S1P lyase at the ER membrane are the final two steps in degradation of sphingolipids, and S1P may bind and alter the function of diverse cytoplasmic and nuclear proteins12; in many cases, the concentration of total tissue S1P has been reported to be high7. Although signaling molecules have in some settings been measured in interstitial fluid extracted by a probe, synthesis of many signaling lipids is induced in response to inflammatory stimuli, making insertion of a needle problematic. For example, the mitogen-activated kinase Erk1/2 phosphorylates sphingosine kinase 1 and increases its catalytic activity13. Moreover, it is not clear how much extracellular lipid is sequestered by proteins or available to bind receptors.
A combination of techniques has given insight into the distribution of S1P, but many questions about S1P’s location and function remain unanswered. There is good evidence that the concentration of S1P is high in lymph and blood compared to the interstitial fluid of lymphoid organs, and this differential guides lymphocytes out of lymphoid organs into circulation7. Two types of experiment support the existence of this gradient. First, mass spectrometry measurements of S1P in blood plasma and lymph fluid report concentrations in the 100 nM-1 μM range, well above the 0.5 nM IC50 of S1P receptor 1 (S1PR1)7,14. Second, S1PR1 is low on the surface of naïve lymphocytes in blood and lymph, while it is abundant on the surface of the same cells in lymphoid organs. S1PR1, like many G protein-coupled receptors, is internalized in response to binding ligand15,16. In the absence of other transcriptional, translational or post-translational changes in receptor expression, differences in surface expression of S1PR1 reflect differences in exposure to S1P. This inference has been validated in many settings for naïve lymphocytes trafficking among lymphoid organs in homeostasis7.
However, these techniques have substantial limitations. First, inflammation induces a fascinating array of transcriptional and post-translational modifications to S1PR1, so in this setting surface expression of S1PR1 no longer simply reflects exposure to S1P7. Hence we have little understanding of how the concentration of extracellular, signaling-available S1P changes over the course of an immune response in lymphoid or non-lymphoid tissues, although S1P has been hypothesized to increase in inflamed tissues and to alter both trafficking and cytokine responses of immune cells17. Second, these techniques offer little insight into the shape of S1P gradients within tissues. S1P has been suggested to orchestrate many cell movements simultaneously, although it can be difficult to reconcile the intricate dance with our crude understanding of S1P distribution.
The spleen is a good example of the problem of measuring S1P in tissues18,19. The spleen can be considered a blood filter. Splenic arterioles terminate in the marginal sinus, which is highly fenestrated. Blood travels out into the marginal zone, where marginal zone B cells and macrophages capture antigen for presentation to lymphocytes in the white pulp. Blood flow continues into the red pulp, where red blood cells are tested for abnormalities before they return to circulation in the venous sinuses. The concentration of S1P is thought to be low in the white pulp, based on the high expression of S1PR1 on the surface of naïve lymphocytes in the spleen. The marginal zone and red pulp are thought to have high S1P, based on the high concentration of S1P in blood and the abundance of red blood cells, which are the main producers of blood S1P20,21. But this simple dichotomy cannot explain many of the migratory steps that S1P is thought to direct within the spleen. S1P receptors have been implicated in confining B and T cells in germinal centers in the B cell follicles within the white pulp, shuttling marginal zone B cells from the white pulp to the marginal zone, directing lymphocyte cell exit from the white pulp to circulation, partitioning dendritic cells between the red pulp bridging channels and marginal zone, and enabling plasma cell and hematopoietic stem cell exit from the red pulp into venous sinuses20,22–26,24.
Here, we report a tool to map signaling-available S1P in tissues, using a strategy that may be widely applicable to GPCR ligands. We use this tool to map S1P in the spleen and define how its distribution is regulated.
Results
Generation of an S1P reporter mouse
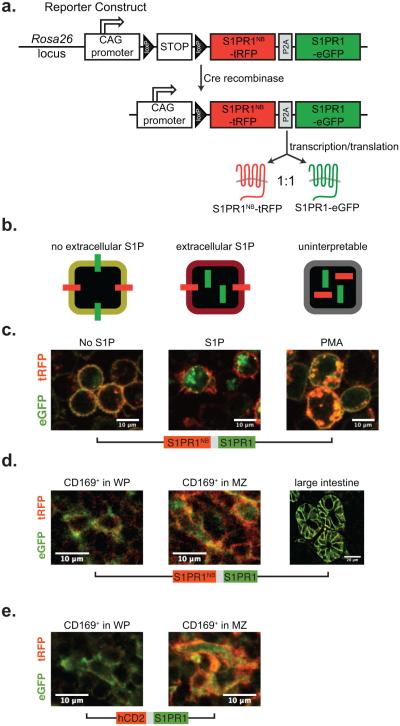
We set out to generate a mouse expressing a reporter that would enable us to map signaling-available S1P (S1P that is both extracellular and free to bind receptors rather than sequestered) in tissues (Fig. 1a,b). The core of the reporter is S1PR1 itself, fused to GFP. In the absence of S1P, S1PR1-GFP should sit on the cell surface; in the presence of S1P, S1PR1-GFP should be internalized in endosomes. As a control, the reporter also encodes a mutant version of S1PR1 with a single amino acid change (R120 to A) in the S1P binding pocket that prevents the receptor from binding S1P27. This mutant S1PR1, which we refer to as S1PR1NB (NB, non-binding), is fused to TagRFP. S1PR1NB should sit on the cell surface regardless of the presence of S1P, and serves three purposes: to mark the plasma membrane, to enable measurement of the ratio of S1PR1-GFP: S1PR1NB-RFP on the plasma membrane, and to flag situations in which there is ligand-independent internalization of S1PR1. The two receptors are separated by a P2A ribosomal skip sequence, so that they are subject to the same translational control. The reporter is knocked into the Rosa26 locus, and transcription is driven by the synthetic CAG promoter28. The reporter is preceded by a floxed transcriptional stop, to avoid potential developmental artifacts caused by overexpression of S1PR1.
Figure 1. Design of the S1P reporter.
(a) Diagram of the S1P reporter construct.
(b) Diagram showing the predicted response to S1P by cells expressing the reporter.
(c) A B cell line (WEHI-231) was retrovirally transduced with the reporter and treated with dimethyl sulfoxide (DMSO), 1 μM S1P or 1 μM phorbol 12-myristate 13-acetate (PMA). Cells were visualized by confocal microscopy. Data are representative of 2 experiments.
(d) Mice were generated expressing the S1P reporter, and organs were visualized by confocal microscopy. Representative CD169+ macrophages deep in the splenic white pulp (left), CD169+ macrophages in the splenic marginal zone (center), and epithelial cells lining large intestine crypts (right). Data are representative of sections from 5 mice for macrophages, and 2 mice for epithelial cells.
(e) A second S1P reporter mouse was made in which human CD2 (hCD2)-TagRFP replaces S1PR1NB-TagRFP, and lymphoid organs were visualized by confocal microscopy. Representative CD169+ macrophages deep in the splenic white pulp (left) and CD169+ macrophages in the splenic marginal zone (center). Data are representative of sections from 7 mice.
Before making a mouse, we validated our strategy in cell culture (Fig. 1c). We retrovirally transduced a B cell lymphoma line with the reporter. As expected, in the absence of S1P, both S1PR1-GFP and S1PR1NB-RFP sat on the cell surface. In the presence of S1P at a concentration comparable to that in blood, S1PR1-GFP was internalized while S1PR1NB-RFP remained on the surface. Upon treatment of the cells with PMA, which activates protein kinase C and in turn causes ligand-independent internalization of many G protein-coupled receptors, both S1PR1-GFP and S1PR1NB-RFP were internalized29. The only situation we encountered in which S1PR1NB-RFP did not flag ligand-independent trafficking of S1PR1 was CD69-mediated internalization (not shown and 30).
