Abstract
Adolescent idiopathic scoliosis (AIS) is a complex inherited spinal deformity whose etiology has been elusive. While common genetic variants are associated with AIS, they explain only a small portion of disease risk. To explore the role of rare variants in AIS susceptibility, exome sequence data of 391 severe AIS cases and 843 controls of European ancestry were analyzed using a pathway burden analysis in which variants are first collapsed at the gene level then by Gene Ontology terms. Novel non-synonymous/splice-site variants in extracellular matrix genes were significantly enriched in AIS cases compared with controls (P = 6 × 10−9, OR = 1.7, CI = 1.4–2.0). Specifically, novel variants in musculoskeletal collagen genes were present in 32% (126/391) of AIS cases compared with 17% (146/843) of in-house controls and 18% (780/4300) of EVS controls (P = 1 × 10−9, OR = 1.9, CI = 1.6–2.4). Targeted resequencing of six collagen genes replicated this association in combined 919 AIS cases (P = 3 × 10−12, OR = 2.2, CI = 1.8–2.7) and revealed a highly significant single-gene association with COL11A2 (P = 6 × 10−9, OR = 3.8, CI = 2.6–7.2). Importantly, AIS cases harbor mainly non-glycine missense mutations and lack the clinical features of monogenic musculoskeletal collagenopathies. Overall, our study reveals a complex genetic architecture of AIS in which a polygenic burden of rare variants across extracellular matrix genes contributes strongly to risk.
Introduction
Scoliosis is a common pediatric musculoskeletal disorder. Approximately 0.3% of all children have scoliosis with a spinal curvature of >20° (Cobb angle) requiring treatment (1), and more than 1 in 10 000 children have severe spine deformity requiring surgery (2). Scoliosis may be explained by genetic syndromes or conditions with distinguishing features in addition to scoliosis, including Marfan syndrome, cerebral palsy and muscular dystrophy (3). However, adolescent idiopathic scoliosis (AIS), which is defined as isolated scoliosis without any underlying diagnosis, represents the largest subset of scoliosis patients.
A genetic predisposition to AIS is evidenced by high rates of concordance in twin studies and increased risk to first-degree relatives of individuals with AIS (4–9). Despite the description of familial cases, genetic linkage studies have been largely unsuccessful in identifying AIS disease genes. In addition, many studies now support a polygenic inheritance model of AIS (9,10). Recently published genome-wide association studies revealed associations of AIS with common variants near neural cell adhesion molecules (11), ladybird homeobox 1 (12) and G-protein-coupled receptor 126 (13), but these explain only a small proportion of disease heritability. We recently demonstrated that rare variants in the genes underlying Marfan syndrome (FBN1) and congenital contractural arachnodactyly (FBN2) contribute to AIS risk and severity (14), suggesting that rare variants may contribute significantly to the genetic architecture of AIS.
Results
Rare variants in extracellular matrix genes collectively influence AIS risk
To comprehensively explore the role of rare genetic variants in AIS susceptibility, exome sequence data were generated for 391 unrelated AIS cases and 843 unrelated controls of European descent. Gene-burden analysis of novel non-synonymous/splice-site variants was performed across all genes, with novel rare variants defined as being absent from dbSNP 141 (excluding variants submitted solely by EVS). Because no single gene surpassed exome-wide significance (Supplementary Material, Fig. S1), we developed a pathway burden analysis framework that, unlike some methods (15), preserves power by utilizing data from all genes, not only those with significant single-gene associations. With this method, variants are first collapsed at the gene level and then by Gene Ontology (GO) term membership.
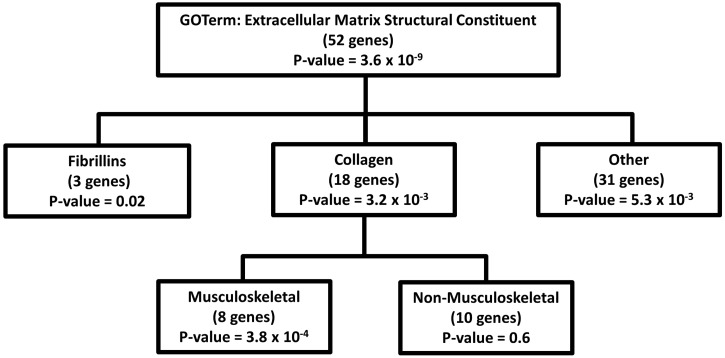
Exome-wide pathway burden analysis yielded a strong association between AIS and novel variants in genes within the GO-terms ‘Extracellular Matrix Structural Constituent’ (P = 6 × 10−9, OR = 1.7 CI = 1.42–2.02) and ‘Extracellular Matrix Disassembly’ (P = 9 × 10−7, OR = 1.58 CI = 1.33–1.88) (Supplementary Material, Table S1) when all 1234 exomes were analyzed. Notably, these and several other top associated GO-terms are highly correlated, often consisting of gene lists that are subsets of one another. While the Extracellular Matrix Structural Constituent GO-term includes only a subset of all known extracellular matrix (ECM) genes, its 52 genes fit into several classes (Supplementary Material, Table S2). Although the majority of genes within the top ECM GO-term displayed a higher frequency of novel variants among AIS cases compared with controls (41/52 genes; binomial-test P-value = 3.6 × 10−5), only fibrillin and collagen genes were significantly associated with AIS risk as individual subgroups (Fig. 1). Even after removal of FBN1 and FBN2 novel variants, which we previously showed are associated with AIS risk (14), ‘Extracellular Matrix Structural Constituent’ remained associated with AIS (P = 5 × 10−6, OR = 1.4 CI = 1.2–1.7), with a significant proportion of the association being driven by novel variants in collagen genes. To ensure that these findings were not due to differences in sequencing coverage, we calculated the average genotype missing rate in cases and in-house controls separately (Supplementary Material, Table S3). These results show consistently low missing rates and comparable numbers of novel variants in both AIS cases and controls among genes within the GO-term ‘Extracellular Matrix Structural Constituent’. Additionally, there was no association between the number of novel synonymous variants between AIS cases and controls, again suggesting that the observed association with non-synonymous variants is not due to bias in capture of sequencing depth (Supplementary Material, Fig. S2).
