Rapid molecular diagnostics (RMDs) can guide the correct β-lactam therapy choice when the prevalence of resistance is known. When an RMD reveals that an isolate of Escherichia coli or Klebsiella pneumoniae is susceptible, the result is likely to be accurate.
Keywords: β-lactams, molecular diagnostics, empiric therapy
Abstract
Background. Rapid molecular diagnostic (RMD) platforms may lead to better antibiotic use. Our objective was to develop analytical strategies to enhance the interpretation of RMDs for clinicians.
Methods. We compared the performance characteristics of 4 RMD platforms for detecting resistance against β-lactams in 72 highly resistant isolates of Escherichia coli and Klebsiella pneumoniae (PRIMERS I). Subsequently, 2 platforms were used in a blinded study in which a heterogeneous collection of 196 isolates of E. coli and K. pneumoniae (PRIMERS II) were examined. We evaluated the genotypic results as predictors of resistance or susceptibility against β-lactam antibiotics. We designed analytical strategies and graphical representations of platform performance, including discrimination summary plots and susceptibility and resistance predictive values, that are readily interpretable by practitioners to inform decision-making.
Results. In PRIMERS I, the 4 RMD platforms detected β-lactamase (bla) genes and identified susceptibility or resistance in >95% of cases. In PRIMERS II, the 2 platforms identified susceptibility against extended-spectrum cephalosporins and carbapenems in >90% of cases; however, against piperacillin/tazobactam, susceptibility was identified in <80% of cases. Applying the analytical strategies to a population with 15% prevalence of ceftazidime-resistance and 5% imipenem-resistance, RMD platforms predicted susceptibility in >95% of cases, while prediction of resistance was 69%–73% for ceftazidime and 41%–50% for imipenem.
Conclusions. RMD platforms can help inform empiric β-lactam therapy in cases where bla genes are not detected and the prevalence of resistance is known. Our analysis is a first step in bridging the gap between RMDs and empiric treatment decisions.
Antibiotic resistance is complex and accelerating in prevalence [1]. The development and implementation of rapid molecular diagnostics (RMDs) is considered a promising strategy to combat the challenge of antibiotic resistance [2]. Especially considering multidrug-resistant (MDR) gram-negative bacteria, the rapid and accurate detection of antibiotic resistance determinants offers the potential to transform clinical practice by informing early appropriate antibiotic therapy; thereby improving clinical outcomes, and shortening the duration and narrowing the spectrum of activity of antibiotic regimens, thus limiting the selection of antibiotic resistant bacteria [3]. RMDs could also streamline development of new antimicrobials by quickly identifying patients infected with antibiotic-resistant bacteria and enriching participant enrollment in clinical trials. Therefore, evaluation of RMDs is a strategic healthcare research priority.
PRIMERS (Platforms for Rapid Identification of MDR-gram negative bacteria and Evaluation of Resistance Studies) are a series of studies launched by the Antibacterial Resistance Leadership Group (ARLG) dedicated to the evaluation of RMDs. Here, the results of PRIMERS I and II are reported. PRIMERS I evaluated the performance characteristics of 4 molecular diagnostic platforms for identifying β-lactam resistance via identification of genes conferring resistance to β-lactam antibiotics in isolates belonging to Enterobacteriaceae (72 strains of highly drug-resistant Escherichia coli and Klebsiella pneumoniae). The 4 platforms evaluated were polymerase chain reaction combined with electrospray ionization mass spectrometry, PCR/ESI-MS [4]; DNA microarrays that detect β-lactamase (bla) genes, Check-Points [5, 6]; a multiplex diagnostic platform that uses allele-specific fluorescently labeled probes to identify genes, molecular beacons (MB) [7]; and a next-generation bench-top sequencing platform, Ion Torrent [8–10]. Analyses focused on the detection of β-lactam resistance, as this family of antibiotics represents the “cornerstone of therapy” for many infections caused by Enterobacteriaceae. In addition, these platforms were chosen because they contained a comprehensive set of molecular probes to investigate β-lactam resistance.