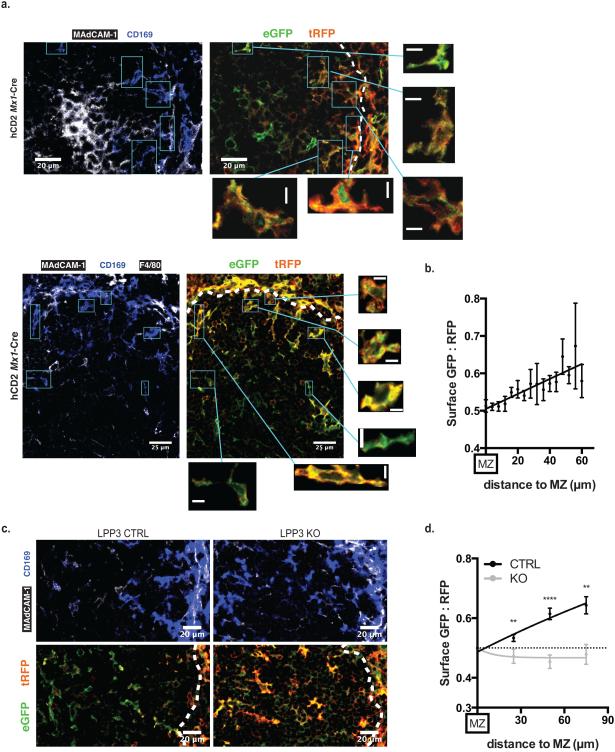
We then generated a mouse expressing the reporter (Fig. 1d, Supplementary Fig. 1a). In our initial experiments, we used Cre driven by the Mx1 promoter to induce transcription of the reporter31. Treatment of pups with polyI:C 3–5 days after birth resulted in widespread reporter expression in cells including subsets of macrophages, endothelial cells, epithelial cells and stromal cells. We were unable to detect the reporter in lymphocytes. As expected, deep in the white pulp of the spleen, cells displayed both S1PR1-GFP and S1PR1NB-RFP on the surface. In the blood-rich marginal zone of the spleen, cells displayed S1PR1NB-RFP on the surface, while much S1PR1-GFP was internalized. In the spleen in homeostasis, we did not detect any instances in which S1PR1NB-RFP failed to localize at the cell surface. Interestingly, neither S1PR1-GFP nor S1PR1NB-RFP trafficked to the luminal side of epithelial cells lining large intestine crypts; although based on S1PR1-GFP alone we might have concluded that S1P was high in the intestinal mucus, S1PR1NB-RFP prevented us from making that erroneous conclusion, and this example validated the need for the S1PR1NB-RFP control in vivo.
We also made a second mouse, in which the plasma membrane protein human CD2-RFP substitutes for S1PR1NB-RFP as an internal control (Fig. 1e, Supplementary Fig. 1b). Human CD2 (hCD2)-RFP lacks 100 amino acids in the hCD2 cytoplasmic tail required for signaling32. The hCD2-RFP reporter was more robustly expressed by many cell types in the spleen than the S1PR1NB-RFP reporter. Therefore, in instances in which the two reporters were qualitatively equivalent, we used the hCD2 reporter for analysis. Reporting cells in the spleen of these animals fell into three categories: those with red membranes, often with green endosomes (detection of endosomes depends on the plane of imaging), for example, CD169+ macrophages in the marginal zone sensing abundant S1P (Fig. 1e); those with yellow membranes, sensing less S1P; those with green membranes (these cells also have hCD2-RFP on the membrane, but appear green due to the microscope and software settings, which were chosen to reveal the greatest range of reporting), for example, CD169+ macrophages deep in the white pulp sensing very little S1P (Fig. 1e, Supplementary Fig. 1b). These observations suggested that the S1P reporter would be an informative tool to measure S1P distribution.
Analysis of S1P distribution in the red pulp
The spleen was an attractive organ for the first application of the S1P reporter. While some aspects of S1P distribution in the spleen are well-established and allow validation of the reporter, many questions remain unanswered. As described above, the white pulp is thought to be S1P-low, while the blood-rich marginal zone and red pulp are thought to have high concentrations of S1P (Supplementary Fig. 1c). In sections, the three regions can be delineated by staining with antibodies to MAdCAM1 and F4/80. The boundary between the marginal zone and the white pulp is marked by MAdCAM1, which is highly expressed by the marginal sinus (and expressed less abundantly on some stromal cells in the B follicle). The red pulp is marked by F4/80, which is highly expressed by red pulp macrophages.
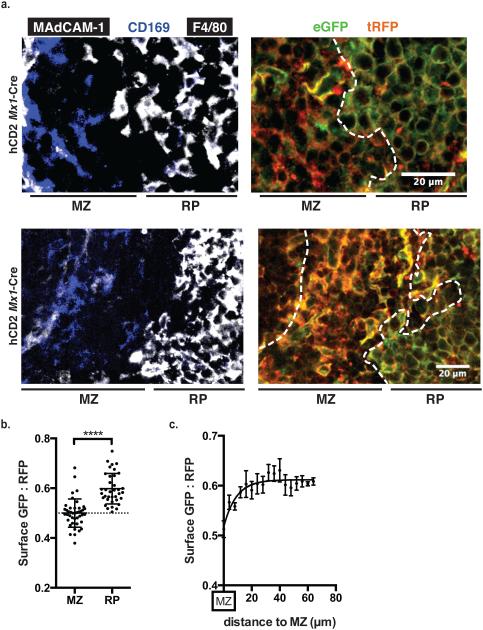
While our S1P reporter mouse confirmed the presence of S1P in the marginal zone, it also revealed, to our surprise, that many regions of the red pulp have little signaling-available S1P. S1PR1-GFP was partially internalized by CD169+ macrophages in the marginal zone, while it was largely on the cell surface of F4/80+ macrophages in the red pulp, particularly in areas of small densely clustered macrophages (Fig. 2a; Supplementary Figs. 2,3a). This observation was reflected in a lower GFP:RFP ratio on the surface of macrophages in the marginal zone than the red pulp (Fig. 2b; Supplementary Fig. 3b,c). Moreover, the decrease in S1P was remarkably sharp at the boundary between the marginal zone and the red pulp, although no physical barrier has been reported between the two regions (Fig. 2a,c). Interestingly, the RFP signal was somewhat more diffuse on some macrophages in the marginal zone than the red pulp; we suspect that this reflects the more elongated shape of macrophages in the marginal zone, which led us more frequently to image a sheet of membrane rather than a crisp edge.
Figure 2. The red pulp has little signaling-available S1P.
(a) Two representative spleen sections from S1P reporter animals (hCD2, Mx1-Cre). MZ, marginal zone. RP, red pulp. For each section we acquired a 4-color image (GFP, RFP, CD169, and F4/80-MAdCAM1), but for clarity we show GFP and RFP stains on the right, and CD169 and F4/80-MAdCAM1 stains on the left. F4/80 and MAdCAM-1 were both stained with Pacific Blue-conjugated antibodies, and both are shown in white. Although the two antibodies are labeled with the same fluorophore, their signals can readily be distinguished by the morphology and location of the stained cells. Data are representative of sections from 7 mice; 3 additional examples are shown in Figures S2 and S3a. High-resolution files of these images are available at Figshare.com, http://figshare.com/s/378978342fdb11e5a4bb06ec4b8d1f61 and http://figshare.com/s/dd3167582fde11e587a606ec4b8d1f61.