Figure 1.
Genes contributing to the association of AIS with the GO-term: ECM structural constituent. All non-fibrillin, non-collagen genes were collapsed and referred to as ‘other’. Associations with specific gene subgroups were performed as for pathway burden analysis, i.e. the number of variants observed across the set of genes were summed and used in a linear regression with AIS status as the outcome variable.
Harboring multiple ECM variants influences clinical systemic features and joint hypermobility
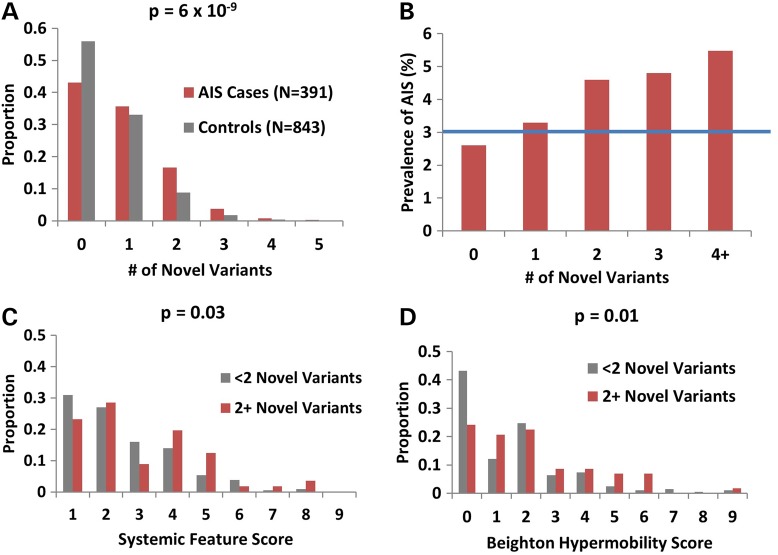
The ECM comprises multiple interacting proteins; therefore, we sought to determine if the association with AIS was due to a polygenic effect of having more than one variant in the 52 ECM genes within this GO-term. On average, significantly more ECM gene novel variants were observed in the AIS cohort compared with controls (0.9 versus 0.6 variants per person, P = 6 × 10−9) (Fig. 2A) with increased risk for every additional novel variant (Fig. 2B). Clinical evaluations revealed that AIS cases with ≥2 novel variants in ECM genes had slightly higher Ghent systemic feature scores (16) (Marfan syndrome diagnostic criteria) (3.0 versus 2.5, P = 0.03) (Fig. 2C) and Beighton joint hypermobility scores (17) (1.7 versus 0.9, P = 0.01) compared with individuals with <2 variants (Fig. 2D). However, very few individuals exhibited scores high enough to warrant screening for connective tissue disorders (Ghent systemic feature score >7 and Beighton score >5) (16,17) and none met criteria for these disorders. These data suggest that the polygenic accumulation of multiple novel variants across a large set of ECM genes, only some of which are contained within the top GO-term, contributes not only to AIS risk but also increases joint hypermobility and other clinical features known to correlate with ECM dysfunction.
Figure 2.
A polygenic burden of novel variants in ECM genes is associated with AIS and correlates with Ghent systemic feature scores and joint hypermobility (A) AIS cases have more non-synonymous variants in genes within the ‘Extracellular Matrix Structural Constituent’ GO-term compared with controls (Fisher's exact test P-value = 6 × 10−9). (B) AIS prevalence increases with every additional novel variant. The blue line represents the population prevalence of 3% for individuals with scoliosis Cobb angle >10°. (C) Ghent systemic feature score and Beighton hypermobility scores are higher in AIS cases with ≥2 novel variants in genes in the ‘Extracellular Matrix Structural Constituent’ GO-term compared with AIS cases with <2 novel variants. None of the individuals in the AIS cohort were diagnosed with any known connective tissue disorder, including Marfan syndrome, Ehlers–Danlos syndrome or Stickler syndrome.