In PRIMERS II, 2 platforms (PCR/ESI-MS and MB) were selected based on the following: the results of PRIMERS I; the ability to modify targeted resistance determinants (add bla genes where needed in the assays); and their ability to identify the bacterial genus and species (a major requirement for clinical implementation). The 2 RMD platforms were tested in a blinded fashion against a heterogeneous collection of 196 susceptible and drug-resistant isolates of E. coli and K. pneumoniae to evaluate whether RMD platforms can discriminate susceptibility vs resistance and thus potentially improve clinical decision-making regarding the selection of empiric antimicrobial therapy.
Next, we developed novel analytical methods that aggregate results of multiple genetic targets in order to discriminate bacterial resistance vs susceptibility for a wide range of β-lactam antibiotics. Analytical strategies were designed to distill high-volume phenotypic and genotypic data into an easy-to-interpret presentation. Analyses also incorporated the prevalence of resistance, given its important impact on the clinical interpretation of diagnostic results. The results account for the potential heterogeneity of resistance rates across geographic areas and time, allowing for broad interpretive applicability. The susceptibility/resistance analysis platform presented here offers a practical paradigm for approaching the interpretation of results obtained from RMD platforms.
METHODS
Antimicrobial Susceptibility Testing and Isolate Selection
Antimicrobial susceptibility testing (AST) was performed on 268 (72 and 196 from PRIMERS I and II, respectively) E. coli and K. pneumoniae isolates using the MicroScan System (Siemens, Tarrytown, New York). Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines [11]. American Type Culture Collection control strains Pseudomonas aeruginosa 27 853 and E. coli 25 922 were used as quality-control strains for susceptibility testing.
Seventy-two isolates were chosen for PRIMERS I. The selected E. coli and K. pneumoniae strains came from heterogeneous geographic locations around the world, were stored in the investigators’ laboratories (R. A. B. and B. N. K.), and were previously determined by AST testing to be resistant to carbapenems and/or expanded-spectrum cephalosporins. These isolates were selected because they expressed a wide variety of β-lactamase (bla) genes, encoding for SHV/TEM extended-spectrum β-lactamases (ESBLs) and non-ESBLs (referred to as wild type [WT] SHV/TEM), as well as OXA, KPC, CMY, CTX-M, ACT, MIR, DHA, NDM, VIM, and IMP β-lactamases [12, 13]. These bla genes were screened on the 4 RMD platforms.
In PRIMERS II, a collection of 196 E. coli and K. pneumoniae isolates (approximately half were susceptible to most commonly used β-lactams, whereas the other half were determined to have carbapenem-resistant and/or expanded-spectrum cephalosporin-resistant phenotypes) were chosen from different geographic regions (northeast Ohio and mid-Atlantic states) to reflect the variation encountered across clinical practices. Isolates were selected based on species and phenotype. The isolates in PRIMERS II were assayed in a blinded fashion in 2 locations. By doing this, the investigators at the different sites running the molecular assays did not have knowledge of the isolate identity, genotype, or susceptibility when they performed the tests.
Analysis of bla Genes Using RMD Platforms
PCR/ESI-MS, MB (PRIMERS I and II), a DNA microarray kit, and a next-generation sequencing platform (PRIMERS I only) were used to identify the genetic determinants of bla-mediated resistance in each isolate (see the Supplementary Materials and Supplementary Table 1 for a detailed description of each method, with specific advantages and disadvantages). The identification of bla genes was validated using established controls [14].
The most common bla gene targets that determine resistance to β-lactam antibiotics were included, as summarized in Table 1.
Table 1.
