Abstract
Sleep duration is implicated in the etiologies of chronic diseases and premature mortality. However, the genetic basis for sleep duration is poorly defined. We sought to identify novel genetic components influencing sleep duration in a multi-ethnic sample. Meta-analyses were conducted of genetic associations with self-reported, habitual sleep duration from seven Candidate Gene Association Resource (CARe) cohorts of over 25 000 individuals of African, Asian, European and Hispanic American ancestry. All individuals were genotyped for ∼50 000 SNPs from 2000 candidate heart, lung, blood and sleep genes. African-Americans had additional genome-wide genotypes. Four cohorts provided replication. A SNP (rs17601612) in the dopamine D2 receptor gene (DRD2) was significantly associated with sleep duration (P = 9.8 × 10−7). Conditional analysis identified a second DRD2 signal with opposite effects on sleep duration. In exploratory analysis, suggestive association was observed for rs17601612 with polysomnographically determined sleep latency (P = 0.002). The lead DRD2 signal was recently identified in a schizophrenia GWAS, and a genetic risk score of 11 additional schizophrenia GWAS loci genotyped on the IBC array was also associated with longer sleep duration (P = 0.03). These findings support a role for DRD2 in influencing sleep duration. Our work motivates future pharmocogenetics research on alerting agents such as caffeine and modafinil that interact with the dopaminergic pathway and further investigation of genetic overlap between sleep and neuro-psychiatric traits.
Introduction
Sleep occupies a substantial portion of our lives and is increasingly being recognized for its role in a number of diseases. Large meta-analyses have confirmed a role of sleep duration in cardiovascular diseases and all-cause mortality (1,2). The indirect and direct impact of sleep disorders and deprivation within the USA has an estimated financial magnitude of hundreds of billions of dollars (3). However, despite intensive study, there are basic gaps in our knowledge of mechanisms controlling sleep, and although habitual sleep duration varies widely across individuals, the factors that influence this are not understood. Sleep is likely to be among the more complicated phenotypes to study genetically due to connections with a broad range of physiologic mechanisms and variation with age (4), gender (5) and ethnicity (6).
Although self-reported sleep duration is heritable with estimates ranging from 9 to 44% (7–9), and there have been prior studies of genetic associations for sleep duration in humans, these studies have been limited to single cohorts (7,8,10,11) or single ethnic groups (12,13). Recently, the CHARGE Consortium identified two genome-significant (P < 5.0 × 10−8) signals near IER3 and PAX8 in European ancestry individuals. Genetic association studies of other complex traits including height have uncovered hundreds of significant loci when analysing hundreds of thousands of individuals (14), suggesting that novel loci significantly associated with sleep duration may emerge with increased sample size.
We identified an opportunity to examine sleep duration genetics across multi-ethnic US populations to further understand the etiology of sleep duration across populations, which may impact the morbidity of cardiovascular and other diseases and provide insights into pharmacogenetic approaches for improving sleep and alertness. Therefore, we conducted a large-scale, multi-ethnic meta-analysis using a customized gene chip (15) to identify novel polymorphisms associated with sleep duration.
Results
The study population comprised 25 465 individuals from seven ethnically diverse cohorts [5842 African Americans (AA); 546 Asian Americans (AsA); 18 026 European Americans (EA) and 1051 Hispanic Americans (HA)], with mean self-reported habitual weekday sleep duration ranging from 6.27 to 7.37 h (Table 1). Heritability estimates for sleep duration based on analysis of family data were h2 = 0.131 (SE 0.022) for EA adults in FHS and 0.043 (0.093) for AA adults in CFS. Heritability estimated using GCTA-based analysis (16) for unrelated individuals and applied to 2058 AAs in CARDIA, JHS, and MESA in whom genome-wide data were available was 0.134 (SE = 0.149).
Table 1.
Characteristics of the multi-ethnic discovery sample
| Ethnic group | Cohort | Median age (IQR) | Percent female | Mean BMI (SD) | Mean self-reported sleep duration (SD) | SNPs ≥ 1% MAF | N |
|---|---|---|---|---|---|---|---|
| African American (AA) | CARDIA | 40 (7) | 59.2 | 30.56 (7.4) | 6.27 (1.4) | 865 755 | 1171 |
| CFSa | 44 (24) | 57.6 | 34.23 (9.7) | 7.12 (1.8) | 858 698 | 524 | |
| CHS | 76 (8) | 66.7 | 28.40 (5.4) | 7.09 (1.7) | 42 283 | 588 | |
| JHS | 49 (14) | 60.9 | 32.28 (7.7) | 6.41 (1.4) | 894 684 | 2168 | |
| MESA | 66 (15) | 54.3 | 30.10 (5.9) | 6.54 (1.4) | 907 215 | 1391 | |
| Asian American (AsA) | MESA | 66 (16) | 49.8 | 24.06 (3.4) | 6.75 (1.2) | 31 213 | 546 |
| European American (EA) | ARIC | 62 (9) | 54.1 | 28.97 (5.2) | 7.20 (1.1) | 33 429 | 4414 |
| CARDIA | 41 (5) | 53.3 | 27.14 (5.9) | 6.77 (1.0) | 34 527 | 1333 | |
| CFSa | 44 (26) | 54.5 | 31.19 (8.2) | 7.19 (1.4) | 34 397 | 552 | |
| CHS | 78 (7) | 59.9 | 26.72 (4.6) | 7.37 (1.4) | 34 312 | 2942 | |
| FHSa | 48 (18) | 53.6 | 27.32 (5.4) | 7.29 (1.1) | 34 228 | 6764 | |
| MESA | 67 (17) | 51.7 | 27.85 (5.2) | 7.01 (1.2) | 34 957 | 2021 | |
| Hispanic American (HA) | MESA | 65 (16) | 52.6 | 29.71 (5.4) | 6.68 (1.4) | 39 936 | 1051 |
Seven studies included 25 465 individuals (5842 AA; 546 AsA; 1051 HA; 18 026 EA). African American N depended on Affymetrix 6.0 and CARe IBC chip coverage of individual SNPs.
aFamily cohort.
European and multi-ethnic meta-analysis identifies DRD2 SNP association with sleep duration
Single SNP genetic analysis was performed in a cohort-specific regression framework adjusting for age, age2, sex, age × sex and 10 population principal components. Ethnicity-specific meta-analyses showed little evidence of genomic inflation (lambda = 0.965–1.022; Supplementary Material, Fig. S1).
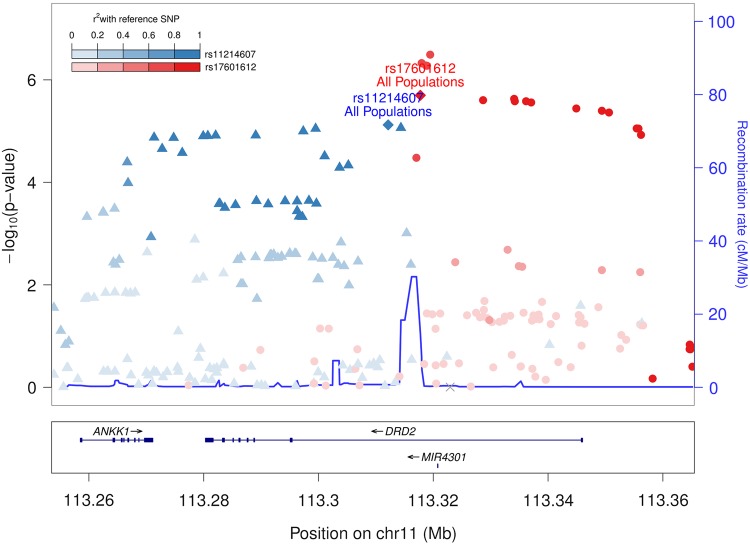
Gene-centric multi-ethnic meta-analysis of 40 520 IBC array SNPs in 25 465 individuals identified the SNP rs17601612 within intron 1 of DRD2 (dopamine D2 receptor) that reached a study-wide significance threshold of P = 1.9 × 10−6 (17) with the derived C allele associated with shorter sleep duration [meta P = 1.83 × 10−6, beta (SE) = −3.66 (0.72) min per C allele; Table 2; Fig. 1]. The association was primarily driven by EAs, our largest ethnic group, and the direction of association was consistent in AsA and HA, but not in AA (Table 2).