Rare variants in musculoskeletal collagen genes are associated with AIS
Because collagens were the most significant gene class within our top AIS-associated ECM GO term (Fig. 1), we expanded our analysis to investigate the frequency of novel variants in all 42 known autosomal collagen genes, only 18 of which were included in the top-ranked ECM GO term. Compared with controls, AIS cases had a greater frequency of novel non-synonymous variants in 14 of the 17 collagen genes known to cause Mendelian musculoskeletal collagenopathies (Table 1) (binomial-test P-value = 0.01), suggesting that only a subset of collagen genes, predominantly those that form highly interconnected networks of fibrils within the ECM (18), contribute to AIS risk. Overall, 32% (126/391) of AIS cases had novel non-synonymous or splice-site variants in these 17 musculoskeletal collagen genes compared with 17% (146/843) of controls (P = 3 × 10−8, OR = 2.0 CI = 1.6–2.7) and 18% (780/4300) of individuals of European descent in the NHLBI Exome Variant Server (EVS) (P = 1 × 10−9, OR = 1.9 CI = 1.6–2.4) (Table 1) (variants listed in Supplementary Material, Table S4). As expected, there was no difference in the combined frequency of novel synonymous variants in the 17 musculoskeletal collagen genes (Supplementary Material, Table S5), or in novel non-synonymous/splice-site variants in the 24 non-musculoskeletal collagen genes (Supplementary Material, Table S6) between AIS cases and controls. We also observed no differences between the frequency of novel non-synonymous/splice-site variants among in-house controls and EVS controls at these collagen genes, suggesting that our analysis pipeline is comparable to that used for EVS individuals and that any differences in ethnicity (i.e. European subgroups) between our cohorts and the EVS cohort are minimal.
Table 1.
AIS is associated with novel variants in musculoskeletal collagen genes
| AIS cases (n = 391) | In-house controls |
EVS controls |
||||
|---|---|---|---|---|---|---|
| n = 843 | P-value | n = 4300 | Odds ratio (95% CI) | P-value | ||
| COL1A1 | 5 (1.3%) | 10 (1.1%) | 0.59 | 27 (0.6%) | 1.9 (0.57–5.1) | 0.2 |
| COL1A2 | 4 (1%) | 6 (0.7%) | 0.44 | 40 (0.9%) | 1.0 (0.26–2.8) | 0.79 |
| COL2A1 | 8 (2%) | 6 (0.7%) | 0.05 | 41 (1%) | 2.0 (0.81–4.4) | 0.07 |
| COL3A1 | 6 (1.5%) | 3 (0.4%) | 0.04 | 32 (0.7%) | 1.9 (0.65–1.14) | 0.14 |
| COL5A1 | 9 (2.3%) | 14 (1.7%) | 0.34 | 78 (1.8%) | 1.2 (0.5–2.4) | 0.57 |
| COL5A2 | 11 (2.8%) | 9 (1.1%) | 0.04 | 47 (1.1%) | 2.4 (1.1–4.8) | 0.02 |
| COL5A3 | 11 (2.8%) | 5 (0.6%) | 0.003 | 47 (1.1%) | 2.4 (1.1–4.8) | 0.02 |
| COL6A1 | 6 (1.5%) | 6 (0.7%) | 0.17 | 54 (1.3%) | 1.1 (0.4–2.7) | 0.65 |
| COL6A2 | 6 (1.5%) | 13 (1.5%) | 0.64 | 75 (1.7%) | 0.84 (0.29–1.89 | 0.84 |
| COL6A3 | 18 (4.6%) | 26 (3.1%) | 0.17 | 118 (2.7%) | 1.6 (0.9–2.6) | 0.09 |
| COL9A1 | 1 (0.3%) | 9 (1.1%) | 0.98 | 34 (0.8%) | 0.3 (0.01–1.8) | 0.36 |
| COL9A2 | 4 (1%) | 1 (0.1%) | 0.04 | 14 (0.3%) | 2.9 (0.7–9.4) | 0.07 |
| COL9A3 | 7 (1.8%) | 8 (0.9%) | 0.19 | 27 (0.6%) | 2.68 (0.98–6.3) | 0.03 |
| COL10A1 | 1 (0.3%) | 3 (0.4%) | 0.8 | 23 (0.5%) | 0.45 (0.01–2.8) | 0.72 |
| COL11A1 | 9 (2.3%) | 8 (0.9%) | 0.07 | 43 (1%) | 2.2 (0.9–4.5) | 0.05 |
| COL11A2 | 11 (2.8%) | 9 (1.1%) | 0.04 | 29 (0.7%) | 3.9 (1.8–8.1) | 5 × 10−4 |
| COL12A1 | 9 (2.3%) | 10 (1.2%) | 0.14 | 51 (1.2%) | 1.8 (0.8–3.8) | 0.11 |
| Total | 126 (32%) | 146 (17%) | 3 × 10−8 | 780 (18%) | 1.9 (1.6–2.4) | 2 × 10−9 |
Shown are minor allele counts for each gene with carrier percentages in parenthesis. P-values were calculated using a two-sided Fisher's exact test. Odds ratios are calculated for AIS cases versus EVS controls. EVS, NHLBI Exome Variant Server. Significant P-values before multiple test correction are in bold.