Genes That Determine Resistance to β-Lactam Antibiotics Targeted in PRIMERS I and II
| Antibiotic | bla Gene |
|---|---|
| Ampicillin | CTX-M-1a, CTX-M-2a, CTX-M-8 & -25a, CTX-M-9a, TEM-WT (TEM-1), TEME104K, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHV-WT (SHV-1), SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP, OXA-48 |
| Ampicillin/Sulbactam | SHV-WT (SHV-1), KPC, NDM, VIM, IMP, OXA-48, CMY-1/MOX, CMY-2/FOX |
| Amoxicillin/Clavulanic | KPC, NDM, VIM, IMP, OXA-48, CMY-1/MOX, CMY-2/FOX |
| Piperacillin/Tazobactam | KPC, NDM, VIM, IMP, OXA-48, CMY-1/MOX, CMY-2/FOX |
| Cefazolin (I) | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP, OXA-48 |
| Cefoxitin (II) | KPC, NDM, VIM, IMP, OXA-48, CMY-1/MOX, CMY-2/FOX |
| Cefotaxime (III) | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP |
| Ceftriaxone (III) | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP |
| Ceftazidime (III) | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP |
| Cefepime (IV) | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC, NDM, VIM, IMP |
| Aztreonam | CTX-M-1, CTX-M-2, CTX-M-8 & -25, CTX-M-9, TEMR164S, TEMR164C, TEMR164H, TEMG238S, TEME240K, TEMA237, SHVG238S, SHVG238A, SHVE240K, CMY-1/MOX, CMY-2/FOX, KPC |
| Ertapenem | KPC, NDM, VIM, IMP, OXA-48 |
| Imipenem | KPC, NDM, VIM, IMP, OXA-48 |
| Meropenem | KPC, NDM, VIM, IMP, OXA-48 |
a Indicates CTX-M group.
Statistical Methods
Each platform evaluated isolates for the presence or absence of the genetic targets (bla genes) that have been associated with resistance, as per Table 1. In operational terms, the platform result was considered “resistant” when any of the targeted genes were found; the result was considered “susceptible” when none of the targets were found. The minimum inhibitory concentrations (MICs) for each strain were used as a “gold standard” to define susceptibility or resistance for each β-lactam antibiotic using CLSI breakpoints and interpretative standards. The platform results were compared with MIC results for each antibiotic.
Discrimination summary plots were used to display the 95% confidence interval (CI) estimates of susceptibility sensitivity, defined as the probability that the platform result is susceptible when the MIC result is susceptible, and of resistance sensitivity, defined as the probability that the platform result is resistant when the MIC result is resistant.
We defined the susceptibility predictive value (SPV) as the probability that an MIC result would indicate susceptibility when the platform result indicates susceptibility, and we defined resistance predictive value (RPV) as the probability that an MIC result would indicate resistance when the platform result indicates resistance. The SPV and RPV are also functions of the prevalence of susceptibility. Since there are temporal and geographic variations in the prevalence of susceptibility, the SPV and RPV were plotted as a function of the prevalence of susceptibility (with 95% confidence bands) to allow for interpretation across the spectrum of prevalence.
The sample size for PRIMERS II (196 isolates) was chosen based on estimating susceptibility and resistance sensitivities with desirable precision. Roughly half of the isolates were expected to be susceptible/resistant and thus available for estimating susceptibility/resistance sensitivities. A sample size of 90 isolates produces a 2-sided 95% CI with a width of 0.13 when the observed susceptibility/resistance sensitivity is 90%.
RESULTS
AST Summary
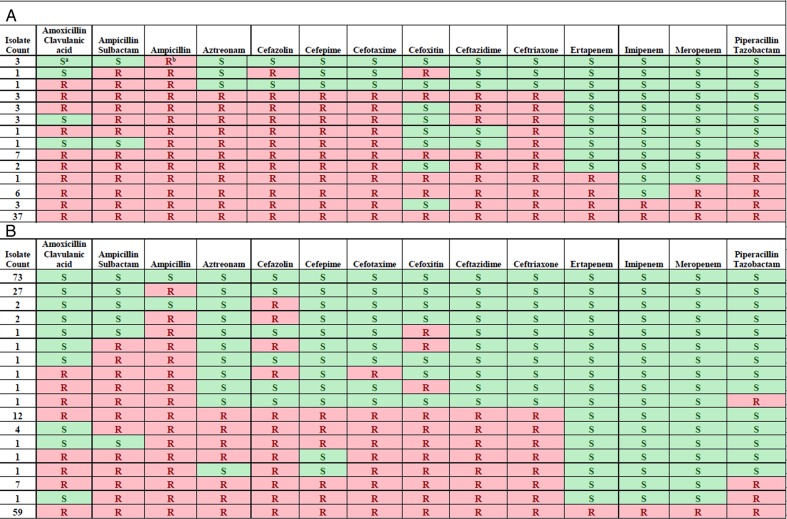
The AST determinations for isolates included in PRIMERS I and II are summarized in Figures 1A and 1B, respectively. In PRIMERS I, 37 isolates were resistant to all cephalosporins and carbapenems tested. In PRIMERS II (Figure 1B), there were 18 unique phenotypic profiles defined by resistance or susceptibility to 14 β-lactam antibiotics, with 73 isolates susceptible to all antibiotics tested, 27 resistant only to ampicillin, and 59 resistant to all β-lactams tested.