(b) Quantification of S1P reporting in marginal zone and red pulp. F4/80+RFP+ and CD169+ RFP+ pixels were identified, marking the surface of reporter-expressing macrophage subsets (F4/80+ primarily in the red pulp, and CD169+ primarily in the marginal zone and white pulp); the ratio of GFP:RFP was measured for each F4/80+RFP+ or CD169+RFP+ pixel; and this ratio was averaged over a defined area. Each MZ point represents the average ratio over an area of at least 2×103 square microns, each RP point represents the average ratio over an area of at least 1×104 square microns, and the graph compiles areas taken from 3 mice. Bars indicate mean, error bars show SEM. Comparisons were performed using a 2-tailed unpaired Student’s t-test; ****, p<0.0001. To account for different microscope settings on different days, for each mouse all ratios were normalized such that the average ratio for the marginal zone was 0.5.
(c) Quantification of S1P reporting in the red pulp at the indicated distances from the marginal zone-red pulp border. 4 μm-thick bands were drawn from the marginal zone-red pulp border, and S1P reporting within each band was calculated as in Fig. 2b. Graph compiles 92 points from 5 sections from a single mouse (without normalization), and is representative of 3 mice. Points indicate mean, error bars show SEM. The data can be fit with the equation y=0.52+0.024*ln(x), where y=surface GFP:RFP and x=distance from the MZ. A Student's t-test of the hypothesis that GFP:RFP does not vary with distance (that the true slope of the curve is zero) yields a probability of <0.0001.
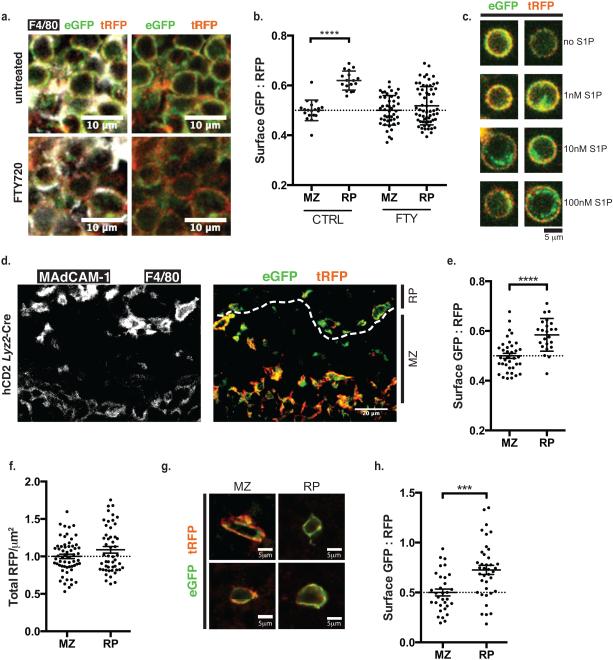
To assess whether red pulp macrophages were faithful reporters of S1P, we took several approaches. First, we treated mice with FTY720, which is phosphorylated in vivo and binds S1PR1 (ref. 7); we found that FTY720 induced internalization of S1PR1-GFP in F4/80+ macrophages within the red pulp (Fig. 3a,b). Second, we purified F4/80+ macrophages from the spleen and incubated them with S1P ex vivo; these cells internalized S1PR1-GFP in response to increasing concentrations of S1P (Fig. 3c). Third, we were concerned that widespread over-expression of S1PR1 might in itself lower concentrations of signaling-available S1P by binding available lipid. But much more restricted expression of the reporter driven by Lyz2-Cre revealed the same pattern of S1P distribution as widespread reporter expression driven by Mx1-Cre (Fig. 3d,e)33. We also quantified the level of reporter expression in marginal zone and red pulp, and found that it was comparable (Fig. 3f). Fourth, we induced expression of the reporter shortly before analysis using Ubc-CreERT2, and observed the same pattern of S1P distribution (Supplementary Fig. 3d). Finally, we validated the pattern using a second cell type. We transduced primary B cells with the S1P reporter and transferred them to wild-type recipients, and found that B cells in the marginal zone internalized S1PR1-GFP more frequently than those in the red pulp (Fig. 3g,h).
Figure 3. Red pulp macrophages faithfully report signaling-available S1P.
(a) A representative section showing red pulp macrophages in a mouse treated with FTY720 and control. Data are representative of sections from 2 pairs of mice.
(b) Quantification of S1P reporting in marginal zone and red pulp of FTY720-treated mice and controls, calculated as in Fig. 2b. Each MZ point represents the average surface GFP:RFP ratio over an area of at least 1×103 square microns, each RP point represents an area of at least 4×103 square microns, and the graph compiles areas taken from 3 control and 2 FTY-treated mice.
(c) F4/80+ splenic macrophages were incubated ex vivo for 45 minutes with the indicated concentrations of S1P. Image shows fluorescence without antibody amplification. Data are representative of 3 experiments (two sets sorted on F4/80, one on CD11b).
(d) A representative spleen section from an S1P reporter mouse (hCD2, Lyz2-Cre). Data are representative of sections from 5 mice; an additional example is shown in Figure S3a.
(e) Quantification of S1P reporting in marginal zone and red pulp of mice expressing the S1P reporter and Lyz2-Cre. RFP+ pixels were identified, marking the surface of reporter-expressing Lyz2-Cre+ cells; the ratio of GFP:RFP was measured for each RFP+ pixel; and this ratio was averaged over a defined area. Each MZ point represents the average ratio over an area of at least 4×102 square microns, each RP point represents an area of at least 1×103 square microns, and the graph compiles areas taken from 2 mice. To account for different microscope settings on different days, for each mouse all ratios were normalized such that the average ratio for the marginal zone was 0.5.
(f) Quantification of reporter density in red pulp and marginal zone. For the areas analyzed in Fig. 2b, the average RFP intensity/μm2 was calculated. To account for different microscope settings on different days, the densities for each mouse were normalized such that the average density for the marginal zone was 1.0.
(g,h) B cells were retrovirally transduced with the S1P reporter (hCD2 version) and transferred into a wild-type recipient. 1 day after transfer, S1P reporting was analyzed. Three experiments were performed, with identical results: one with CD19+ B cells from a wild-type donor, one with CD19+ B cells from a Cd69−/− donor, and one with B220+CD23hiCD1dlo follicular B cells from a Cd69−/− donor. The transduced cells were >94% B220+. (g) Representative B cells in the marginal zone and red pulp from the two Cd69−/− donors. (h) Quantification of reporting by B cells in marginal zone and red pulp. For each B cell, the ratio of GFP:RFP was measured for each RFP+ pixel, and this ratio was averaged over the cell. Each point represents one cell, and cells were included from 3 experiments. To account for different microscope settings on different days, for each mouse all ratios were normalized such that the average ratio for the marginal zone was 0.5.
For all panels, bars indicate mean, error bars show SEM. Comparisons were performed using a 2-tailed unpaired Student’s t-test; ***, p<0.001; ****, p<0.0001.