Replication of musculoskeletal collagen gene-burden associations
To replicate our collagen association results, we sequenced a subset of six musculoskeletal collagen genes (COL2A1, COL3A1, COL5A2, COL6A3, COL11A1 and COL11A2) that showed associations or large numbers of carriers in the initial screen. These genes were sequenced in an independent cohort of 435 AIS cases of European descent (Cohort II) using our BAC-based Multiplex Direct Genomic Selection (MDiGS) targeted re-sequencing method (19). Genotypes called using this resequencing method were extensively validated previously (14,19) and we validated 35/35 randomly selected novel non-synonymous variants observed across the six sequenced genes using Sanger sequencing. The proportion of musculoskeletal collagen novel non-synonymous variant carriers in the AIS cohort II (13%; 58/435) was similar to the AIS exome discovery cohort I (16%; 63/391) and much higher than controls (7%; 371/5143) (Table 2). In order to maximize power, all available family members of variant carriers were Sanger sequenced and included in the analysis of the combined sample after controlling for relatedness using famSKAT. In the combined data set, novel variants in these six resequenced collagen genes occurred at a significantly higher frequency in AIS cases (121/826; 15%) (P = 3 × 10−12, OR = 2.2, CI = 1.8–2.7).
Table 2.
Replication of AIS association with novel variants in six resequenced collagen genes
| Controls (n = 5143) | Cohort I (Exomes) |
Cohort II (MDiGS) |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AIS cases (n = 391) | Odds ratio (95% CI) | P-value | AIS cases (n = 435) | Odds ratio (95% CI) | P-value | AIS cases (n = 919) | Odds ratio (95% CI) | P-value | ||
| COL2A1 | 47 (0.9%) | 8 (2%) | 2.3 (0.9–4.8) | 0.06 | 7 (1.6%) | 1.8 (0.7–4.0) | 0.19 | 15 (1.8%) | 2.0 (1.0–3.6 | 0.02 |
| COL3A1 | 35 (0.7%) | 6 (1.5%) | 2.3 (0.77–5.5) | 0.07 | 4 (0.9%) | 1.0 (0.2–3.2) | 1 | 10 (1.1%) | 1.8 (0.8–3.7) | 0.12 |
| COL5A2 | 49 (1%) | 11 (2.8%) | 3.0 (1.4–5.8) | 2 × 10−3 | 6 (1.4%) | 1.5 (0.5–3.4) | 0.44 | 17 (2.1%) | 2.2 (1.2–3.8) | 0.01 |
| COL6A3 | 151 (2.9%) | 18 (4.6%) | 1.6 (0.90–2.6) | 0.07 | 21 (4.8%) | 1.7 (0.99–2.6) | 0.04 | 39 (4.7%) | 1.6 (1.1–2.3) | 0.01 |
| COL11A1 | 51 (1%) | 9 (2.3%) | 2.3(1.0–4.8) | 0.04 | 8 (1.8%) | 1.9 (0.76–4.0) | 0.13 | 17 (2.1%) | 2.1 (1.1–3.7) | 0.01 |
| COL11A2 | 38 (0.7%) | 11 (2.8%) | 3.8 (1.8–7.7) | 5 × 10−4 | 12 (2.8%) | 3.8 (1.8–7.4) | 4 × 10−4 | 23 (2.8%) | 4.3 (2.6–7.2) | 6 × 10−9 |
| Total | 371 (7%) | 63 (16%) | 2.3 (1.7–3.1) | 3 × 10−8 | 58 (13%) | 2.0 (1.5–2.7) | 7 × 10−5 | 121 (15%) | 2.2 (1.8–2.7) | 3 × 10−12 |
Shown are minor allele carriers for novel variants with percentages in parentheses. P-values were calculated using Fisher's exact test. For the combined analysis, famSKAT was used to incorporate genotype information from all available AIS cases and their relatives (total n = 919). All P-values are versus combined in-house and EVS controls (n = 5143).