Table 2.
Ethnicity-specific and multi-ethnic meta-analysis of significant (P < 1.9 × 10−6) and suggestive (P < 1 × 10−5) signals of association with habitual sleep duration
| Population | SNP | Gene | Derived allele | DAF | Beta (SE), min | P | Direction | Phet | N |
|---|---|---|---|---|---|---|---|---|---|
| European Americans | rs17601612 | DRD2 | C | 0.37–0.39 | −3.66 (0.72) | 7.52 × 10−7 | −−−−−− | 0.085 | 18 022 |
| rs11214607 | DRD2 | G | 0.15–0.16 | 4.08 (1.02) | 4.37 × 10−5 | ++++++ | 0.9 | 18 024 | |
| Asian Americans | rs17601612 | DRD2 | C | 0.028 | −16.14 (13.56) | 0.233 | − | N/A | 546 |
| rs11214607 | DRD2 | G | 0.38 | 2.04 (4.32) | 0.64 | + | N/A | 546 | |
| Hispanic Americans | rs17601612 | DRD2 | C | 0.28 | −8.46 (3.90) | 0.031 | − | N/A | 1051 |
| rs11214607 | DRD2 | G | 0.30 | 5.46 (3.96) | 0.169 | + | N/A | 1050 | |
| African Americans | rs17601612 | DRD2 | C | 0.12–0.15 | 2.46 (2.34) | 0.30 | −++++ | 0.7 | 5546 |
| rs11214607 | DRD2 | G | 0.07–0.09 | 3.60 (3.00) | 0.23 | +−+++ | 0.9 | 5546 | |
| Discovery meta-analysis | rs17601612 | DRD2 | C | 0.028–0.39 | −3.32 (0.70) | 1.83 × 10−6 | −−−−−−−−−++++ | 0.054 | 25 165 |
| rs11214607 | DRD2 | G | 0.072–0.38 | 4.08 (0.90) | 8.26 × 10−6 | +++++++++−+++ | 1.00 | 25 166 | |
| Replicationa | rs17601612 | DRD2 | C | 0.37–0.39 | −2.05 (1.41) | 0.15 | +−−− | 0.55 | 5 529 |
| rs11214607 | DRD2 | G | 0.16 | 3.64 (1.94) | 0.061 | +++− | 0.44 | 5 529 | |
| Combined meta-analysis | rs17601612 | DRD2 | C | 0.028–0.39 | −3.07 (0.63) | 9.75 × 10−7 | −− | 0.54 | 30 694 |
| rs11214607 | DRD2 | G | 0.072–0.38 | 4.04 (0.90) | 7.76 × 10−6 | ++ | 0.60 | 20 695 |
DAF, derived allele frequency range in component studies.
aProxy rs2471854 with pairwise r2 = 0.90 in EUR used for replication studies.
Figure 1.
Regional association plot of DRD2 Sleep duration multi-ethnic association results for the discovery sample. −log10 P-value of the multi-ethnic (AA + AsA + EA + EA) association results is plotted on the y-axis, and chromosomal position is plotted on the x-axis encompassing the DRD2 locus. Individual directly genotyped SNPs are binned based on the stronger linkage disequilibrium relationship with either rs17601612 (blue) or rs11214607 or (red). Note the recombination hotspot separating the two independent association signals. LD was calculated based on 1000 Genomes European population data.
Regional association testing conditional on rs17601612 revealed a second independent signal of association at this locus [lead SNP rs11214607 G Punadjusted = 8.26 × 10−6, beta (SE) = 4.08 (0.90) min, Pconditional = 8.8 × 10−4, beta (SE) = 2.91 (0.88) min; Fig. 1]. This SNP was largely uncorrelated with rs17601612 (r2 < 0.05) in 1000 Genome African (AFR), East Asian (ASN) and European (EUR) samples, although weak correlation was observed in CARe EA cohorts (r2 < 0.075).
Effect estimates did not appreciably change after adjustment for covariates that may influence sleep duration through independent or pleiotropic pathways (e.g. depression, obesity, smoking and alcohol; Supplementary Material, Table S1). No SNPs previously reported to be associated with sleep duration present on the IBC array were nominally significantly associated in our study (Supplementary Material, Table S2).
Replication of lead SNPs from both DRD2 signals was attempted in independent cohorts comprised primarily of elderly individuals of European American descent: MAP + ROS, MrOS, SOF (mean ages 83.2, 76.7, 83.9; n = 1705, 2348, 1502, respectively) (18–23). Stronger statistical significance was seen in meta-analysis of the discovery and replication studies (rs17601612 P = 9.75 × 10−7; rs11214607 P = 7.76 × 10−6; Table 2). Haplotype analyses confirmed two independent signals (Supplementary Material, Table S3). In contrast, no generalizability was observed in a Korean cohort (age 40–69, n = 8842) with a different pattern of linkage disequilibrium at the locus [rs17601612 beta (SE) 0.44 (0.19), P = 0.02; rs11214607 beta (SE) 0.013 (0.02), P = 0.51].
Correlated functional SNPs in DRD2 region: bioinformatics data
Gene-based association analysis using VEGAS (24) confirmed DRD2 as the only significantly associated gene in Europeans after correction for multiple testing (P = 8.0 × 10−6; Padj = 0.042; Supplementary Material, Table S4). The two next best non-significant genes (ANKK1 and TTC12) are located near DRD2.
To localize genic regions with putative functional significance, we performed multi-ethnic regional fine mapping by 1000 Genomes Project (1KG) imputation. The most significant association was observed for the imputed SNP rs4274224 (imputation quality score r2 ≥ 0.967, P = 3.22 × 10−7, 1000 Genomes EUR r2 with rs17601612 = 0.648; Supplementary Material, Table S5). The most significant SNP from the secondary association signal was rs11214607 (P = 7.56 × 10−6). Twenty-three candidate regulatory SNPs were in high LD (EUR r2 ≥ 0.95) with one of the top SNPs contributing to each of the two association signals in DRD2 [Supplementary Material, Table S6 (25–27)]. If these SNPs contribute additive effects on multiple regulatory elements, then transferability of these effects may differ across individual haplotypes and populations.
DRD2 sleep duration SNP association with polysomnography-determined traits
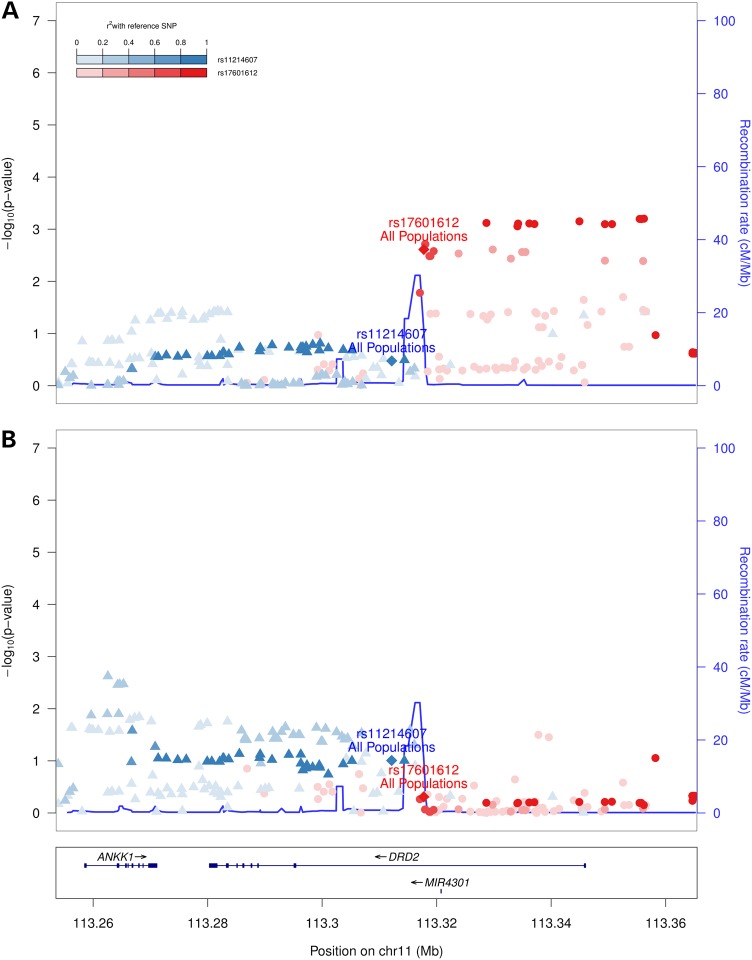
As exploratory analyses to provide insight into mechanisms influencing association with sleep duration, we tested for association of the two independent DRD2 signals with 15 additional polysomnographic and 4 self-reported sleep traits across 6 domains (Fig. 2, Supplementary Material, Table S7). Consistent with the primary results, and despite a limited available sample size, the strongest results varied between both signals. rs17601612 C was most significantly associated with objectively measured shorter sleep latency [i.e. time to fall asleep after ‘lights off’; beta (SE) −2.08 (0.69) min, P = 2.4 × 10−3, n = 2096, Fig. 2A], while rs11214607 G displayed trends with indices of sleep depth [i.e. lower Stage 1 percentage; −0.19 (0.12) percent, P = 0.099, n = 3740, Fig. 2B].