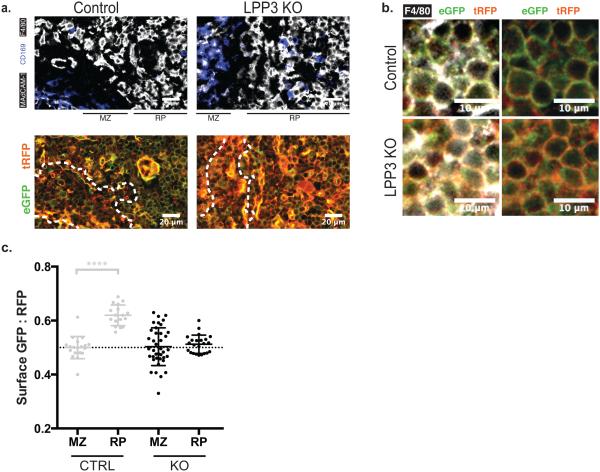
To determine how the boundary between the marginal zone and red pulp was maintained in the absence of a physical barrier, we considered the possibility that S1P-degrading enzymes in the red pulp rapidly remove S1P entering from the marginal zone. Six S1P-degrading enzymes are known (Supplementary Fig. 3e)11. The lipid phosphate phosphatases are of particular interest, as they sit on the plasma membrane with their active sites in the extracellular space, making them well-positioned to shape S1P gradients. We had previously found that lipid phosphate phosphatase 3 (LPP3) maintains low concentrations of S1P in the thymus, and here we asked whether LPP3 had any function in the spleen34. We bred our S1P reporter onto an LPP3-deficient background (Ppap2bf/f: Reporter+: Mx1-Cre+, treated with polyI:C 3–5 days after birth)35, and found that in the absence of LPP3 S1P rises in the red pulp (Fig. 4a). Importantly, internalization of S1PR1-GFP was evident both when we compared broad views of the red pulp (Fig. 4a) and when we specifically compared F4/80+ macrophages in control and LPP3-deficient animals (Fig. 4b). The increased S1P was reflected in a lower ratio of GFP:RFP on the surface of macrophages in the red pulp of LPP3-deficient mice compared to controls (Fig. 4c).
Figure 4. LPP3 maintains low red pulp S1P.
(a) Representative sections of spleen from an LPP3-deficient mouse and control. Data are representative of 3 pairs of mice.
(b) Representative sections of spleen from an LPP3-deficient mouse and control, focusing on F4/80+ macrophages. Data are representative of 3 pairs of mice.
(c) Quantification of S1P reporting in marginal zone and red pulp of LPP3-deficient mice and controls. Calculations were performed as in Fig. 2b. Each MZ point represents the average surface GFP:RFP ratio over an area of at least 1.4×103 square microns, each RP point represents an area of at least 3×103 square microns, and the graph compiles areas taken from 3 control mice and 2 LPP3-deficient mice. The mice were analyzed in groups of three (control, LPP3-deficient, and FTY720-treated control), so the controls are the same in Fig. 3b and 4c; the repeated control data is shown in grey in Fig. 4c. Bars indicate mean, error bars show SEM. Comparisons were performed using a 2-tailed unpaired Student’s t-test; ****, p<0.0001.
The finding that S1P is low in the red pulp may explain the observations that plasma cells and hematopoietic stem cells require S1PR1 to exit the red pulp into the venous sinuses25,26; these cells may simply be following an S1P gradient out of the organ. The requirement for LPP3 to separate the marginal zone from the red pulp cements the importance of LPP3 in regulating signaling-available S1P in vivo.
Analysis of S1P distribution in the white pulp
Our S1P reporter mouse confirmed that there is little signaling-available S1P deep in the white pulp; CD169+ macrophages and other cell types in this location had little internalized S1PR1-GFP (Fig. 5a, Supplementary Figs. 4,5). By contrast, CD169+ macrophages and other cell types in the white pulp at the border of the marginal sinus were clearly sensing S1P (Fig. 5a, Supplementary Figs. 4,5). S1P sensing decayed gradually with distance from the marginal zone in the white pulp (Fig. 5b). Although we cannot formally distinguish whether cells adjacent to the marginal sinus sense S1P because S1P has entered the white pulp or because these cells extend processes into the marginal zone, our results demonstrate that cells in the white pulp do sense S1P. This observation is consistent with structural studies that have revealed gaps in the marginal sinus wall, which is formed by anastomosing vessels36,37 – these gaps may facilitate extension of processes into the marginal zone or leak of S1P into the white pulp. The observation is also consistent with the finding that S1P receptor 2 (which antagonizes cell migration towards S1P) expressed by germinal center B and T cells maintains the integrity of germinal center clusters22,23. We found that LPP3 regulates S1P sensing in the white pulp as well as the red pulp; in LPP3-deficient mice S1P was sensed by cells farther from the marginal sinus than in control animals (Fig. 5c,d). These findings suggest that S1P can be sensed by cells in the white pulp, and that LPP3 regulates the range of S1P availability.
Figure 5. Cells within the white pulp sense S1P near the marginal sinus.
(a) Two representative spleen sections. Scale bar for individual cells is 5 μm. Data are representative of sections from 12 mice, 7 Mx1-Cre and 5 Lyz2-Cre. 4 additional examples are shown in Figures S4 and S5. High-resolution files of these images are available at Figshare.com, http://figshare.com/s/54f80e76309411e5b67d06ec4b8d1f61 and http://figshare.com/s/a26624e0309411e5a91d06ec4bbcf141.
(b) Quantification of S1P reporting in the white pulp at the indicated distances from the marginal zone-white pulp border. 4 μm-thick bands were drawn from the marginal zone-white pulp border, and the average surface GFP:RFP ratio within each band was calculated as in Fig. 2b. Graph compiles 84 points from 5 sections from a single mouse (without normalization), and is representative of 3 mice. Points indicate mean, error bars show SEM. The data can be fit with the equation y=0.54+0.002*x, where y=surface GFP:RFP and x=distance from the MZ. A Student's t-test of the hypothesis that GFP:RFP does not vary with distance (that the true slope of the curve is zero) yields a probability of <0.0001.
(c) Representative section of spleen from an LPP3-deficient mouse and control. Data are representative of 3 pairs of mice.
(d) Quantification of S1P reporting in the white pulp at the indicated distances from the marginal zone-white pulp border in LPP3-deficient mice and controls. 30 μm-thick bands were drawn from the marginal zone-white pulp border, and the average surface GFP:RFP ratio within each band was calculated as in Fig. 2b. Graph compiles 2 LPP3-deficient and 3 control mice, 1-5 sections per mouse. Error bars show SEM. Comparisons were performed using a 2-tailed unpaired Student’s t-test; **, p<0.01; ****, p<0.0001.
The role of LPP3 in marginal zone B cell shuttling
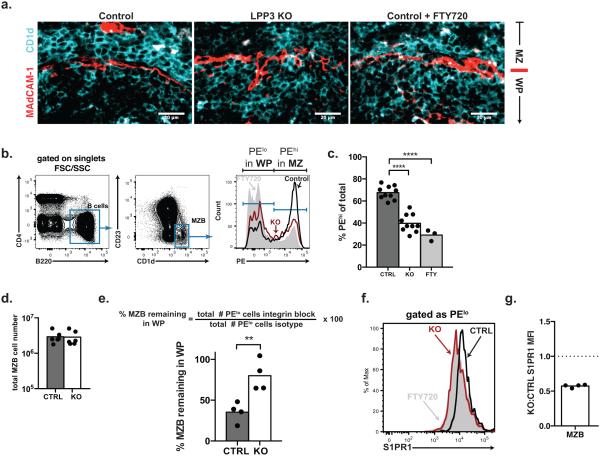
The finding that LPP3 regulates the distribution of signaling-available S1P in the spleen suggested that immune cell migration may be disrupted in LPP3-deficient mice. To test this, we analyzed the effect of LPP3 on marginal zone B cell (MZB) shuttling. We chose this step because it is early in the immune response and hence unlikely to be confounded by effects of LPP3 on multiple cell types, and because the role of S1P gradients in MZB shuttling has not been addressed directly. Specifically, we tested the hypothesis that LPP3 is required for MZB to return efficiently from the B cell follicles, where they deposit complement-coated antigen for B cell surveillance, to the marginal zone, where they pick up more cargo from the blood. S1pr1−/− MZB and MZB in mice treated with FTY720 remain in the follicles, presumably because they cannot follow the S1P gradient back to the marginal zone38. We predicted that in the presence of increased S1P in the B cell follicles of LPP3-deficient mice, the attraction of the marginal zone would be blunted, and MZB would remain longer in the B follicles before returning to the marginal zone.