Novel variants in COL11A2 increase risk of AIS
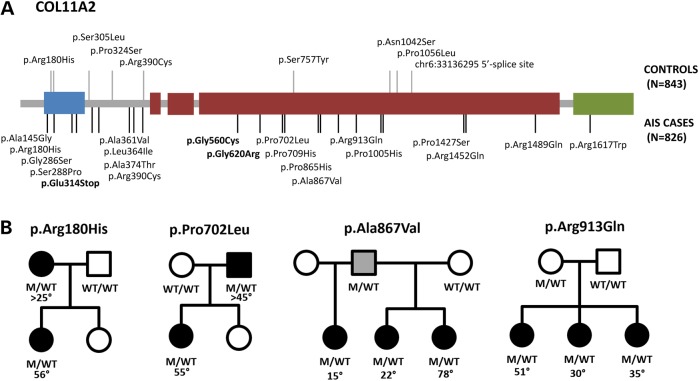
When burden analysis was performed on each of the six musculoskeletal collagen genes individually in the replication cohort, a higher frequency of novel variants was seen in AIS cases compared with controls, with strongest replication of the association with COL11A2 (P = 4 × 10−4, OR = 3.8, CI = 1.8–7.4) (Table 2). For the combined sample, COL11A2 (P = 2 × 10−9, OR = 3.8, CI = 2.2–6.6) well surpassing the exome-wide significant P-value threshold of 2.5 × 10−6 required to correct for multiple comparisons for 20 000 genes. COL11A2 encodes a fibrillar collagen that has previously been associated with rare cases of dominant and recessively inherited osteochondrodysplasia, otospondylomegaepiphyseal (OSMED) as well as non-syndromic sensorineural hearing impairment (20–22). COL11A2 mutations are also a minor cause of Stickler syndrome, having been described in only a few cases (20,23). As the majority of pathogenic mutations in collagen genes reside within the triple-helical regions critical for trimerization, we compared the frequency of variants within the triple-helical domain and the non-helical domains separately. We find that both the triple-helical and non-triple-helical regions harbor increased numbers of novel non-synonymous variants in AIS cases compared with controls (P = 0.003 and 2.5 × 10−4, respectively). In addition to the higher frequency of COL11A2 novel variants in AIS cases compared with controls (Fig. 3A), support for COL11A2 in isolated AIS pathogenesis is provided by the segregation of COL11A2 non-glycine missense variants with AIS in four families (Fig. 3B), and absence of cleft palate, Robin sequence, facial dysmorphism, hearing loss or other features of Stickler syndrome in all of our AIS cases and their family members (Supplementary Material, Table S7). Because few of our AIS COL11A2 variants were insertions, deletions, nonsense or glycine-altering missense variants that are typically associated with these more severe disorders, our data suggest a genotype–phenotype correlation, with missense variants being associated with an isolated AIS phenotype.
Figure 3.
Novel COL11A2 variants are enriched in AIS cases and segregate with AIS in families. (A) Novel non-synonymous/splice-site variants observed in AIS cases (black lines on bottom) and in-house controls (grey lines on top). Glycine altering variants within the triple-helical region are bolded. Blue, thrombospondin domains; green, C-terminal propeptides. (B) Segregation of COL11A2 variants in families with multiple affected individuals. Black is braced or surgically treated AIS. Grey is joint hypermobility.
Novel variants in ECM genes observed in AIS are less deleterious than mutations observed in Mendelian collagenopathies
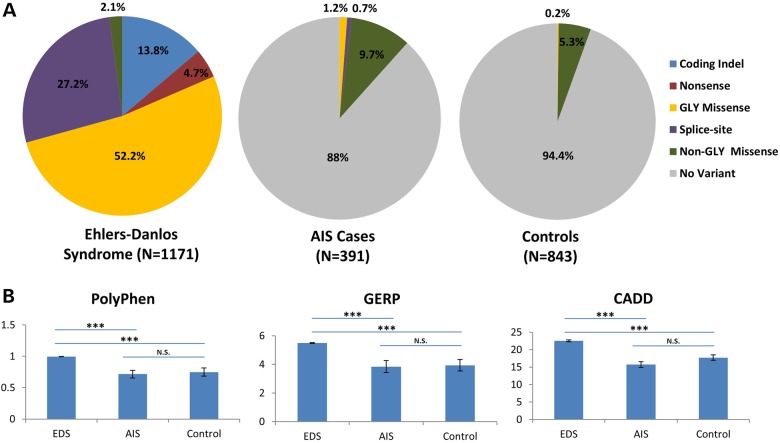
Further support for a genotype–phenotype correlation is provided by the smaller proportion of coding-indels, nonsense, splice-site altering or glycine-altering missense mutations within the triple-helical regions of six collagen genes associated with Ehlers–Danlos syndrome (COL1A1, COL1A2, COL3A1, COL5A1, COL5A2 and COL5A3) among exome-sequenced AIS cases (17%; 8/46) compared with those reported in Ehlers–Danlos syndrome (95%; 606/641) (http://www.le.ac.uk/ge/collagen/) (P < 10−120) (Fig. 4A). In fact, non-glycine missense mutations drive the association between novel variants in musculoskeletal collagen genes and AIS, with the association remaining strong even when only non-glycine missense variants are included in the analysis (P = 3 × 10−6 versus in-house controls and P = 2 × 10−8 versus EVS). Furthermore, functional algorithms predicted novel collagen variants in AIS cases to be less damaging on average compared with EDS mutations, and similarly damaging as novel variants identified in our in-house controls (Fig. 4B and Supplementary Material, Fig. S3). Clinical features also support a correlation of less damaging variants with isolated AIS, as none of our AIS carriers had clinical features associated with the Mendelian collagenopathies (Supplementary Material, Table S7). Our demonstration that variants observed in AIS are less damaging than those found in Mendelian disease is also consistent with complex polygenic inheritance in which an accumulation of minor variants contributes to disease risk.
Figure 4.