Figure 2.
Regional association plot of DRD2 multi-ethnic association results for sleep latency and Stage 1 sleep. Combined AA–EA results are shown for each trait: (A) sleep latency and (B) Stage 1 sleep. Phenotypes were collected on a subset of Sleep Heart Health Study I (ARIC, CHS, FHS) and CFS participants. rs17601612 and rs11214607 results are listed in Supplementary Material, Table S7. The axes and linkage disequilibrium pattern are as presented in Figure 1.
Association of a schizophrenia genetic risk score with sleep duration
Several candidate associations have been reported for European proxy SNPs of the second DRD2 signal (represented by rs11214607) with various neurobehavioral traits (Supplementary Material, Table S8), including differences in long/short DRD2 isoform ratios with distinct pre- and post-synaptic isoform localization and roles (28,29). Notably, a recent study reported genome-wide significant association of 108 loci with schizophrenia in GWAS from individuals of largely European ancestry, including multiple variants within DRD2 (rs17601612, P = 1.71 × 10−8) (30). The lead SNP (rs2514218, P = 4.09 × 10−10) is in strong LD with rs17601612 (EUR r2 = 0.74), suggesting a pleiotropic effect, with the rs17601612 C protective for schizophrenia and associated with short sleep duration in our study. While several SNPs were outside the region that was successfully imputed in our study, rs17601612 and 10 additional genome-significant SNPs for schizophrenia were imputed in our sleep duration study (rs7948028, rs4337071, rs4630328, rs11601054, rs61902787, rs7110440, rs35277073, rs61902807, rs7121986, rs6589377). These SNPs showed suggestive association to sleep duration in AA–AsA–EA–HA cohorts with P < 1.0 × 10−5 but were not significant and showed opposite direction for most African American cohorts, as observed for rs17601612. Given this potential pleiotropic association, and given that disturbances in sleep, including total sleep time and sleep latency, have been associated with schizophrenia in meta-analysis (31), we constructed a weighted genetic risk score of the 12/108 schizophrenia variants (or r2 > 0.8 proxies in CEU) that were genotyped on the IBC array and tested this genetic risk score for association with sleep duration in EA. The schizophrenia genetic risk score was associated with longer sleep duration with and without including the lead DRD2 variant rs17601612 [beta (SE) 0.86(0.24) min per allele, P = 2.4 × 10−4, beta (SE) 0.54(0.25) min/allele, P = 0.03; n = 18 026].
Discussion
In this study, we have demonstrated a novel association between sleep duration and common polymorphisms within the dopamine receptor DRD2 in over 25 000 individuals from a multi-ethnic US population. While genome-wide significance for each association signal was not reached, several lines of evidence provide strong support that these associations are valid: (i) conditional analysis reveals two independent DRD2 association signals with opposite effect directions, each in linkage disequilibrium with putatively functional SNPs, (ii) a consistent direction of allelic effects is observed in at least three of four ethnic groups, (iii) gene-based analysis confirms significant association of DRD2, (iv) suggestive association is observed with objective sleep traits including effects on sleep latency and sleep architecture and (v) the lead SNP is in high LD with a recently identified genetic variant for schizophrenia, another neurobehavioral phenotype. Our findings, including secondary analysis of polysomnographic sleep measures, were robust to alternative statistical models that considered potential sources of phenotype heterogeneity. However, available external replication was limited, requiring some caution in generalization of findings from our large European American sample to multiple ethnic groups.
DRD2 has a wide range of influence, including over 80 human biological processes observed in the Gene Ontology (32). The literature provides strong biological support for gene-level effects of DRD2 on sleep and circadian rhythms phenotypes, dating to 1917 (33,34). The dopamine receptor agonist apomorphine has been shown to induce yawning in rats, which was blocked by treatment with a preferential D2 antagonist (35). A common medicine for promoting alertness for disorders of hypersomnolence, including narcolepsy, is modafinil, which enhances wakefulness, at least in part, through effects on dopamine transporter (DAT) function (36) and DRD2/DRD3 receptor availability as measured by [11-C]raclopride-binding potential (37). Dat knockout mice display increased sleep fragmentation and decreased NREM sleep time relative to wild-type littermates (36). Both Drd2 pre-synaptic autoreceptors and post-synaptic heteroreceptors influence dopamine expression levels in mice (38). Drd2-binding efficiency effects may be due to DAT/DRD2 interactions and/or DRD2 modulation of DAT cell surface expression through the MAPK pathway (29,39,40). Drd2 knockout mice display significant differences in wakefulness, NREM sleep and sleep latency (41).
Secondary analyses of polysomnographic indices also showed variation in objective measures of sleep. Our primary DRD2 signal associated with shorter sleep latency that may reflect a greater homeostatic drive concurrent with reduced sleep duration, while the second independent DRD2 signal tended to be associated with improved sleep quality as measured by decreased time in stage N1 and may indicate a positive effect on both sleep duration and sleep depth.
There are a number of pathways by which DRD2 may influence sleep (as well as alertness), including through a complex involving the adenosine receptor ADORA2A (a target of caffeine) (42–44). Both proteins are prominently expressed in the human striatum (45). Caffeine ingestion increases [11-C]raclopride binding to DRD2/DRD3 in the human ventral striatum (46). Two weeks of increasing melatonin exposure have been shown to increase the affinity of Drd2 receptors in rat striatum by 48%, with minimal impact on receptor density (47). A single night of sleep deprivation reduced raclopride binding in the human striatum (48). Drd2 is linked to the rhythmicity of the key circadian gene Per2 in the rat striatum (49). It is possible that neuroinflammatory processes and sleep inter-relate given a central neuroinflammation role for Drd2 (50,51). Interestingly, rs17601612 and the 5 SNPs with the lowest P-values in our primary fine-mapped DRD2 association signal overlap an enhancer region in astrocytes (Supplementary Material, Table S6; rs4245146, rs4245147, rs4936271, rs4936272, rs4274224; P = 3.22 × 10−7–5.31 × 10−7). DRD2 also forms complexes with a number of other receptors including NTS1, which impacts mammalian sleep and circadian phase-shifting (52–54). DRD2 affects clock output through altered ARNTL/CLOCK complex activity (55). The secondary signal splice-site SNP rs1076560 affects phosphorylation of the circadian gene GSK3B and has been modestly associated with eyes-closed waking electroencephalographic activity (56,57). Despite this rich literature implicating DRD2 in sleep and circadian rhythms, this is the first large-scale genetic analysis to show an association between DRD2 variants and human sleep.