If MZB were retained longer in the B follicles, this would result in an increased fraction of MZB in the follicles as opposed to the marginal zone. By histology, we observed a redistribution of MZB from the marginal zone to the follicles in LPP3-deficient mice (Ppap2bf/fMx1-Cre+, treated with polyI:C 3–5 days after birth)(Fig. 6a). To quantify the redistribution, we took advantage of a technique to label selectively blood-exposed cells39 (Supplementary Fig. 6a). We injected mice intravenously with a PE-conjugated antibody to CD45, which does not cross endothelial barriers in 5 min. We sacrificed the mice 5 min later and quantified the fraction of MZB that were PE+ and hence in the marginal zone. We found that 40% of MZB were in the marginal zone in LPP3-deficient mice, while 68% were in the marginal zone in littermate controls (Fig. 6b,c). FTY720 served as a positive control for MZB redistribution to the follicles; based on down-modulation of S1PR1 on bulk follicular B cells, we expected the effect of FTY720 to be more severe than loss of LPP3 (Fig. 6a–c, and not shown). The total number of MZB did not vary between LPP3-deficient mice and controls (Fig. 6d).
Figure 6. MZB are displaced into the B cell follicle in LPP3-deficient mice.
(a) Representative immunofluorescence of MZB (CD1dhi, cyan) and marginal sinus lining cells (MAdCAM1+, red) in a splenic cross-section of an LPP3-deficient mouse (Ppap2bf/f;Mx1-Cre treated with polyI:C at d3-5 after birth), littermate control, and littermate control treated with FTY720. Image is representative of 2 sets of mice.
(b,c) LPP3-deficient mice, littermate controls, and littermate controls treated with FTY720 were injected intravenously with a PE-conjugated antibody to CD45. Animals were sacrificed 5 minutes later, and MZB were analyzed by flow cytometry. Because of their size, PE-conjugated antibodies do not leave the blood and enter tissues for at least 5 minutes after injection. CD45-PE positive cells are blood exposed in the marginal zone, while PE-negative cells are protected in the B cell follicle. (b) Gating strategy. (c) Compilation of 10 experiments with 10 pairs of LPP3-deficient and control mice. In 2 experiments, 1-2 littermate controls were treated with FTY720. Bars indicate mean. Comparisons were performed using an unpaired, 2-tailed Student’s t test; ****, p<0.0001.
(d) Total number of MZB in the spleen of LPP3-deficient mice and littermate controls. Bars indicate mean. Compilation of 6 experiments.
(e) Groups of 4 littermates, 2 LPP3-deficient and 2 control mice, were analyzed together. 1 pair of LPP3-deficient and control mice was injected with blocking antibodies to integrins α4 and αL, which releases MZB from the marginal zone. The other pair was either untreated or treated with isotype control antibodies. All 4 mice were sacrificed 2.5 hours after antibody injection. The % MZB residing in the white pulp (WP) for at least 2.5 hours was calculated as 100*(# cells in WP of mice with integrin blockade)/(# cells in WP of mice without blockade). Compilation of 4 experiments. Bars indicate mean. Comparisons were performed using a paired 2-tailed Student’s t test; **, p<0.01.
(f) Representative surface S1PR1 staining on splenic MZB in the B cell follicle (PElo). Grey histogram is negative control (MZB from an FTY720-treated mouse). Note that the background is very high; we suspect that although the S1PR1 stain on MZB from LPP3-deficient mice overlaps with the stain on MZB from FTY720-treated controls, there is nonetheless more surface S1PR1 remaining on cells in LPP3-deficient animals than FTY720-treated controls.
(g) The ratio of surface S1PR1 mean fluorescence intensity (MFI) on MZB in LPP3-deficient mice to surface S1PR1 on MZB in their littermate controls. Compilation of 4 experiments.
An increased fraction of MZB in the follicles could reflect MZB staying too long in the follicles, as hypothesized. Alternatively, it could reflect MZB staying too briefly in the marginal zone. To test whether MZB were retained longer in the follicles of LPP3-deficient mice, we measured their residence time40 (Fig. 6e, Supplementary Fig. 6b). We analyzed mice in groups of four littermates – two LPP3-deficient mice, and two controls. One pair of LPP3-deficient and control animals received a control injection, which gave us a “time zero” measurement of the number of MZB in the B cell follicles. The other pair of LPP3-deficient and control animals was injected with blocking antibodies to integrins. This antibody blockade releases MZB from the marginal zone, and they are carried by flow into circulation; over time MZB are also lost from the follicles, as they exit into the marginal zone and are carried out of the spleen40. 2.5 hours after injection, we sacrificed the animals and quantified the number of MZB in the B cell follicles. We found that in LPP3-deficient animals, 79% of MZB remained in the B cell follicles after integrin blockade, while in controls only 34% remained (Fig. 6e). This finding indicated that MZB were retained longer in the B cell follicles in LPP3-deficient mice.
We also predicted that MZB themselves in the B cell follicles would have reduced expression of surface S1PR1 in the absence of LPP3, as they would be exposed to elevated S1P. Although MZB themselves do not detectably express the S1P reporter, and the endogenous S1PR1 stain on MZB has very high background, we consistently observed reduced endogenous S1PR1 on the surface of MZB in the follicles of LPP3-deficient mice (Fig. 6f,g).
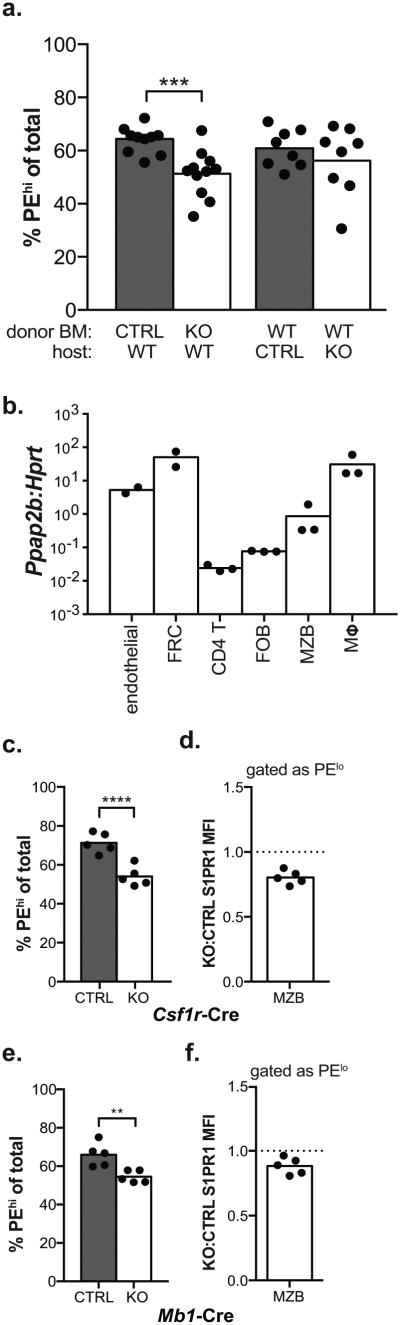
We then asked which cells required LPP3 to facilitate MZB shuttling. We began by making bone marrow chimeras, in which either radiation-sensitive hematopoietic cells were LPP3-deficient or radiation-resistant cells were LPP3-deficient. We found that LPP3 in the two compartments was largely redundant to regulate MZB positioning, although there was a statistically significant redistribution of MZB to the B cell follicles when LPP3 was lost on radiation-sensitive hematopoietic cells (Fig. 7a). Among the hematopoietic cells that we assayed, F4/80hi macrophages expressed abundant LPP3 transcript (Fig. 7b). We hypothesized that this high LPP3 expression might be shared by other macrophage subsets (which could not be cleanly purified for qPCR), and that macrophages sitting in or adjacent to the marginal zone might limit S1P penetration into the follicle. We deleted LPP3 in macrophages using Csf1r-Cre41, and found that this resulted in a redistribution of MZB to the B cell follicles and reduction in surface S1PR1 expression on MZB in the follicles (Fig. 7c,d). The development of Cre recombinase strains that specifically target splenic macrophage subsets will be invaluable to dissect the mechanism. Interestingly, MZB cells themselves also expressed detectable amounts of LPP3, and deletion of LPP3 in B cells using Mb1-Cre also resulted in a small, but consistent, redistribution of MZB to the B cell follicles (Fig. 7e,f)42. Mixed bone marrow chimeras suggested that the role of LPP3 in B cells was not cell-intrinsic, and instead reflected collective degradation of the lipid; when only 10% of B cells lacked LPP3 and 90% of B cells were wild-type, the LPP3-deficient MZB shuttled normally (Supplementary Fig. 6c). These findings demonstrate that LPP3 expression by multiple cell types regulates marginal zone B cell shuttling.