Novel collagen variants identified in AIS cases are less damaging than those reported in Ehlers–Danlos syndrome. (A) Variant class distribution of novel coding variants observed in six Ehlers–Danlos syndrome (EDS) genes (COL1A1, COL1A2, COL3A1, COL5A1, COL5A2 and COL5A3) in AIS cases (n = 46), EVS controls (n = 47) or reported in EDS (n = 1171) (http://www.le.ac.uk/ge/collagen/). (B) Three functional prediction algorithms report EDS variants to be more deleterious than variants in AIS cases or controls. ***P < 10−16.
Discussion
ECM proteins are the most abundant proteins in the human body (24) and have already been linked to many monogenic human diseases. Although our data show that specific ECM genes, including COL11A2, contribute more significantly to AIS risk than others, what is equally striking is the combined impact of variants in ECM genes as a class on AIS susceptibility. Not only do the majority of genes within this group contribute to AIS risk, but risk increases proportionally with the number of variants within an individual. Demonstration of the collective effect of multiple rare variants across ECM genes on AIS risk fits with a polygenic burden disease model first described nearly 100 years ago by Fisher (25). Furthermore, as monogenic collagenopathies are associated with a spectrum of skeletal features, including clubfoot, cleft palate, hip dysplasia and pectus excavatum in addition to scoliosis, we hypothesize that the model of polygenic inheritance of rare ECM variants shown here for AIS will also apply to these isolated disorders, with each gene contributing different proportions.
In conclusion, genome-wide pathway burden analysis of exome sequence data identifies ECM genes as a major class of genes contributing to the polygenic inheritance of AIS. More specifically, novel coding variants in musculoskeletal collagen genes increase AIS risk by >2-fold with those in COL11A2 being most strongly associated. Importantly, while damaging mutations in collagen genes have been long known to cause Mendelian disorders, our study demonstrates a role for less damaging mutations in isolated AIS consistent with complex and polygenic inheritance. These findings are of particular clinical importance because accurate prediction of scoliosis risk is needed to enable early intervention and personalized treatment strategies for AIS.
Methods
AIS subjects
Exome sequencing cohort (cohort I) comprised AIS cases recruited from St Louis Children's Hospital and Shriners Hospital for Children—St Louis. The average age of these patients was 20, 83% were female and the average Cobb angle was 54°. Cohort II comprised AIS cases recruited from St Louis Children's Hospital, Shriners Hospital for Children, Texas Scottish Rite Hospital, University of Colorado and University of Iowa. The average age of these patients was 20, 82% were female and the average Cobb angle was 52°. All patients had Cobb angles ≥10° and were of European ancestry. Patients with developmental delay, multiple congenital anomalies or known underlying medical disorders or connective tissue disorder were excluded. Beighton hypermobility scores (17) and Ghent scores (16) were also recorded for patients recruited from St Louis Children's Hospital and Shriners Hospital for Children—St Louis.
Control cohorts
In-house controls consisted of unrelated healthy individuals or patients ascertained for conditions other than scoliosis (i.e. Alzheimer's disease or Amyotrophic Lateral Sclerosis). As the population prevalence of scoliosis with spinal curves ≥10° is ∼3%, we estimate that at most 24 control individuals in our in-house control cohort would have scoliosis to the extent required to be considered a case. All in-house controls were of European ancestry. Additional control data were derived from the National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project [(Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA (URL: http://evs.gs.washington.edu/EVS/, accessed February 2015)].
Sequencing analysis and validation
AIS cases and in-house control DNA were both captured with Agilent and sequenced at GTAC using similar methodologies. For both cases and controls, next-generation sequencing reads were aligned to hg19 human reference sequence (Genome Reference Consortium Human Build 37) using Novoalign software (Novocraft Technologies, Selangor, Malaysia). For both exome and MDiGS sequencing data, only individuals with >95% of captured nucleotide positions covered with at least eight sequencing reads were used in subsequent analyses. Additionally, variant were only included in subsequent analyses if >80% of both cases and controls were covered >8× at that position. Variant calling was done using SAMtools (26) for each individual separately and merged using vcftools (27). Variants were annotated using SeattleSeq Annotation 137 (28). Additionally, all low-quality variants (phred-scaled quality score <30 or genotype quality score <75) were excluded from analyses. Novel missense/nonsense/splice-site changes in collagen genes were validated via Sanger sequencing using an ABI 3730 Sequencer (Life Technologies, Carlsbad, CA, USA).
Targeted resequencing
Targeted resequencing of six collagen genes (COL2A1, COL3A1, COL5A2, COL6A3, COL11A1 and COL11A2) was performed using the Multiplexed Direct Genomic Selection (MDiGS) method as previously described (19). Six bacterial artificial chromosomes (BACs) spanning the coding regions of COL2A1 [RP11-805I19 (chr12:48236980–48428284)], COL5A2 and COL3A1 [RP11-844M2 (chr2:189805929–189986514)], COL6A3 [RP11-66F3 (chr2:238145641–238332665)], COL11A1 [RP11-644O22 (chr1:103285556–103479392) and RP11-483E5 (chr1:103471921–103612378)] and COL11A2 [CTD-2054I15 (chr6:33092626–33234941)] were purchased from either Millipore or BACPAC and used as baits. Indexed samples were pooled in batches of 48–96 prior to capture. BACs were biotinylated using nick translation with biotin-16-dUTP (Roche). Pooled samples were then hybridized with biotinylated BACs and sequenced on one lane of a MiSeq (Illumina).