The SNPs we identified are likely to have functional effects. DRD2 receptor availability, density and/or location vary with different secondary signal DRD2 splice SNP (rs1076560 and rs2283265) variant isoforms (28,58). The pre-synaptic short isoform variant has differential effects on DRD2 autoreceptor function and striatal release of GABA and glutamate (reviewed in Ref. 29). We show novel associations between sleep duration and these SNPs (Supplementary Material, Table S8). Other prominent SNPs include rs1800497 (Taq1A, AsA–EA–HA P = 2.29 × 10−5) and rs1079597 (Taq1B, AsA–EA–HA P = 9.81 × 10−6) from the secondary signal, which have been associated with human striatum receptor density differences (59,60). rs1800497 is a missense ANKK1 polymorphism located outside of DRD2. Its effects on DRD2 are suggested to be mechanistically related to the two splice SNPs (28). SNPs corresponding to our primary signal are in low LD with rs1800497 (1000 Genomes AFR, ASN and EUR r2 < 0.01), are physically proximal (<1500 bp) and individually overlap epigenetic enhancer evidence from 9–51 cell lines (Supplementary Material, Table S6). A study of anxiety disorders and clock genes identified rs4245146 as the most significant SNP across all assayed genes (61). rs4274224, the most significant imputed SNP from our primary signal, was implicated in depression and associated with dorsolateral prefrontal cortex activation during anticipation of reward and stress-induced dopamine release (62,63). SNPs corresponding to our primary signal were recently associated with schizophrenia (30), including rs17601612 and rs61902807 (located within DRD2 and overlapping promoter histone marks in 12 brain cell lines and 22 additional cell lines in a region of mammalian conservation). The schizophrenia-associated SNPs extend beyond the range of our imputed genotypes. These convergent cellular and imaging finding studies suggest multiple functional effects for DRD2 SNPs, many of which likely relate to sleep–wake traits, that should be investigated further in future studies.
The effect size of each allele, while relatively modest, is within the range of other reports of genetic associations for sleep duration (10,13). The size of the effect approximately doubles when considering the combined contributions of both DRD2 loci. Small effect sizes are generally found for individual loci in other complex traits such as height (14). It is also likely that random misclassification introduced by use of questionnaire-based information on sleep duration (rather than objective data) attenuated the strength of actual associations. Future corroboration with objective sleep data is likely to improve estimates of effect sizes.
Study strengths include the relatively large sample size from multiple ethnic groups. Subtle population stratification effects were controlled using genomic control and 10 population principal components. Detailed polysomnography data in a subset of the sample amplified the primary questionnaire-based results, suggesting that the top SNPs for sleep duration also were associated with sleep latency and sleep architecture. Fine mapping by imputation provided insights into possible physiological mechanisms, including numerous SNPs in regulatory regions, which are suggested to have ‘pervasive involvement’ in human disease (64).
Weaknesses of this study include the use of a self-reported measure of sleep duration, which is the trait most available for analysis of large numbers of individuals. Although likely contributing to random misclassification, self-reported data have been shown to predict health outcomes in numerous epidemiological studies (1,2). Sleep duration is likely influenced by multiple environmental, social and biological factors. Although we controlled for many covariates while examining our DRD2 associations, we did not have detailed psychiatric or mood data to further explore pleiotropic relationships. Of particular interest was the observed association between a schizophrenia genetic risk score and longer sleep duration, a phenotype that is poorly understood, but often occurs in individuals with mood disorders or psychiatric diseases and can predict a number of adverse cardiometabolic health outcomes. Research aimed at dissecting the potential shared neurological bases for sleep and psychiatric traits may help to clarify the extent to which variation in sleep traits with chronic diseases reflect causal associations, or rather, are manifestations of disturbances in overlapping pathways. Genotyping with the IBC chip allowed dense coverage of ∼2000 genes suspected to play a role in primarily cardiovascular disease, but did not provide adequate coverage of regulatory regions or gene deserts. Although consistency across cohorts and populations were observed for some associations, in other cases there was heterogeneity, which may have resulted from any number of causes. External replication data were limited to elderly cohorts. Sleep duration, human DRD2 H3K4me3 epigenetic marks, mRNA levels and DRD2/3 receptor availability are all impacted by age (4,65–67). Epistasis with other dopamine-related polymorphisms is possible (40,68,69). While further studies are required to determine the generalizability limits of our findings, given the plausible mechanisms, past findings and the importance of DRD2 across related phenotypes, future mechanistic investigation into the role of this gene in sleep regulation is warranted.
In summary, the findings of this study highlight the importance of a gene involved in a number of neurological processes in also influencing sleep duration, a trait that impacts many health outcomes such as obesity and cardiometabolic diseases. Although further replication of our results is required, future studies that address the influence of DRD2 on physiology may be advanced by also considering the influence of DRD2 on sleep-related outcomes. DRD2 and dopamine functional interactions with caffeine and modafinil suggest that these variants are likely to play an important role in inter-individual differences in drug efficacy, thus providing a new avenue for pharmocogenetics research in sleep and alerting medications.
Materials and Methods
Participating studies
The discovery sample included data from over 25 000 individuals of European, African, Hispanic and Asian ancestries from seven cohorts participating in the Candidate gene Association Resource (CARe) Consortium (70): Atherosclerosis Risk In Communities (ARIC), Coronary Artery Risk Development in Young Adults (CARDIA), Cleveland Family Study (CFS), Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Jackson Heart Study (JHS) and Multi-Ethnic Study of Atherosclerosis (MESA). The Sleep Heart Health Study (SHHS) further contributed ARIC, CHS and FHS subsets from Visit 1 (1995–98) (71). Population characteristics are displayed in Table 1. Children under the age of 18 were omitted from analyses. Replication cohorts included the Rush Memory and Aging Project + Religious Orders Study (MAP/ROS), Osteoporotic Fractures in Men (MrOS), the Study of Osteoporotic Fractures (SOF) and the Korean Genome and Epidemiology Study (KoGES). Local IRB approval was obtained for the CARe study.
The Atherosclerosis Risk in Communities Study (ARIC) was primarily designed to investigate cardiovascular risk factors and atherosclerosis across four communities: Forsyth County, NC; Jackson, MS; suburban Minneapolis, MN and Washington County, MD. Cohort component visits began in 1987. All sleep duration data were obtained through the SHHS exam, which included a subgroup of EA individuals recruited from the Minneapolis (n = 1000) and Washington County (n = 750) sites. Sleep duration information was obtained through a SHHS Sleep Habits Questionnaire (SHQ) administered to EAs.
The Coronary Artery Risk Development in Young Adults (CARDIA) study longitudinally examines cardiovascular health in four communities: Birmingham, AL; Chicago, IL; Minneapolis, MN and Oakland, CA. Examinations have occurred from 1985 to 2010. All AA and EA results were obtained from a sleep questionnaire administered in 2005–06 (Year 20).
The Cleveland Family Study (CFS) is a family-based longitudinal study designed to examine the genetic basis of Obstructive Sleep Apnea (OSA) in AAs and EAs studied between 1990 and 2006. Index probands with confirmed OSA were recruited, along with additional family members and neighborhood control families from Northeast Ohio. Data were used from the last examination available for each individual. The final wave of data also included overnight in-laboratory polysomnography. Sleep duration was assessed from a standardized questionnaire (72).
The Cardiovascular Health Study (CHS) assessed adults aged 65 or greater through a series of examinations from 1989 to 1999, with field centers in Washington County, MD; Allegheny County, PA; Sacramento County, CA and Winston-Salem, NC. Of these subjects, 3280 individuals from Allegheny, Sacramento and Washington Counties participated in the SHHS and had data from the Sleep Habits Questionnaire available for this analysis. Additional phenotype and covariate data obtained from CHS visits in years 9–11 were used for 1424 CHS individuals who did not participate in the SHHS examination.
The Framingham Heart Study (FHS) began data collection in 1948, with a primary focus on identifying factors contributing to cardiovascular disease. Based in Framingham MA, the study includes second- and third-generation cohorts from which current study data were obtained. Approximately 700 second-generation individuals (Offspring cohort) were recruited for SHHS polysomnography. The majority of Offspring information was obtained from the Sleep Habits Questionnaire (n = 2148), with remaining data (n = 758) obtained from questionnaires administered at FHS Exams 7 (1998–2001) and 8 (2005–08). Generation 3 data (n = 3875) were obtained from Exam 1 (2002–05).
The Jackson Heart Study (JHS) is investigating factors influencing cardiovascular disease in individuals and families in Jackson, MS. All data used in the present analyses were obtained from Exam 1 from 2004, with JHS data analyzed independently of other original ARIC site data.
The Multi-Ethnic Study of Atherosclerosis (MESA) is investigating the characteristics related to the progression of subclinical to clinical cardiovascular disease, with an emphasis on understanding any possible ethnic, age and gender differences in the risk factors for and the progression of subclinical cardiovascular disease. MESA is following individuals from six communities: Baltimore, MD; Chicago, IL; Los Angeles, CA; New York, NY; Minneapolis/St Paul, MN and Winston-Salem, NC. Examinations began in 2000 with five exams completed and follow-up ongoing. The current data derive from questionnaires administered in Exam 4 (2004–05).