Figure 7. Hematopoietic LPP3 is required for efficient MZB shuttling.
(a) Lethally irradiated wild-type mice were reconstituted with LPP3-deficient or littermate control bone marrow, or lethally irradiated LPP3-deficient or littermate control mice were reconstituted with wild-type bone marrow. The fraction of MZB residing in the marginal zone was quantified as in Fig. 6b. Pooled data from 11 pairs of mice with 5 pairs of bone marrow donors for loss of radiation-sensitive hematopoietic LPP3, and 8 pairs of mice with 3 wild-type bone marrow donors for loss of radiation-resistant LPP3.
(b) Expression of LPP3 (Ppap2b) mRNA relative to Hprt mRNA by sorted subsets of spleen cells, assessed by RT-qPCR. Endothelial: CD31+CD45−gp38−, fibroblastic reticular cells (FRC): CD31−CD45−gp38+, CD4 T cells (CD4 T): B220−CD4+, follicular B cells (FOB): B220+CD4−CD21/35loCD23hi, marginal zone B cells (MZB): B220+CD4−CD21/35hiCD23lo, F4/80hi macrophages (Mφ): F4/80hiB220−. Each point represents a separate sort and RT-qPCR reaction.
(c) The fraction of MZB residing in the marginal zone of Ppap2bf/f;Csf1r-Cre mice or littermate controls. Pooled data from 5 pairs of mice.
(d) The ratio of surface S1PR1 MFI of PElo Ppap2bf/f;Csf1r-Cre MZB to S1PR1 MFI of PElo littermate control MZB. Pooled data from 5 pairs of mice.
(e) The fraction of MZB residing in the marginal zone of Ppap2bf/f;Mb1-Cre mice or littermate controls. Pooled data from 5 pairs of mice.
(f) The ratio of surface S1PR1 MFI of PElo Ppap2bf/f;Mb1-Cre MZB to S1PR1 MFI of PElo littermate control MZB. Pooled data from 5 pairs of mice.
For all panels, bars indicate mean. Comparisons were done using paired, 2-tailed Student’s t test; **, p<0.01; ***, p<0.001, ****, p<0.0001.
Discussion
We have developed a tool to map signaling-available S1P in tissues, using a strategy that may be widely applicable to small molecules, lipids, and peptides that signal through surface receptors. We have used this tool to map splenic S1P. As expected, S1P is abundant in the marginal zone, and low deep in the white pulp. Surprisingly, the concentration of S1P is low in many regions of the blood-rich red pulp, particularly areas dense with F4/80+ macrophages. Moreover, cells in the white pulp adjacent to the marginal sinus sense substantial S1P. These observations reconcile some of the apparently contradictory roles of S1P in cell trafficking within the spleen. Finally, we found that LPP3 maintains low splenic S1P, and enables efficient return of shuttling marginal zone B cells from the white pulp to the marginal zone.
We expect that the S1P reporter will be a model for detection of other elusive signaling molecules in vivo. For example, these principles could be used to understand how the distribution of 7α-25 hydroxycholesterol changes upon inflammation, to locate TGF-β released from the inhibitory latency-associated protein, to define the shape of LPA gradients that induce melanoma metastasis, and to determine the range of formyl peptides released from dying cells43. Two elegant and complementary mouse models that identify cells that have experienced S1PR1 signaling have recently been described; one replaces endogenous S1PR1 with S1PR1-eGFP, and the other modifies endogenous S1PR1 using the Tango system44,45. The reporter we present here differs in that we measure distribution of S1P itself. The S1P reporter distinguishes ligand-dependent and -independent receptor activation, enables measurements of S1P in areas where the other four S1P receptors (S1PR2-S1PR5) dominate, and allows measurement of differential S1P signaling across a cell. Together, the receptor and ligand reporters provide a powerful set of tools to dissect the role of extracellular lipid.
The S1P reporter revealed exquisitely tight regulation of signaling available S1P. Surprisingly, the amount of signaling-available S1P cannot be predicted by the presence of blood, despite the micromolar concentrations of S1P in plasma, or by the presence of red blood cells, the primary producers of blood S1P. Many regions of the red pulp have little signaling available S1P, although the reticular meshwork of the red pulp has a higher hematocrit than peripheral blood46. The concentration of S1P drops sharply at the boundary between the marginal zone and the red pulp, although there is no known physical barrier between the two compartments. Yet cells in the B cell follicle, in proximity to the marginal sinus, are able to detect S1P. The tight control of S1P gradients may explain how a single lipid can simultaneously direct many cell movements, and emphasizes the need for the S1P reporter.
LPP3 plays an important role in shaping S1P gradients in the spleen, as well as the thymus and cerebellum34,47. We predict that LPP3 regulates the migration of many cell types beyond MZB, and future work will test this hypothesis. It will also be fascinating to understand the different roles of LPP3 in different subsets of cells, and how LPP3 expression and S1P gradients are altered in inflammation.
Ultimately, the ability to map the distribution of signaling lipids will offer deep mechanistic insight into how and when these signals regulate immune function.
Methods
Mice
Ppap2bf/f35, Mx1-Cre31, Lyz2-Cre33, Ubc-CreERT248, Cd69−/−49, Csf1r-Cre41, Mb1-Cre42, and Ubc-GFP50 mice have been previously described. CD45.1+ congenic B6 mice were obtained from the National Cancer Institute. S1P reporter mice were generated in the transgenic facility at the University of California San Diego Moores Cancer Center. Mice were used at 8–20 weeks of age, and female and male mice were used depending on availability (we observed no sex differences in any of the parameters measured). No animals were excluded from analysis unless they were clearly sick (hunched, low body weight). To induce Mx1-Cre, 3–5 day old mice received a single intraperitoneal injection of 50–70 μl pI-pC (GE Healthcare) at a concentration of 2 mg/ml in PBS. All mice in each litter (LPP3-deficient mice and controls) were treated identically. To induce Ubc-CreERT2, adult mice received daily intraperitoneal injections of 200 μg of tamoxifen for 5 days, and mice were analyzed 5 days after the last tamoxifen dose. For bone marrow chimeras, recipients were lethally irradiated by either two doses of 6.6 Gray separated by 3 h or a single 13 Gray dose from a cesium source, followed by intravenous transfer of 2–10 × 106 bone marrow cells. Chimeras were analyzed at least 8 weeks after transplantation. Mice were housed in specific pathogen-free conditions at the Skirball Institute Animal Facility. All animal experiments were performed in accordance with protocols approved by the New York University Institutional Animal Care and Use Committee.
S1P reporter design
The S1P reporter is as described in Fig. 1a. We further tagged S1PR1NB-RFP with HA (YPYDVPDYA) and S1PR1-GFP with FLAG (DYKDDDDK). The tags were placed 17 amino acids from the N-terminus of the receptors, because the N-terminus is prone to cleavage. A signal sequence (MDSKGSSQKGSRLLLLLVVSNLLLCQGVVSD) preceding S1PR1NB-RFP and S1PR1-GFP was retained from previous constructs that were well-expressed. The nucleotide sequences for the two receptors were codon diversified and optimized for mouse. The targeting vector was as described in 28.