Figure 1.
Phenotypic profiles derived from antimicrobial susceptibility testing of isolates included in (A) PRIMERS I and (B) PRIMERS II. Notes: S highlighted with green denotes a susceptible interpretation for the drug, based on Clinical and Laboratory Standards Institute (CLSI) breakpoints. R highlighted with red denotes a resistant (or intermediate) interpretation, based on CLSI breakpoints.
PRIMERS I
The genotype:phenotype correlations that form the basis for the interpretation of the RMD platforms are presented in Table 1. PRIMERS I, using these resistance markers, was designed to compare the 4 platforms with respect to the ability to identify resistance and susceptibility to β-lactams (ie, the probability that the platform result reveals a bla gene that would confer resistance when MIC shows resistance). The performance against 14 β-lactam antibiotics (3 of which are β-lactam/β-lactamase inhibitor combinations) were estimated with 95% CIs for each platform.
The performance of each of the 4 platforms was similar for all 14 antibiotics (detailed results are provided in Supplementary Figure 2). In general, the 4 platforms (based on genotype) were able to predict resistance to β-lactams. Resistance sensitivity was best for first- and third-generation cephalosporins. In contrast, resistance sensitivity for certain carbapenems and β-lactam/β-lactamase combinations was suboptimal. To illustrate, we found that imipenem, ceftazidime, and cefepime resistance sensitivities were >95%; for piperacillin/tazobactam, resistance sensitivities were ≤80% for all 4 RMD platforms. The results of PRIMERS I established that we could reliably identify bla genotypes. The PCR/ESI-MS and MB platforms were then selected for further study in PRIMERS II because the resistance determinants that they target can be expanded and because of their ability to identify the bacterial genus and species (an important consideration for clinical implementation).
PRIMERS II
PRIMERS II was designed to obtain precise estimates of resistance sensitivities, susceptibility sensitivities, SPVs, and RPVs to the 14 antibiotics in a blinded fashion. Both susceptible and resistant isolates were selected to reflect clinical practice.
To begin our analysis, we first compared the ability of each of the 2 platforms to detect different bla genes. The PCR/ESI-MS platform identified 50 unique genetic profiles in the 196 isolates. As this was a highly heterogeneous collection that mimics what clinical laboratories are likely to encounter, the most common genetic profile was the absence of resistance determinants (no bla gene detection in 75 isolates) followed by identification of only WT blaSHV-1 in 24 isolates (Table 2). In contrast, the MB platform identified 30 unique genetic profiles, with the most common profiles being absence of gene detection (121 isolates) and identification of KPC only (9 isolates; Table 3). The MB platform lacked 2 common bla genes (WT blaTEM-1 and WT blaSHV-1), which resulted in fewer unique genetic profiles as a whole.
Table 2.