Pathway burden analysis
Only novel variants were included in pathway burden analysis. First, non-synonymous/splice-site variants within a gene were collapsed to obtain the number of novel (not present in dbSNP 141 or submitted by EVS only) variants per gene. The numbers of variants within groups of genes are then summed based on membership within a given GO-term and used as the dependent variable in a linear regression. GO-terms were obtained from the UniProt Knowledgebase (http://www.uniprot.org/help/uniprotkb).
Statistical analysis
To reduce the risk of population stratification, only cases and controls with principal components confirmed European ancestry were included. Principal components were calculated using EIGENSTRAT (29) from whole-exome SNP data using all common (MAF > 10%) SNPs or all common SNPs present within the MDIGs captured regions for individuals with collagen resequencing data only to ensure the accuracy of self-reported race (Supplementary Material, Fig. S4). For statistical analysis of exome sequencing data, affected individuals from cohorts I and II (discovery and replication cohorts) were compared with exome controls, EVS controls or combined exome and EVS controls. For individual genes and for collapsed collagen association analyses, Fisher's exact tests were performed using R to compare the frequency of novel variants in cases and controls. The Wilcoxon rank-sum tests were performed using R to compare the average Ghent systemic feature score and Beighton hypermobility score among AIS cases who are carriers of 2+ or <2 novel non-synonymous/splice-site variants in the GO-term: ECM Structural Constituent gene set. The prevalence of AIS among carriers of differing numbers of novel non-synonymous/splice-site variants in musculoskeletal collagen genes or novel non-synonymous variants musculoskeletal collagen genes was calculated assuming a population prevalence of AIS of 3% (1) according to the formula P(AIS | # of variants) = P(# of variants | AIS) × P(AIS)/P(# of variants).
Supplementary Material
Funding
This research was supported by Shriners Hospital for Children Research Grants, the University of Missouri Spinal Cord Injury Research Program, the Victor McKusick Fellowship of the Marfan Foundation (DA), and the Children's Discovery Institute of St Louis Children's Hospital and Washington University (CAG) and by the National Institutes of Health (NICHD award R01HD052973), the TSRHC Research Fund, Crystal Charity Ball and Scoliosis Research Society (CAW). The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant #UL1RR024992 from the National Center for Research Resources (NCRR), a component of the NIH (National Institute of Health) and NIH Roadmap for Medical Research. Computations were performed using the facilities of the Washington University Center for High Performance Computing, which were partially funded by NIH grants 1S10RR022984-01A1 and 1S10OD018091-01. C.C. and A.G. contributed exomes for use as controls funded under grants R01-AG044546, R01-AG035083 and the Alzheimer Association (NIRG-11-200110). This research was conducted while C.C. was a recipient of a New Investigator Award in Alzheimer's disease from the American Federation for Aging Research. Additional control samples were obtained from the NHLBI GO ESP and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the BroadGOSequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). The Research reported in this publication was also supported by the Washington University Institute of Clinical and Translational Sciences grant UL1 TR000448 from the National Center for Advancing Translational Sciences (NCATS) of the NIH.
Supplementary Material
Acknowledgements
We thank the patients and their families for their role in this work. We thank Drs Scott Luhmann, Lawrence Lenke and Keith Bridwell for allowing us to recruit patients from their clinics at Washington University. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis.
Conflict of Interest statement. None declared.