Mouse treatments
For intravenous PE labeling, mice were injected in the tail vein with 2 μg PE anti-CD45.2 (clone 104) and/or 2 μg PE anti-CD45.1 (clone A20) in 100 μl PBS, and euthanized precisely 5 min later. The spleen was quickly harvested and processed through a 70 μm cell strainer in 5 ml ice-cold staining buffer (1× PBS, 2% fetal bovine serum, 0.5 mM EDTA, 0.1% sodium azide, pH 7.3).
For integrin blockade, mice were injected intravenously with 100 μg anti-α4 mAb (clone PS/2) and 100 μg anti-αL mAb (clone M17/4), both from BioXCell. Rat IgG2a (clone 2A3, anti-trinitrophenol) and rat IgG2b (clone LTF-2, anti-keyhole limpet hemocyanin), also from BioXCell, were used as isotype controls.
Mice were treated with various doses of FTY720 (Cayman Chemical). For marginal zone B cell shuttling experiments, mice were treated with 1 mg/kg FTY720 intraperitoneally the night before sacrifice. For immunofluorescence experiments, mice were treated with either 1 mg/kg intravenously 6 h prior to sacrifice or 3 mg/kg intraperitoneally 18 h before sacrifice followed by 2 mg/kg intravenously 6 h before sacrifice.
Confocal microscopy of cultured cells
The coding region of the S1P reporter (S1PR1NB-TagRFP version) was cloned into the pMXs retroviral vector; Plat-E packaging cells were used to generate retrovirus; and WEHI-231 cells were transduced with viral supernatant. These cells were not recently authenticated or tested for mycoplasma. F4/80+ macrophages were isolated from spleen by mechanical disruption followed by positive selection using an EasySep mouse biotin selection kit (Stemcell Technologies) according to the manufacturer’s instructions. Borosilicate coverglass chambers (Nunc) were prepared by 2 h incubation at 37 °C with 1 μg/ml human fibronectin (Corning) in PBS. Cells were plated in the chambers and incubated with the indicated concentration of S1P (Sigma) or 1 μM phorbol 12-myristate 13-acetate (PMA) (Sigma) diluted in RPMI media (HyClone) with 0.5% fatty acid-free BSA (Calbiochem) for 45 min at 37 °C. Following incubation, cells were fixed in 1% PFA (Electron Microscopy). Cells were visualized by confocal microscopy (without antibody amplification) using a Zeiss LSM710 inverted confocal microscope using a 63× oil immersion objective and ZEN 2010 software. Images were processed with ImageJ v1.49.
Confocal microscopy of tissue sections
Mice were lethally anesthetized and fix-perfused with 1% PFA in PBS. Organs were harvested; fixed in 4% PFA for 1 h at room temperature (22-25 °C) with gentle shaking; dehydrated overnight in 30% sucrose in 1× PBS at 16 °C with gentle shaking; embedded in OCT (Sakura); and snap-frozen in dry ice-cold 2-methylbutane. 8–20 μm sections were cut, fixed with ice cold acetone for 10–15 min, and air dried. All stains were performed at room temperature (22-25 °C). Sections were permeabilized with 0.5% Triton X-100 in PBS for 10 min, washed and incubated for 10–30 min with 0.1% Triton X-100 5% normal goat and 5% normal donkey serum, washed and incubated for 1 h with chicken anti-GFP (Abcam) and rabbit anti-TagRFP (Evrogen), and washed and incubated with Alexa488 goat anti-chicken and Alexa647 donkey anti-rabbit (both from Jackson Immunoresearch). Additional stains were with antibodies listed below; Avidin/Biotin blocking kit (Vector Laboratories) was used with biotinylated antibodies. Slides were mounted with either G-Fluoromount (Southern Biotech) or Mowiol 4-88 (Sigma). Slides were visualized using a Zeiss 710 inverted confocal microscope with 25×, 40× or 63× oil immersion objectives and ZEN 2010 software. Images were processed with ImageJ v1.49. All direct comparisons (e.g. FTY-treated vs. control, LPP3-deficient vs. control) were stained and imaged the same day with the same settings.
Antibodies
Rat anti-mouse S1PR1 (clone 713412) was from R&D. Donkey anti-rat biotin was from Jackson Immunoresearch. Fluorochrome-conjugated anti-CD1d (clone 1B1), CD4 (clone RMA 4-5), CD19 (clone 6D5), CD21/35 (clone 7E9), CD23 (clone B3B4), CD31 (clone MEC13.3), CD45 (clone 30F11), CD45.1 (clone A20), CD45.2 (clone 104), B220/CD45RO (clone RA3-6B2), CD169 (clone 3D6.112), F4/80 (clone BM8), MAdCAM-1 (clone MECA-367), and streptavidin were purchased from BioLegend.
Retroviral transduction of primary B cells
The coding region of the S1P reporter (hCD2-TagRFP version) was cloned into the pMXs retroviral vector, and Plat-E packaging cells were used to generate retrovirus. B cells were purified from lymph nodes and spleen by cell sorting (gated as B220+CD23hiCD1dlo) or magnetic bead separation (positive selection for CD19 using an EasySep biotin selection kit, StemCell Technologies). The purified B cells were activated overnight with 25 μg/ml LPS (Sigma); transduced with a retrovirus encoding the S1P reporter by spinfection (1258g, 90 min, 32 °C); incubated overnight with 25 μg/ml LPS; transduced once more with the retrovirus; rested overnight without LPS; and transferred intravenously into wild-type recipients. The following morning, recipients were sacrificed and spleens were analyzed by confocal microscopy.
Flow cytometry
Lymphocytes were isolated by mechanical disruption and filtration through a 70 μm cell strainer and enumerated with a cell counter (Beckman Coulter) set to detect nuclei between 3.5 and 7.5 μm. Splenic stromal cells were isolated as previously described34. Cells were stained and analyzed on an LSRII (BD) or sorted on a MoFlo (Beckman Coulter) or FACSAria (BD). Data were analyzed with FlowJo v. 8.7 & v. 10.0.8 (Tree Star).
Quantitative RT-PCR
To quantify messenger RNA abundance in cells, total RNA was extracted from sorted cell populations using TRIZOL (Invitrogen) according to the manufacturer’s instructions. Before reverse transcription, RNA was treated with DNase I (Invitrogen). The RNA was converted to cDNA with the Superscript III Firstrand Synthesis System (Invitrogen), according to the manufacturer’s instructions, using a mix of oligo-dT and random hexamers as primers. Real-time quantitative PCR was performed on a Roche Lightcycler according to the manufacturer’s instructions. All primer pairs were tested for linear amplification over two orders of magnitude. Primers used were the following: Hprt sense, 5′-AGGTTGCAAGCTTGCTGGT-3′; Hprt antisense, 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′; Ppap2b sense, 5′-GGATCGTCATCGCCATCCTG-3′; Ppap2b antisense, 5′-AAAGGAAGCATCCCACTTGC-3′.
Code availability
All programming was done in ImageJ v1.49 (NIH). The colocalization programs are included as Supplementary text.
Statistics
Samples were compared using Student’s 2-tailed t-test, paired as appropriate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Schwab lab, J. Cyster (University of California-San Francisco), T. Schmidt (University of California-San Francisco), J. Green (University of California-San Francisco), D. Littman, M. Dustin, S. Koralov, and T. Lu (Weill Cornell Medical College) for helpful discussions. We are grateful to J. Cyster and L. Pitt for critical reading of the manuscript. This research was supported by grants to SRS from NIH (AI085166) and the Pew Charitable Trusts.