Genetic Profile of Isolates Studied in PRIMERS II According to Polymerase Chain Reaction Combined With Electrospray Ionization Mass Spectrometry
| Isolate Count | TEM-WT | TEME104K | TEMR164S | TEMR164C | TEMR164H | TEMG238S | TEMG240K | TEMA237 | SHV-WT | SHVG156D | SHVG238S | SHVG238A | SHVE240K | CTX- M-1a | CTX- M-2a | CTX-M -8a &-25a | CTX-M-9a | KPC | NDM | VIM | IMP | CMY-2/ FOX | CMY-1/ /MOX | OXA-48 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 75 | ○b | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ●c | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ● | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 24 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 2 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 2 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 2 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 2 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | |
| 2 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | |
| 1 | ○ | ○ | ● | ○ | ○ | ● | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | |
| 1 | ○ | ● | ○ | ○ | ○ | ● | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 5 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 9 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 3 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | |
| 4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | |
| 4 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ○ | ● | ○ | ○ | ○ | ● | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 6 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | |
| 3 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ● | ○ | ○ | ○ | ● | ● | ● | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | |
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ● | |
| 3 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ○ | |
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ |
Genotypic profile derived from antimicrobial susceptibility testing in PRIMERS II. Results from PCR/ESI-MS.
Abbreviation: PCR/ESI-MS, polymerase chain reaction combined with electrospray ionization mass spectrometry.
a Indicates CTX-M group.
b ○ denotes the platform tested negative for the genotype.
c ● denotes the platform tested positive for the genotype.
Table 3.
Genetic Profile of Isolates Studied in PRIMERS II According to Molecular Beacons
| Isolate Count | TEM-WT | TEME104K | TEMR164S | TEMR164C | TEMR164H | TEMG238S | TEMG240K | TEMA237 | SHV-WT | SHVG156D | SHVG238S | SHVG238A | SHVE240K | CTX- M-1a | CTX- M-2a | CTX-M -8a &-25a | CTX-M-9a | KPC | NDM | VIM | IMP | CMY-2/ FOX | CMY-1/ /MOX | OXA-48 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 121 | ○b | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ●c | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ● | ○ | ○ | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 4 | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 5 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 5 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 2 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 3 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ||||
| 9 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 6 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| 2 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ||||
| 5 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ||||
| 7 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ||||
| 6 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ● | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ||||
| 1 | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | ○ | ● | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ||||
| 1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ |
Genotypic profile derived from antimicrobial susceptibility testing in PRIMERS II.
a Indicates CTX-M group.
b ○ denotes the platform tested negative for the genotype.
c ● denotes the platform tested positive for the genotype.
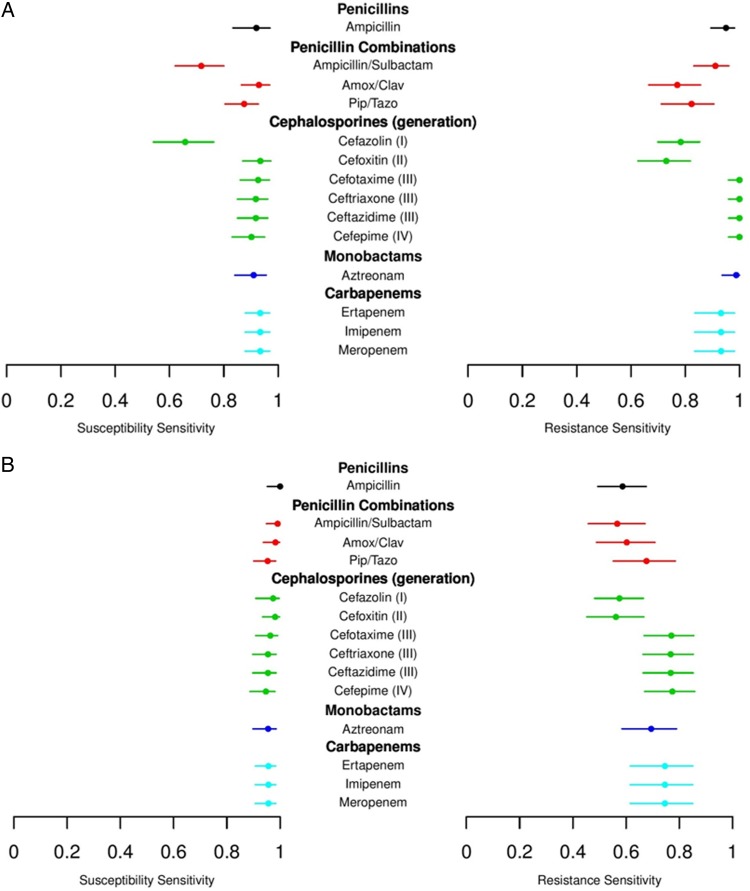
Estimates of the susceptibility and resistance sensitivities were displayed using discrimination summary plots (Figure 2A for PCR/ESI-MS and Figure 2B for MB). For the PCR/ESI-MS platform, the susceptibility sensitivities were >90% for expanded-spectrum cephalosporins and carbapenems but approximately 70% for ampicillin/sulbactam and cefazolin. The resistance sensitivities were >90% for later-generation cephalosporins, carbapenems, and ampicillin but were approximately 80% or lower for penicillin combinations and early-generation cephalosporins. In a comparison similar to the first analysis, the susceptibility sensitivities of the MB platform were >90% for all antibiotics tested, while the resistance sensitivities ranged from approximately 60% to 80%, with slightly higher results for later-generation cephalosporins and carbapenems than for early-generation cephalosporins and penicillin combinations.