The Osteoporotic Fractures in Men Study (MrOS) (21,22) is a prospective cohort of 5994 males who were aged 65 or older at the baseline visit. Recruitment occurred between 2000 and 2002 in six communities: Birmingham, AL; Minneapolis, MN; Palo Alto, CA; Monongahela Valley, PA; Portland, OR and San Diego, CA. An ancillary study to MrOS, Outcomes of Sleep Disorders in Older Men, enrolled 3135 participants in an examination that included administration of sleep questionnaires, actigraphy and polysomnography between 2003 and 2005. The Study of Osteoporotic Fractures (SOF) is an analogous study involving 9704 women aged 65 and older (23). The study commenced in 1986 with recruitment in Baltimore, MD; Monongahela Valley, PA; Minneapolis, MN and Portland, OR. Standardized sleep measures were collected from 4727 SOF participants at the eighth clinic visit, which took place between 2002 and 2004.
The Rush Memory and Aging Project (MAP) (18) is a longitudinal investigation of genetic and environmental risk factors for chronic conditions of old age in community-dwelling older individuals (19) recruited from the Chicago, IL region beginning in 1997. The current replication data are derived from participants who underwent actigraphy testing, which was added in 2005.
The Religious Orders Study (ROS) is a prospective study investigating the association of risk factors for Alzheimer's disease, mild cognitive impairment and other neurocognitive outcomes (20). Examinations of members of over 40 religious communities from multiple ethnicities (88% non-Hispanic EAs) began in 1994.
The Korean Genome and Epidemiology Study (KoGES) (73,74) was designed to investigate chronic diseases within an industrial city (Ansan) and a rural community (Ansung) in South Korea. This population-based study began studying adults aged 40–69 in 2001. The initial baseline examination comprised 2497 females and 2523 males.
Phenotype and covariate definition
The primary phenotype was a measure of self-reported habitual nightly weekday sleep duration interrogated using single questions from standardized questionnaires [n = 21 373 individuals; largely using or adapting the Pittsburgh Sleep Quality Index question ‘During the past month, how many hours of actual sleep did you get at night? (This may be different than the number of hours you spend in bed.)’ (75)] or calculated from self-reported sleep onset and offset times via the SHHS Sleep Habits Questionnaire [n = 5641 individuals (71)] and harmonized to provide comparable data across cohorts. Exact questionnaire text is provided in Supplementary Material, Table S9. Implausible values (sleep duration ≤3 h or ≥14 h) or values of sleep onset between 4 AM and 6 PM (indicating possible shift work) were excluded from analysis. Data were extracted from multiple visits to maximize information. When sleep duration was reported from multiple visits, exams that included more detailed questionnaires (e.g. bed and wake times) were prioritized over exams with less complete information.
Missing and discordant gender information was inferred based on X chromosome SNP heterozygosity checks performed within PLINK (76). For secondary analyses to examine the effects of environmental exposures and comorbidities on the DRD2 SNPs (Supplementary Material, Table S1), alcohol, Type 2 diabetes, hypertension, retirement status and smoking covariates were expressed as binary variables. Depression status was determined by harmonizing within-cohort single-question ordinal scales typically considering the last 1–4 weeks (Supplementary Material, Table S10). Snoring (a symptom of obstructive sleep apnea) was dichotomized according to frequency (≥3 times, ‘several nights’, or ‘often’ per week).
Polysomnographic data [available from SHHS (71) and CFS (77)] and self-reported sleep traits used the same visit as the sleep duration questionnaire collection whenever possible. Traits across six domains were tested with the fine-mapped SNPs as exploratory analyses to provide insight into the primary sleep duration results: sleepiness (Epworth Sleepiness Scale); sleep apnea (minimum oxygen saturation across the night, apnea hyponea index within NREM); sleep latency; sleep timing [weekend average sleep (when reported), latency to first REM episode, self-reported sleep midpoint (weekday and weekend), time in bed, total sleep time]; arousability [sleep maintenance efficiency, arousal index (across the night, within NREM, within REM), wake after sleep onset]; and sleep stage percentages (Stages 1, 2, 3 + 4 and REM) (AA + EA n = 2096–9332).
Genotyping and quality control
Genotyping was performed by the Broad Institute and included an Affymetrix 6.0 chip for AAs and a customized IBC candidate gene chip on all individuals. The IBC chip targeted 2016 loci of cardiovascular, metabolic, inflammatory and sleep interest based on literature searches, pathway analyses, mouse eQTL data and multiple large GWAS. Coverage of these regions was designed to be dense and cosmopolitan. A large number of low MAF SNPs were included, and r2 performance at lower MAF thresholds was generally equal to the Illumina 1 M array in the three HapMap populations (15).
Quality control for CARe has previously been reported (70). Briefly, for the Illumina IBC array, SNPs were called by Beadstudio and for the Affymetrix 6.0 array by Birdseed. SNPs in linkage disequilibrium (r2 > 0.3) were pruned, and Eigenstrat (78) was used to compute 10 principal components on the subset of individuals passing quality control for use as covariates and to remove population outliers. Samples were also excluded for cryptic relatedness and sample call rate <90% (PLINK). SNPs were excluded for SNP call rate <95%, MAF <1%, heterozygous haploid genotypes and discordant alleles on both arrays. Genotypes with a minor allele frequency (MAF) ≥1% were analyzed.
Statistical analysis
The primary model for large-scale genetic analysis and GWAS of habitual sleep duration as a quantitative trait included adjustment for age, age2, sex, age × sex and 10 population principal components (70). Secondary models were constructed to examine the effects of environmental exposures and comorbidities on the DRD2 SNPs (Supplementary Material, Table S1): Model 2 (alcohol, smoking, years of education and retirement status); and Model 3 (BMI, BMI2, depression, diabetes, hypertension and snoring), with each higher order model including prior variables. Covariate-adjusted residual values for sleep duration were calculated in each cohort using R (79). Individual cohort analyses (sub-divided by ancestry) were performed using an additive model within PLINK (76) and GWAF (80) for non-family and family cohorts. A fixed-effect, inverse variance-weighted meta-analysis was performed in METAL using a standard error model with genomic control applied at the meta-analysis level (81). A P-value of 1.9 × 10−6 was considered IBC array-wide significant (17), while the genome-wide significance threshold was used for AA analyses (P = 5.0 × 10−8). Ethnicity-specific specific QQ and Manhattan plots are provided in Supplementary Material, Figure S1. LocusZoom (82) was used for figure visualizations. Gene-based association analysis was performed using VEGAS (24), and Bonferroni correction for 5275 annotated genes with n = 2 or n > 2 SNPs was applied to determine the statistical significance threshold (9.48 × 10−6).
DRD2 locus analysis
Conditional linear regression was performed using GCTA (83). Twenty-five IBC SNPs were located within 50 kb of rs17601612. The minimal Hardy–Weinberg equilibrium P-value for any SNP in this region was 0.0001 (CARDIA AAs, rs7131440). DRD2 fine mapping imputation was performed in MACH and Minimac (84) using 1000 Genomes Phase 1 (Release 3) (85) reference haplotypes. Individual cohort genotypes (minimal MAF 1%) were phased across all of Chromosome 11 with a 10 MB segment used for imputation. MAF-dependent Rsq quality score cutoffs (<10% MAF: 0.75, <20%: 0.70, <30%: 0.66, <40%: 0.60, <50%: 0.55) were based on guidelines (86) that found a specificity of 98.1% and a sensitivity of 96.4%. Linkage disequilibrium was calculated using Haploview (87) for directly assayed SNPs and vcftools (88) for European 1000 Genomes SNPs. Genomic control was not used on fine-mapping results due to the strengthened, non-normal distribution of P-values within the region. As such, P-values are not directly comparable between assayed and imputed results. Imputed SNP haplogroup membership was assessed via EUR r2 with the top EA assayed SNPs rs2471854 and rs17601612. Secondary analysis tested association of 20 imputed SNPs at the DRD2 locus with 15 phenotypes assessed by polysomnography and 4 questionnaire-based measures described above. Epigenetic and transcription factor binding site regions were based on HaploReg v3 (26) annotations of ENCODE (25,89–91) and NIH Roadmap Epigenomics (27) data using an imputed model generated on 14 February 2015. 1000 Genomes Phase 1 EUR r2 cutoffs of 0.95 were used for candidate functional SNPs, as calculated internally using vcftools.