Footnotes
AUTHOR CONTRIBUTIONS
W.D.R.P. performed experiments, including microscopy, and analyzed data; V.F. performed quantitative image analysis; D.E.-A. provided Ppap2bf/f mice; M.C. provided assistance with microscopy and image analysis; W.D.R.P, V.F., and S.R.S. designed the study and wrote the manuscript; and S.R.S. supervised the work.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Sadik CD, Luster AD. Lipid-cytokine-chemokine cascades orchestrate leukocyte recruitment in inflammation. Journal of leukocyte biology. 2012;91:207–215. doi: 10.1189/jlb.0811402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanda H, et al. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nature immunology. 2008;9:415–423. doi: 10.1038/ni1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Z, et al. Constitutive lymphocyte transmigration across the basal lamina of high endothelial venules is regulated by the autotaxin/lysophosphatidic acid axis. Journal of immunology. 2013;190:2036–2048. doi: 10.4049/jimmunol.1202025. [DOI] [PubMed] [Google Scholar]
- 4.Kabashima K, et al. Thromboxane A2 modulates interaction of dendritic cells and T cells and regulates acquired immunity. Nature immunology. 2003;4:694–701. doi: 10.1038/ni943. [DOI] [PubMed] [Google Scholar]
- 5.Gatto D, Brink R. B cell localization: regulation by EBI2 and its oxysterol ligand. Trends in immunology. 2013;34:336–341. doi: 10.1016/j.it.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Cyster JG, Dang EV, Reboldi A, Yi T. 25-Hydroxycholesterols in innate and adaptive immunity. Nature reviews. Immunology. 2014;14:731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- 7.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annual review of immunology. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 8.Groom JR, et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37:1091–1103. doi: 10.1016/j.immuni.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Saba JD, Hla T. Point-counterpoint of sphingosine 1-phosphate metabolism. Circ Res. 2004;94:724–734. doi: 10.1161/01.RES.0000122383.60368.24. [DOI] [PubMed] [Google Scholar]
- 12.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitson SM, et al. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. The EMBO journal. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandala S, et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 15.Liu CH, et al. Ligand-induced trafficking of the sphingosine-1-phosphate receptor EDG-1. Mol Biol Cell. 1999;10:1179–1190. doi: 10.1091/mbc.10.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwab SR, et al. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 17.Chi H. Sphingosine-1-phosphate and immune regulation: trafficking and beyond. Trends in pharmacological sciences. 2011;32:16–24. doi: 10.1016/j.tips.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mebius RE, Kraal G. Structure and function of the spleen. Nature reviews. Immunology. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 19.Groom AM, Schmidt EE. Splenic Microcirculatory Blood Flow and Function with Respect to Red Blood Cells. In: Bowdler AJ, editor. The Complete Spleen: Structure, Function, and Clinical Disorders. Humana Press; 2002. pp. 23–50. IC. [Google Scholar]
- 20.Arnon TI, Cyster JG. Blood, sphingosine-1-phosphate and lymphocyte migration dynamics in the spleen. Current topics in microbiology and immunology. 2014;378:107–128. doi: 10.1007/978-3-319-05879-5_5. [DOI] [PubMed] [Google Scholar]
- 21.Pappu R, et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science. 2007;316:295–298. doi: 10.1126/science.1139221. [DOI] [PubMed] [Google Scholar]
- 22.Green JA, et al. The sphingosine 1-phosphate receptor S1P(2) maintains the homeostasis of germinal center B cells and promotes niche confinement. Nature immunology. 2011;12:672–680. doi: 10.1038/ni.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriyama S, et al. Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. The Journal of experimental medicine. 2014;211:1297–1305. doi: 10.1084/jem.20131666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czeloth N, et al. Sphingosine-1 phosphate signaling regulates positioning of dendritic cells within the spleen. Journal of immunology. 2007;179:5855–5863. doi: 10.4049/jimmunol.179.9.5855. [DOI] [PubMed] [Google Scholar]
- 25.Kabashima K, et al. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. The Journal of experimental medicine. 2006;203:2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortez-Retamozo V, et al. Angiotensin II drives the production of tumor-promoting macrophages. Immunity. 2013;38:296–308. doi: 10.1016/j.immuni.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrill AL, et al. Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. The Journal of biological chemistry. 2000;275:39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 28.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 29.Watterson KR, et al. Dual regulation of EDG1/S1P(1) receptor phosphorylation and internalization by protein kinase C and G-protein-coupled receptor kinase 2. The Journal of biological chemistry. 2002;277:5767–5777. doi: 10.1074/jbc.M110647200. [DOI] [PubMed] [Google Scholar]
- 30.Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. The Journal of biological chemistry. 2010;285:22328–22337. doi: 10.1074/jbc.M110.123299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 32.Hahn WC, Bierer BE. Separable portions of the CD2 cytoplasmic domain involved in signaling and ligand avidity regulation. The Journal of experimental medicine. 1993;178:1831–1836. doi: 10.1084/jem.178.5.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 34.Breart B, et al. Lipid phosphate phosphatase 3 enables efficient thymic egress. The Journal of experimental medicine. 2011;208:1267–1278. doi: 10.1084/jem.20102551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escalante-Alcalde D, Sanchez-Sanchez R, Stewart CL. Generation of a conditional Ppap2b/Lpp3 null allele. Genesis. 2007;45:465–469. doi: 10.1002/dvg.20314. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt EE, MacDonald IC, Groom AC. Comparative aspects of splenic microcirculatory pathways in mammals: the region bordering the white pulp. Scanning microscopy. 1993;7:613–628. [PubMed] [Google Scholar]
- 37.Schmidt EE, MacDonald IC, Groom AC. Microcirculation in mouse spleen (nonsinusal) studied by means of corrosion casts. Journal of morphology. 1985;186:17–29. doi: 10.1002/jmor.1051860103. [DOI] [PubMed] [Google Scholar]
- 38.Cinamon G, et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nature immunology. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 39.Cinamon G, Zachariah MA, Lam OM, Foss FW, Jr., Cyster JG. Follicular shuttling of marginal zone B cells facilitates antigen transport. Nature immunology. 2008;9:54–62. doi: 10.1038/ni1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnon TI, Horton RM, Grigorova IL, Cyster JG. Visualization of splenic marginal zone B-cell shuttling and follicular B-cell egress. Nature. 2013;493:684–688. doi: 10.1038/nature11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng L, et al. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. The American journal of pathology. 2010;176:952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muinonen-Martin AJ, et al. Melanoma Cells Break Down LPA to Establish Local Gradients That Drive Chemotactic Dispersal. PLoS biology. 2014;12:e1001966. doi: 10.1371/journal.pbio.1001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kono M, et al. Sphingosine-1-phosphate receptor 1 reporter mice reveal receptor activation sites in vivo. The Journal of clinical investigation. 2014;124:2076–2086. doi: 10.1172/JCI71194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahalan SM, et al. Actions of a picomolar short-acting S1P(1) agonist in S1P(1)-eGFP knock-in mice. Nature chemical biology. 2011;7:254–256. doi: 10.1038/nchembio.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald IC, Schmidt EE, Groom AC. The high splenic hematocrit: a rheological consequence of red cell flow through the reticular meshwork. Microvascular research. 1991;42:60–76. doi: 10.1016/0026-2862(91)90075-m. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Juarez A, et al. Expression of LPP3 in Bergmann glia is required for proper cerebellar sphingosine-1-phosphate metabolism/signaling and development. Glia. 2011;59:577–589. doi: 10.1002/glia.21126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell stem cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murata K, et al. CD69-null mice protected from arthritis induced with anti-type II collagen antibodies. Int Immunol. 2003;15:987–992. doi: 10.1093/intimm/dxg102. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cellular immunology. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.