Figure 2.
Estimates of the susceptibility and resistance sensitivities displayed using discrimination summary plots. Results for (A) polymerase chain reaction combined with electrospray ionization mass spectrometry and for (B) molecular beacons.
Next, we asked how one applies these findings to different clinical scenarios that have varying prevalences of β-lactam resistance. The predictive values of the results obtained from the RMD platforms were evaluated using SPV and RPV plots. Supplementary Figures 2A–H present SPV and RPV plots for imipenem, ceftazidime, piperacillin/tazobactam, and cefepime derived from PCR/ESI-MS and MB. Plots for the remaining antibiotics are provided in Supplementary Figures 2A, 2B, and 2I. These analyses indicate that in a clinical setting where, for instance, the prevalence of ceftazidime-resistant K. pneumoniae is 15% (ie, 85% susceptibility), the SPVs of PCR/ESI-MS and MB for ceftazidime are approximately 100% and 96%, respectively (see Supplementary Figures 2C and 2D, where the prevalence of MIC = susceptible on the horizontal axis is equal to 0.85). In a region where the prevalence of carbapenem resistance is 5%, the SPVs of PCR/ESI-MS and MB are approximately 100% and 99% for imipenem, respectively. In contrast, for that same situation, RPVs of PCR/ESI-MS and MB are 69% and 73% for ceftazidime and 41% and 50% for imipenem, respectively.
DISCUSSION
We developed an analytical strategy that integrates microbiologic data and genetic analyses to demonstrate that RMD platforms can accurately identify bla genotypes that confer β-lactam resistance in E. coli and K. pneumoniae and assist with appropriate decision-making regarding β-lactam selection. Such informed decision-making, especially when it relates to empiric therapy, could potentially have important clinical impact by improving patient outcomes and by reducing the emergence of resistance, the risk of unnecessary toxicity, and healthcare costs [3, 15].
Our analyses also show that it is possible to transform the β-lactam resistance genotypic data into an empiric decision-making tool that would be useful to the clinician through SPV and RPV plots. High SPVs demonstrated by both platforms show that, given a susceptible result, clinicians can be confident in prescribing antibiotics with a narrow spectrum of activity and hence meet the goals of a stewardship program [16]. The lower estimates of RPV when the prevalence of resistance is modest, on the other hand, highlight the difficulty of obtaining high levels of confidence in resistant results under some clinical settings. Additional work is needed to more completely understand what it means when a gene associated with antibiotic resistance is detected and an isolate tests susceptible to that antibiotic.