Schizophrenia genetic risk score analysis
We identified proxies with r2 > 0.8 in CEU for 12/108 schizophrenia sentinel SNPs on the IBC array. We constructed a genetic risk score weighted by the scaled schizophrenia effect estimate for each SNP and evaluated combined association of the genetic risk score with sleep duration in the EA meta-analysis, as implemented in GTX (92).
Replication analysis
Four primarily European American and 1 Korean cohort were used for replication and generalizability of top SNPs for both signals. Meta-analysis was performed using a fixed-effects inverse variance model in METAL.
Data availability
Meta-analysis results are freely available and have been posted to https://sleepgenetics.org/downloads
Supplementary Material
Funding
B.E.C. is supported by National Institutes of Health grants (T32-HL007901-16, R01-HL113338-02). R.S. is supported by NIH R21 HL121728. Centralized CARe resources for the following eight cohorts were supported through a National Institutes of Health grant to the Broad Institute (N01-HC-65226).
Atherosclerotic Risk in Communities (ARIC): The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C, R01HL087641, R01HL59367 and R01HL086694); a National Human Genome Research Institute contract (U01HG004402); and National Institutes of Health contract (HHSN268200625226C). Infrastructure was partly supported by Grant Number (UL1RR025005), a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Cardiovascular Health Study (CHS): This research was supported by contracts (HHSN268201200036C, HHSN268200800007C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086) and grant (HL080295) from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided by (AG023629) from the National Institute on Aging.
Cleveland Family Study (CFS): National Institutes of Health grants to Case Western Reserve University (RO1 HL46380-01-16, M01-RR-00080).
Coronary Artery Risk in Young Adults (CARDIA): The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of Alabama at Birmingham (HHSN268201300025C, HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C) and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging and an intra-agency agreement between the National Institute on Aging and the National Heart, Lung, and Blood Institute (AG0005).
Framingham Heart Study (FHS): National Institutes of Health grants to Boston University (N01-HC-25195, R01-HL-092577, R01-HL-076784, R01-AG-028321).
Jackson Heart Study (JHS): JHS is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities.
Multi-Ethnic Study of Atherosclerosis (MESA): MESA is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with MESA investigators. Support for MESA is provided by contracts (N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and CTSA UL1-RR-024156). Funding for CARe genotyping was provided by the National Heart, Lung, and Blood Institute contract (N01-HC-65226).
Sleep Heart Health Study (SHHS): National Institutes of Health grants to Johns Hopkins University (U01 HL064360), Case Western University (U01 HL063463), University of California, Davis (U01 HL053916), University of Arizona (U01 HL053938), University of Minnesota (relocated in 2006 to University of Arizona) (U01 HL053934), University of Pittsburgh (U01 HL077813), Boston University (U01 HL053941), MedStar Research Institute (U01 HL063429), Johns Hopkins University (U01 HL053937).
Korean Genome and Epidemiology Study (KoGES): This study was provided with biospecimens and data from the Korean Genome Analysis Project (4845-301), the Korean Genome and Epidemiology Study (2001-347-6111-221, 2002-347-6111-221) and Korea Biobank Project (4851-307, KBP-2013-25) that were supported by the Korea Center for Disease Control and Prevention, Republic of Korea.
Religious Orders Study (ROS): The Religious Orders Study is supported by grants from the National Institute on Aging: P30AG10161, R01AG15819, R01AG24871, R01AG26147, R01AG26916, P01AG09466, P01AG14449; K08AG00849, K23AG23675, the Illinois Department of Public Health, the Elsie Heller Brain Bank Endowment Fund, and the Robert C. Borwell Chair of Neurological Sciences.
Rush Memory and Aging Project (MAP): The Rush Memory and Aging Project is supported by the National Institutes of Health grants R01AG017917, P30AG010161, R01AG015819, R01AG024480, R01NS078009, R01AG036836 and R01AG030146.
Osteoporotic Fractures in Men (MrOS) Study: The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Institute on Aging, the National Center for Research Resources, and NIH Roadmap for Medical Research under the following grant numbers: (U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810 and UL1 RR024140). The National Heart, Lung, and Blood Institute provides funding for the MrOS Sleep ancillary study ‘Outcomes of Sleep Disorders in Older Men’ under the following grant numbers: (R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838 and R01 HL070839). The National Institute of Arthritis and Musculoskeletal and Skin Diseases provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number (RC2ARO58973).
Study of Osteoporotic Fractures (SOF): The Study of Osteoporotic Fractures (SOF) is supported by National Institutes of Health funding. The National Institute on Aging provides support under the following grant numbers: (R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574 and R01 AG027576). The SOF Sleep Study is supported by National Institutes of Health funding. The National Institute on Aging provides support under the following grant numbers: (R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, R01 AG027576 and R01 AG026720).
Supplementary Material
Acknowledgements
The authors acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. The authors thank the staff and participants of the ARIC study for their important contributions. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. This manuscript has been reviewed by CARDIA for scientific content. They thank the investigators and staff of the San Francisco Coordinating Center at California Pacific Medical Center for their assistance in both the MrOS and SOF studies.
Conflict of Interest statement. B.E.C., D.J.G., D.A.B., A.S.B., S.G.B., P.L.De J., T.F., S.A.G., E.K.L., D.S.L., S.K.L., S.R.P., N.M.P., R.S., C.S., J.W., P.C.Z. and X.Z. and H.K. have nothing to disclose. The following reports are deemed by the corresponding author to not be a direct conflict of interest: D.S.E., G.J.T. and S.R. and W.C.J. report grants from NIH relevant to the conduct of the study. A.S.L. reports grants from Canadian Institutes of Health Research, from Heart and Stroke Foundation of Ontario, personal fees from UCB Pharma Inc., and personal fees from Merck and Co., outside the submitted work. K.L.S. reports personal fees from Merck and Co, outside of the submitted work. K.Y. reports personal fees from Novartis and Pfizer as a consultant, serves on DSMBs for Takeda, Inc. and an NIH sponsored Study and on the Beeson Scientific Advisory Committee.