References
- 1.Rogala E.J., Drummond D.S., Gurr J. (1978) Scoliosis: incidence and natural history. A prospective epidemiological study. J. Bone Joint Surg. Am., 60, 173–176. [PubMed] [Google Scholar]
- 2.Martin C.T., Pugely A.J., Gao Y., Mendoza-Lattes S.A., Ilgenfritz R.M., Callaghan J.J., Weinstein S.L. (2014) Increasing hospital charges for adolescent idiopathic scoliosis in the United States. Spine, 39, 1676–1682. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein S.L., Dolan L.A., Cheng J.C., Danielsson A., Morcuende J.A. (2008) Adolescent idiopathic scoliosis. Lancet, 371, 1527–1537. [DOI] [PubMed] [Google Scholar]
- 4.Gorman K.F., Julien C., Moreau A. (2012) The genetic epidemiology of idiopathic scoliosis. Eur. Spine J., 21, 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadley Miller N. (2000) Spine update: genetics of familial idiopathic scoliosis. Spine, 25, 2416–2418. [DOI] [PubMed] [Google Scholar]
- 6.Kesling K.L., Reinker K.A. (1997) Scoliosis in twins. A meta-analysis of the literature and report of six cases. Spine, 22, 2009–2014; discussion 2015. [DOI] [PubMed] [Google Scholar]
- 7.Miller N.H. (2007) Genetics of familial idiopathic scoliosis. Clin. Orthop. Relat. Res., 462, 6–10. [DOI] [PubMed] [Google Scholar]
- 8.Riseborough E.J., Wynne-Davies R. (1973) A genetic survey of idiopathic scoliosis in Boston, Massachusetts. J. Bone Joint Surg. Am., 55, 974–982. [PubMed] [Google Scholar]
- 9.Ward K., Ogilvie J., Argyle V., Nelson L., Meade M., Braun J., Chettier R. (2010) Polygenic inheritance of adolescent idiopathic scoliosis: a study of extended families in Utah. Am. J. Med. Genet. A, 152A, 1178–1188. [DOI] [PubMed] [Google Scholar]
- 10.Kruse L.M., Buchan J.G., Gurnett C.A., Dobbs M.B. (2012) Polygenic threshold model with sex dimorphism in adolescent idiopathic scoliosis: the Carter effect. J. Bone Joint Surg. Am., 94, 1485–1491. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S., Gao X., Londono D., Devroy S.E., Mauldin K.N., Frankel J.T., Brandon J.M., Zhang D., Li Q.Z., Dobbs M.B. et al. (2011) Genome-wide association studies of adolescent idiopathic scoliosis suggest candidate susceptibility genes. Hum. Mol. Genet., 20, 1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi Y., Kou I., Takahashi A., Johnson T.A., Kono K., Kawakami N., Uno K., Ito M., Minami S., Yanagida H. et al. (2011) A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat. Genet., 43, 1237–1240. [DOI] [PubMed] [Google Scholar]
- 13.Kou I., Takahashi Y., Johnson T.A., Takahashi A., Guo L., Dai J., Qiu X., Sharma S., Takimoto A., Ogura Y. et al. (2013) Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat. Genet., 45, 676–679. [DOI] [PubMed] [Google Scholar]
- 14.Buchan J.G., Alvarado D.M., Haller G.E., Cruchaga C., Harms M.B., Zhang T., Willing M.C., Grange D.K., Braverman A.C., Miller N.H. et al. (2014) Rare variants in FBN1 and FBN2 are associated with severe adolescent idiopathic scoliosis. Hum. Mol. Genet., 23, 5271–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman B.T., Huang da W., Tan Q., Guo Y., Bour S., Liu D., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. (2007) DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics, 8, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeys B.L., Dietz H.C., Braverman A.C., Callewaert B.L., De Backer J., Devereux R.B., Hilhorst-Hofstee Y., Jondeau G., Faivre L., Milewicz D.M. et al. (2010) The revised Ghent nosology for the Marfan syndrome. J. Med. Genet., 47, 476–485. [DOI] [PubMed] [Google Scholar]
- 17.Beighton P., Horan F. (1969) Orthopaedic aspects of the Ehlers–Danlos syndrome. J. Bone Joint Surg. Br., 51, 444–453. [PubMed] [Google Scholar]
- 18.Ricard-Blum S. (2011) The collagen family. Cold Spring Harb. Perspect. Biol., 3, a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarado D.M., Yang P., Druley T.E., Lovett M., Gurnett C.A. (2014) Multiplexed direct genomic selection (MDiGS): a pooled BAC capture approach for highly accurate CNV and SNP/INDEL detection. Nucleic Acids Res., 42, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melkoniemi M., Brunner H.G., Manouvrier S., Hennekam R., Superti-Furga A., Kaariainen H., Pauli R.M., van Essen T., Warman M.L., Bonaventure J. et al. (2000) Autosomal recessive disorder otospondylomegaepiphyseal dysplasia is associated with loss-of-function mutations in the COL11A2 gene. Am. J. Hum. Genet., 66, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vikkula M., Mariman E.C., Lui V.C., Zhidkova N.I., Tiller G.E., Goldring M.B., van Beersum S.E., de Waal Malefijt M.C., van den Hoogen F.H., Ropers H.H. et al. (1995) Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell, 80, 431–437. [DOI] [PubMed] [Google Scholar]
- 22.McGuirt W.T., Prasad S.D., Griffith A.J., Kunst H.P., Green G.E., Shpargel K.B., Runge C., Huybrechts C., Mueller R.F., Lynch E. et al. (1999) Mutations in COL11A2 cause non-syndromic hearing loss (DFNA13). Nat. Genet., 23, 413–419. [DOI] [PubMed] [Google Scholar]
- 23.Brunner H.G., van Beersum S.E., Warman M.L., Olsen B.R., Ropers H.H., Mariman E.C. (1994) A Stickler syndrome gene is linked to chromosome 6 near the COL11A2 gene. Hum. Mol. Genet., 3, 1561–1564. [DOI] [PubMed] [Google Scholar]
- 24.Lehninger A.L. (1975) Biochemistry: The Molecular Basis of Cell Structure and Function. Worth Publishers, New York. [Google Scholar]
- 25.Fisher R.A. (1918) The correlation between relatives on the supposition of Mendelian inheritance. Trans. Roy. Soc. Edinb., 52, 399–433. [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and Genome Project Data Processing, S. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E. et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature, 461, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.