As a synthesis of the phenotypic and genotypic data derived from this study, we developed discrimination summary plots to provide clinicians and researchers with a succinct summary of the overall performance of a platform (and genotypes tested) in identifying susceptibility and resistance. Interestingly, these plots illustrate that the identification of susceptibility or resistance is better for certain β-lactams than for others (eg, carbapenems vs first-generation cephalosporins). While resistance sensitivities were high for most antibiotics, there were exceptions. For example, for imipenem and ceftazidime, there was >0.90 probability that the PCR/ESI-MS platform accurately identifies resistance when the AST confirms E. coli or K. pneumoniae resistance to these 2 agents. In contrast, examining piperacillin/tazobactam, a common agent used by clinicians to treat gram-negative bacterial infections in the clinical setting, we saw that there was approximately 0.80 probability of being accurate in identifying resistance.
How does a clinician rationalize these findings? Treating bacteria that possess multiple β-lactamases or finding β-lactamase genes that are not inactivated by tazobactam (eg, AmpCs) represent unexpected challenges when applying RMDs to resistant pathogens. The implications of the discrepancy between genotype and phenotype for the determination of resistance to piperacillin/tazobactam in E. coli and K. pneumoniae, and the clinical consequences of this error in particular, merit further study, especially for the treatment of bacteremia caused by ESBL producing E. coli and K. pneumoniae.
Most concerning, we discovered that there were isolates of K. pneumoniae that did not have detectable carbapenemase genes, yet were carbapenem resistant by AST. Further molecular studies suggest that these strains have mutations in outer membrane porins (OMPs) and possess either AmpCs or blaCTX-M genes [17]. OMPs are protein channels by which antibiotics enter K. pneumoniae (eg, OmpK35 and OmpK36). We also found E. coli isolates containing blaVIMs that did not test as carbapenem resistant by AST. We suspect that E. coli strains carrying blaVIMs do not express resistance to imipenem as readily as when blaVIM is harbored in P. aeruginosa. We speculate that this may be a mechanism of “silent dissemination” and may pose an even greater threat to future infection control programs. These examples point out genotype/phenotype mismatches that might result in diagnostic errors and inappropriate treatment. However, RMDs that detect “silent genes” can offer a unique advantage over conventional microbiology testing.
How does one reconcile differences when interpreting the results of RMD platforms? Failing to identify susceptibility may result in overtreatment. In this case, clinicians would be using broad-spectrum antibiotics needlessly. The consequences of this may be the further propagation of resistance by applying undesired selective pressure, increased costs, and possibly increased toxicity. On the other hand, failing to identify resistance bears more direct consequences for the patient, who may receive ineffective or suboptimal antibiotic treatment. Here, failure to treat the patient with effective drugs may even result in death, especially in the context of critical illness [18].
There are important limitations in this study. Notably, clinical studies will be needed to expand these observations and test the methods developed. Nevertheless, we believe that this work provides an analytical foundation for such future investigations. To have maximum impact, a platform must be easy to use and maintain, generate reliable results that can be readily interpreted by microbiology laboratory personnel and clinicians, be expandable to include novel genetic mechanisms of resistance, and be cost efficient [3]. Our analyses demonstrated that each of the RMD platforms tested perform similarly well. In a “real-world” setting, these platforms would be directly applied to clinical samples, as described previously [19]. Refining the performance characteristics of the platforms on clinical samples with multiple resistance genes would require further study in a prospective trial. To date, these challenges have not been met. Therefore, future considerations to differentiate the utility of the various platforms will include cost, complexity of testing, turnaround time, how the technologies scale (eg, incorporation of new genotypes), their adaptability to various clinical settings, and endurance of obsolescence. It is further anticipated that continuous surveillance will be needed to refine estimates of the prevalence of antibiotic resistance and to capture the full repertoire of bla genes relevant for resistance, as new and more complex genotypes will continue to emerge and disseminate among bacteria of clinical interest.
In summary, PRIMERS I and II create a new analytical paradigm with which to evaluate RMDs as a tool in clinical decision-making. We show that RMDs demonstrate great promise in guiding the selection of β-lactam antibiotics. More importantly, when the prevalence of β-lactam resistance genes is considered, SPVs are consistently high, indicating that clinicians can act on the susceptibility result with high confidence (ie, the risk of the most important error in treating with a truly ineffective or inactive β-lactam is negligible), providing the opportunity for more efficient antibiotic use in practice. In contrast, when RPVs are modest, our analysis shows that susceptibility may be present despite a platform result that indicates resistance. The impact of the prevalence of resistance on the interpretation of RMD results illustrates the importance of having robust methods that will be able to conduct resistance surveillance programs in different settings to help inform the clinical use of these platforms [20].