References
- 1.Cappuccio F.P., Cooper D., D'Elia L., Strazzullo P., Miller M.A. (2011) Sleep duration predicts cardiovascular outcomes: a systematic review and meta-analysis of prospective studies. Eur. Heart J., 32, 1484–1492. [DOI] [PubMed] [Google Scholar]
- 2.Cappuccio F.P., D'Elia L., Strazzullo P., Miller M.A. (2010) Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep, 33, 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colten H.R., Altevogt B.M. (ed). (2006) Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 4.Klerman E.B., Dijk D.-J. (2008) Age-related reduction in the maximal capacity for sleep—implications for insomnia. Curr. Biol., 18, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goel N., Kim H., Lao R.P. (2005) Gender differences in polysomnographic sleep in young healthy sleepers. Chronobiol. Int., 22, 905–915. [DOI] [PubMed] [Google Scholar]
- 6.Durrence H.H., Lichstein K.L. (2006) The sleep of African Americans: a comparative review. Behav. Sleep. Med., 4, 29–44. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb D.J., O'Connor G.T., Wilk J.B. (2007) Genome-wide association of sleep and circadian phenotypes. BMC Med. Genet., 8 (Suppl. 1), S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne E.M., Gehrman P.R., Medland S.E., Nyholt D.R., Heath A.C., Madden P.A., Hickie I.B., Van Duijn C.M., Henders A.K., Montgomery G.W. et al. (2013) A genome-wide association study of sleep habits and insomnia. Am. J. Med. Genet. B Neuropsychiatr. Genet., 162B, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath A.C., Kendler K.S., Eaves L.J., Martin N.G. (1990) Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep, 13, 318–335. [DOI] [PubMed] [Google Scholar]
- 10.Ollila H.M., Utge S., Kronholm E., Aho V., Van Leeuwen W., Silander K., Partonen T., Perola M., Kaprio J., Salomaa V. et al. (2012) TRIB1 constitutes a molecular link between regulation of sleep and lipid metabolism in humans. Transl. Psychiatry, 2, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ollila H.M., Kettunen J., Pietiläinen O., Aho V., Silander K., Kronholm E., Perola M., Lahti J., Räikkönen K., Widen E. (2014) Genome-wide association study of sleep duration in the Finnish population. J. Sleep Res., 23, 609–618. [DOI] [PubMed] [Google Scholar]
- 12.Allebrandt K.V., Amin N., Müller-Myhsok B., Esko T., Teder-Laving M., Azevedo R.V., Hayward C., van Mill J., Vogelzangs N., Green E.W. et al. (2013) A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol. Psychiatry, 18, 122–132. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb D.J., Hek K., Chen T.H., Watson N.F., Eiriksdottir G., Byrne E.M., Cornelis M., Warby S.C., Bandinelli S., Cherkas L. et al. (2015) Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol. Psychiatry, 20, 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N. et al. (2008) Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE, 3, e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena R., Elbers C.C., Guo Y., Peter I., Gaunt T.R., Mega J.L., Lanktree M.B., Tare A., Castillo B.A., Li Y.R. et al. (2012) Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet., 90, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. (2012) Overview and findings from the Rush Memory and Aging Project. Curr. Alzheimer Res., 9, 646–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim A.S., Yu L., Costa M.D., Leurgans S.E., Buchman A.S., Bennett D.A., Saper C.B. (2012) Increased fragmentation of rest-activity patterns is associated with a characteristic pattern of cognitive impairment in older individuals. Sleep, 35, 633–640B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. (2012) Overview and findings from the Religious Orders Study. Curr. Alzheimer Res., 9, 628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orwoll E., Blank J.B., Barrett-Connor E., Cauley J., Cummings S., Ensrud K., Lewis C., Cawthon P.M., Marcus R., Marshall L.M. et al. (2005) Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp. Clin. Trials, 26, 569–585. [DOI] [PubMed] [Google Scholar]
- 22.Blank J.B., Cawthon P.M., Carrion-Petersen M.L., Harper L., Johnson J.P., Mitson E., Delay R.R. (2005) Overview of recruitment for the osteoporotic fractures in men study (MrOS). Contemp. Clin. Trials, 26, 557–568. [DOI] [PubMed] [Google Scholar]
- 23.Cummings S.R., Black D.M., Nevitt M.C., Browner W.S., Cauley J.A., Genant H.K., Mascioli S.R., Scott J.C., Seeley D.G., Steiger P. et al. (1990) Appendicular bone density and age predict hip fracture in women. The Study of Osteoporotic Fractures Research Group. JAMA, 263, 665–668. [PubMed] [Google Scholar]
- 24.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., AMFS Investigators, Hayward N.K., Montgomery G.W., Visscher P.M. et al. (2010) A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol., 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Bertolino A., Fazio L., Blasi G., Rampino A., Romano R., Lee M.L., Xiao T., Papp A., Wang D. et al. (2007) Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc. Natl Acad. Sci. USA, 104, 20552–20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Mei C., Ramos M., Iitaka C., Borrelli E. (2009) Getting specialized: presynaptic and postsynaptic dopamine D2 receptors. Curr. Opin. Pharmacol, 9, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schizophrenia Working Group of the Psychiatric Genomics Consortium. (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chouinard S., Poulin J., Stip E., Godbout R. (2004) Sleep in untreated patients with schizophrenia: a meta-analysis. Schizophr. Bull., 30, 957–967. [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas C.J. (1917) A prompt emergency hypnotic. New York Med. J., 106, 1032. [Google Scholar]
- 34.Corsini G.U., Del Zompo M., Manconi S., Piccardi M.P., Onali P.L., Mangoni A. (1977) Evidence for dopamine receptors in the human brain mediating sedation and sleep. Life Sci., 20, 1613–1618. [DOI] [PubMed] [Google Scholar]
- 35.Depoortère R., Bardin L., Rodrigues M., Abrial E., Aliaga M., Newman-Tancredi A. (2009) Penile erection and yawning induced by dopamine D2-like receptor agonists in rats: influence of strain and contribution of dopamine D2, but not D3 and D4 receptors. Behav. Pharmacol., 20, 303–311. [DOI] [PubMed] [Google Scholar]
- 36.Wisor J.P., Nishino S., Sora I., Uhl G.H., Mignot E., Edgar D.M. (2001) Dopaminergic role in stimulant-induced wakefulness. J. Neurosci., 21, 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkow N.D., Fowler J.S., Logan J., Alexoff D., Zhu W., Telang F., Wang G.J., Jayne M., Hooker J.M., Wong C. et al. (2009) Effects of modafinil on dopamine and dopamine transporters in the male human brain: clinical implications. JAMA, 301, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anzalone A., Lizardi-Ortiz J.E., Ramos M., De Mei C., Hopf F.W., Iaccarino C., Halbout B., Jacobsen J., Kinoshita C., Welter M. et al. (2012) Dual control of dopamine synthesis and release by presynaptic and postsnyaptic dopamine D2 receptors. J. Neurosci., 26, 9023–9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeman P., Guan H.C., Hirbec H. (2009) Dopamine D2High receptors stimulated by phencyclidines, lysergic acid diethylamide, salvinorin A, and modafinil. Synapse, 63, 698–704. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan D., Pinsonneault J.K., Papp A.C., Zhu H., Lemeshow S., Mash D.C., Sadee W. (2013) Dopamine transporter DAT and receptor DRD2 variants affect risk of lethal cocaine abuse: a gene–gene–environment interaction. Transl. Psychiatry, 3, e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu W.M., Xu X.H., Yan M.M., Wang Y.Q., Urade Y., Huang Z.L. (2010) Essential role of dopamine D2 receptor in the maintenance of wakefulness, but not in homeostatic regulation of sleep, in mice. J. Neurosci., 30, 4382–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zahniser N.R., Simosky J.K., Mayfield R.D., Negri C.A., Hanania T., Larson G.A., Kelly M.A., Grandy D.K., Rubinstein M., Low M.J. et al. (2000) Functional uncoupling of adenosine A(2A) receptors and reduced response to caffeine in mice lacking dopamine D2 receptors. J. Neurosci., 20, 5949–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciruela F., Burgueño J., Casadó V., Canals M., Marcellino D., Goldberg S.R., Bader M., Fuxe K., Agnati L.F., Lluis C. et al. (2004) Combining mass spectrometry and pull-down techniques for the study of receptor heteromerization. Direct epitope-epitope electrostatic interactions between adenosine A2A and dopamine D2 receptors. Anal. Chem., 76, 5354–5363. [DOI] [PubMed] [Google Scholar]
- 44.Navarro G., Aymerich M.S., Marcellino D., Cortés A., Casadó V., Mallol J., Canela E.I., Agnati L., Woods A.S., Fuxe K. et al. (2009) Interactions between calmodulin, adenosine A2A, and dopamine D2 receptors. J. Biol. Chem., 284, 28058–28068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen Human Brain Atlas Study [Internet]. Allen Institute for Brain Science, Seattle, WA: ©2015. http://human.brain-map.org/ 25 October 2015, last access date. [Google Scholar]
- 46.Kaasinen V., Aalto S., Någren K., Rinne J.O. (2004) Dopaminergic effects of caffeine in the human striatum and thalamus. Neuroreport, 15, 281–285. [DOI] [PubMed] [Google Scholar]