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. The authors thank Ms C. Lascols and the Study for Monitoring Antimicrobial Resistance Trends for sharing isolates with R. A. B.
Disclaimer. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health (NIH) or the Department of Veterans Affairs. S. R. E. and R. A. B. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases of the NIH (award number UM1AI104681). Funds and facilities were provided to R. A. B. by the Cleveland Department of Veterans Affairs. F. P. is supported by the Clinical and Translational Science Collaborative of Cleveland (award number UL1TR000439).
Potential conflicts of interest. T. H., C. M., R. S., and D. J. E. are salaried employees of Ibis Biosciences, Abbott. B. N. K. consults for Pfizer and Abbott. R. A. B. has received grants from AstraZeneca, Merck, Rib-X (Melinta), and Steris. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nathan C, Cars O. Antibiotic resistance—problems, progress, and prospects. New Engl J Med 2014; 371:1761–3. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin Infect Dis 2013; 56:1445–50. [DOI] [PubMed] [Google Scholar]
- 3.Caliendo AM, Gilbert DN, Ginocchio CC et al. . Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57(suppl 3):S139–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin CD, Howe GB, Sampath R et al. . Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn Microbiol Infect Dis 2009; 63:403–8. [DOI] [PubMed] [Google Scholar]
- 5.Endimiani A, Hujer AM, Hujer KM et al. . Evaluation of a commercial microarray system for detection of SHV-, TEM-, CTX-M-, and KPC-type beta-lactamase genes in gram-negative isolates. J Clin Microbiol 2010; 48:2618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endimiani A, Hujer KM, Hujer AM et al. . Are we ready for novel detection methods to treat respiratory pathogens in hospital-acquired pneumonia? Clin Infect Dis 2011; 52(suppl 4):S373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Mediavilla JR, Endimiani A et al. . Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J Clin Microbiol 2011; 49:579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Chavda KD, Findlay J et al. . Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob Agents Chemother 2014; 58:4196–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Chavda KD, Melano RG et al. . Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 2014; 58:2871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleo FR, Chen L, Porcella SF et al. . Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 2014; 111:4988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. In: 22nd Informational Supplement Wayne, PA: Clinical and Laboratory Standards Institute, 2012. [Google Scholar]
- 12.Lascols C, Hackel M, Hujer AM et al. . Using nucleic acid microarrays to perform molecular epidemiology and detect novel beta-lactamases: a snapshot of extended-spectrum beta-lactamases throughout the world. J Clin Microbiol 2012; 50:1632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lascols C, Hackel M, Marshall SH et al. . Increasing prevalence and dissemination of NDM-1 metallo-beta-lactamase in India: data from the SMART study (2009). J Antimicrob Chemother 2011; 66:1992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endimiani A, Hujer AM, Perez F et al. . Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J Antimicrob Chemother 2009; 63:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers HF, Bartlett JG, Bonomo RA et al. . Antibacterial resistance leadership group: open for business. Clin Infect Dis 2014; 58:1571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer KA, Perez KK, Forrest GN, Goff DA. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 2014; 59(suppl 3):S134–45. [DOI] [PubMed] [Google Scholar]
- 17.Poulou A, Voulgari E, Vrioni G et al. . Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J Clin Microbiol 2013; 51:3176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girometti N, Lewis RE, Giannella M et al. . Klebsiella pneumoniae bloodstream infection: epidemiology and impact of inappropriate empirical therapy. Medicine (Baltimore) 2014; 93:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacconi A, Richmond GS, Baroldi MA et al. . Improved sensitivity for molecular detection of bacterial and Candida infections in blood. J Clin Microbiol 2014; 52:3164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien TF, Stelling J. Integrated multilevel surveillance of the world's infecting microbes and their resistance to antimicrobial agents. Clin Microbiol Rev 2011; 24:281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.