- 47.Hamdi A. (1998) Melatonin administration increases the affinity of D2 dopamine receptors in the rat striatum. Life Sci., 63, 2115–2120. [DOI] [PubMed] [Google Scholar]
- 48.Volkow N.D., Wang G.J., Telang F., Fowler J.S., Logan J., Wong C., Ma J., Pradhan K., Tomasi D., Thanos P.K. et al. (2008) Sleep deprivation decreases binding of [11C]raclopride to dopamine D2/D3 receptors in the human brain. J. Neurosci., 28, 8454–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hood S., Cassidy P., Cossette M.P., Weigl Y., Verwey M., Robinson B., Stewart J., Amir S. (2010) Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J. Neurosci., 30, 14046–14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wisor J.P., Schmidt M.A., Clegern W.C. (2011) Evidence for neuroinflammatory and microglial changes in the cerebral response to sleep loss. Sleep, 34, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao W., Zhang S.Z., Tang M., Zhang X.H., Zhou Z., Yin Y.Q., Zhou Q.B., Huang Y.Y., Liu Y.J., Wawrousek E. et al. (2013) Suppression of neuroinflammation by astrocytic dopamine D2 receptors via αB-crystallin. Nature, 494, 90–94. [DOI] [PubMed] [Google Scholar]
- 52.Fuxe K., Marcellino D., Woods A.S., Giuseppina L., Antonelli T., Ferraro L., Tanganelli S., Agnati L.F. (2009) Integrated signaling in heterodimers and receptor mosaics of different types of GPCRs of the forebrain: relevance for schizophrenia. J. Neural Transm., 116, 923–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitzpatrick K., Winrow C.J., Gotter A.L., Millstein J., Arbuzova J., Brunner J., Kasarskis A., Vitaterna M.H., Renger J.J., Turek F.W. (2012) Altered sleep and affect in the neurotensin receptor 1 knockout mouse. Sleep, 35, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer-Spasche A., Reed H.E., Piggins H.D. (2002) Neurotensin phase-shifts the firing rate rhythm of neurons in the rat suprachiasmatic nuclei in vitro. Eur. J. Neurosci., 16, 339–344. [DOI] [PubMed] [Google Scholar]
- 55.Yujnovsky I., Hirayama J., Doi M., Borrelli E., Sassone-Corsi P. (2006) Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc. Natl Acad. Sci. USA, 103, 6386–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blasi G., Napolitano F., Ursini G., Taurisano P., Romano R., Caforio G., Fazio L., Gelao B., Di Giorgio A., Iacovelli L. et al. (2011) DRD2/AKT1 interaction on D2 c-AMP independent signaling, attentional processing, and response to olanzapine treatment in schizophrenia. Proc. Natl Acad. Sci. USA, 108, 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koehler S., Wacker J., Odorfer T., Reif A., Gallinat J., Fallgatter A.J., Herrmann M.J. (2011) Resting posterior minus frontal EEG slow oscillations is associated with extraversion and DRD2 genotype. Biol. Psychol., 87, 407–413. [DOI] [PubMed] [Google Scholar]
- 58.Moyer R.A., Wang D., Papp A.C., Smith R.M., Duque L., Mash D.C., Sadee W. (2011) Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology, 36, 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson J., Thomas N., Singleton A., Piggott M., Lloyd S., Perry E.K., Morris C.M., Perry R.H., Ferrier I.N., Court J.A. (1997) D2 dopamine receptor gene (DRD2) Taq1 A polymorphism: reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics, 7, 479–484. [DOI] [PubMed] [Google Scholar]
- 60.Jönsson E.G., Nöthen M.M., Grünhage F., Farde L., Nakashima Y., Propping P., Sedvall G.C. (1999) Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol. Psychiatry, 4, 290–296. [DOI] [PubMed] [Google Scholar]
- 61.Sipilä T., Kananen L., Greco D., Donner J., Silander K., Terwilliger J.D., Auvinen P., Peltonen L., Lönnqvist J., Pirkola S. et al. (2010) An association analysis of circadian genes in anxiety disorders. Biol. Psychiatry, 67, 1163–1170. [DOI] [PubMed] [Google Scholar]
- 62.Peciña M., Mickey B.J., Love T., Wang H., Langenecker S.A., Hodgkinson C., Shen P.H., Villafuerte S., Hsu D., Weisenbach S.L. et al. (2013) DRD2 polymorphisms modulate reward and emotion processing, dopamine neurotransmission and openness to experience. Cortex, 49, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nyman E.S., Sulkava S., Soronen P., Miettunen J., Loukola A., Leppä V., Joukamaa M., Mäki P., Järvelin M.R., Freimer N. et al. (2011) Interaction of early environment, gender and genes of monoamine neurotransmission in the aetiology of depression in a large population-based Finnish birth cohort. BMJ Open, 1, e000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheung I., Shulha H.P., Jiang Y., Matevossian A., Wang J., Weng Z., Akbarian S. (2010) Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl Acad. Sci. U S A, 107, 8824–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothmond D.A., Weickert C.S., Webster M.J. (2012) Developmental changes in human dopamine neurotransmission: cortical receptors and terminators. BMC Neurosci., 13, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volkow N.D., Tomasi D., Wang G.J., Telang F., Fowler J.S., Logan J., Benveniste H., Kim R., Thanos P.K., Ferré S. (2012) Evidence that sleep deprivation downregulates dopamine D2R in ventral striatum in the human brain. J. Neurosci., 32, 6711–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goel N., Banks S., Lin L., Mignot E., Dinges D.F. (2011) Catechol-O-methyltransferase Val158Met polymorphism associates with individual differences in sleep physiologic responses to chronic sleep loss. PLoS ONE, 6, e29283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reuter M., Peters K., Schroeter K., Koebke W., Lenardon D., Bloch B., Hennig J. (2005) The influence of the dopaminergic system on cognitive functioning: a molecular genetic approach. Behav. Brain Res., 164, 93–99. [DOI] [PubMed] [Google Scholar]
- 70.Musunuru K., Lettre G., Young T., Farlow D.N., Pirruccello J.P., Ejebe K.G., Keating B.J., Yang Q., Chen M.H., Lapchyk N. et al. (2010) Candidate gene association resource (CARe): design, methods, and proof of concept. Circ. Cardiovasc. Genet., 3, 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quan S.F., Howard B.V., Iber C., Kiley J.P., Nieto F.J., O'Connor G.T., Rapoport D.M., Redline S., Robbins J., Samet J.M. et al. (1997) The Sleep Heart Health Study: design, rationale, and methods. Sleep, 20, 1077–1085. [PubMed] [Google Scholar]
- 72.Kump K., Whalen C., Tishler P.V., Browner I., Ferrette V., Strohl K.P., Rosenberg C., Redline S. (1994) Assessment of the validity and utility of a sleep-symptom questionnaire. Am. J. Respir. Crit. Care Med., 150, 735–741. [DOI] [PubMed] [Google Scholar]
- 73.Shin C., Abbott R.D., Lee H., Kim J., Kimm K. (2004) Prevalence and correlates of orthostatic hypotension in middle-aged men and women in Korea: the Korean Health and Genome Study. J. Hum. Hypertens., 18, 717–723. [DOI] [PubMed] [Google Scholar]
- 74.Kim Y.J., Go M.J., Hu C., Hong C.B., Kim Y.K., Lee J.Y., Hwang J.Y., Oh J.H., Kim D.J., Kim N.H. et al. (2011) Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet., 43, 990–995. [DOI] [PubMed] [Google Scholar]
- 75.Buysse D.J., Reynolds C.F. III, Monk T.H., Berman S.R., Kupfer D.J. (1989) The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res., 28, 193–213. [DOI] [PubMed] [Google Scholar]
- 76.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Redline S., Tishler P.V., Tosteson T.D., Williamson J., Kump K., Browner I., Ferrette V., Krejci P. (1995) The familial aggregation of obstructive sleep apnea. Am. J. Respir. Crit. Care Med., 151, 682–687. [DOI] [PubMed] [Google Scholar]
- 78.Price A.L., Patterson N.J., Pleng R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 79.R Development Core Team. (2007) R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0 http://www.R-project.org. [Google Scholar]
- 80.Chen M.H., Yang Q. (2010) GWAF: an R package for genome-wide association analyses with family data. Bioinformatics, 26, 580–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of ANthropometric Traits (GIANT) Consortium, DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.1000 Genomes Project Consortium. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson T.A., Fujita H., Hara K., Tsunoda T.. Optimized Rsq thresholds for quality control of MACH/minimac imputed genotypes [abstract]. Presented at the 12th International Congress of Human Genetics/61st Annual Meeting of The American Society of Human Genetics, October 12, 2011, Montreal, Canada. [Google Scholar]
- 87.Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265 . [DOI] [PubMed] [Google Scholar]
- 88.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y. et al. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res., 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yip K.Y., Cheng C., Bhardwaj N., Brown J.B., Leng J., Kundaje A., Rozowsky J., Birney E., Bickel P., Snyder M. et al. (2012) Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol., 13, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dastani Z., Hivert M.F., Timpson N., Perry J.R., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.P. et al. (2012) Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet., 8, e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Meta-analysis results are freely available and have been posted to https://sleepgenetics.org/downloads