Abstract
Nitric oxide (NO) is a key regulator of skeletal muscle function and metabolism, including vasoregulation, mitochondrial function, glucose uptake, fatigue and excitation–contraction coupling. The main generator of NO in skeletal muscle is the muscle-specific form of neuronal nitric oxide synthase (nNOSμ) produced by the NOS1 gene. Skeletal muscle nNOSμ is predominantly localized at the sarcolemma by interaction with the dystrophin protein complex (DPC). In Duchenne muscular dystrophy (DMD), loss of dystrophin leads to the mislocalization of nNOSμ from the sarcolemma to the cytosol. This perturbation has been shown to impair contractile function and cause muscle fatigue in dystrophic (mdx) mice. Here, we investigated the effect of restoring sarcolemmal nNOSμ on muscle contractile function in mdx mice. To achieve this, we designed a modified form of nNOSμ (NOS-M) that is targeted to the sarcolemma by palmitoylation, even in the absence of the DPC. When expressed specifically in mdx skeletal muscle, NOS-M significantly attenuates force loss owing to damaging eccentric contractions and repetitive isometric contractions (fatigue), while also improving force recovery after fatigue. Expression of unmodified nNOSμ at similar levels does not lead to sarcolemmal association and fails to improve muscle function. Aside from the benefits of sarcolemmal-localized NO production, NOS-M also increased the surface membrane levels of utrophin and other DPC proteins, including β-dystroglycan, α-syntrophin and α-dystrobrevin in mdx muscle. These results suggest that the expression of NOS-M in skeletal muscle may be therapeutically beneficial in DMD and other muscle diseases characterized by the loss of nNOSμ from the sarcolemma.
Introduction
Nitric oxide is directly involved in many aspects of skeletal muscle function, including vasomodulation (1), mitochondrial biogenesis (2), glucose uptake (3,4), suppression of oxidative stress (5), activation of cyclic GMP synthesis (6) and modulation of excitation–contraction coupling via nitrosylation of the ryanodine receptor (7). As nitric oxide (NO) regulates several important functions in healthy muscle, it is not surprising that perturbations of NO production and cellular localization are associated with muscle diseases, including the muscular dystrophies.
In skeletal muscle fibers, NO is predominantly produced by two isoforms of neuronal nitric oxide synthase (nNOS) in response to binding of Ca++/calmodulin (CaM). These isoforms, nNOSµ and nNOSβ, are both products of the NOS1 gene. nNOSµ is very similar to the brain form, nNOSα, but contains the muscle-specific µ insert of 34 residues, located between the oxygenase and reductase catalytic domains, and immediately adjacent to the CaM-binding site (8). nNOSµ is primarily associated with the sarcolemma through interaction with dystrophin and α-syntrophin (9,10). nNOSβ differs from nNOSα in that it lacks the amino terminal PDZ scaffolding domain (11). In skeletal muscle, nNOSβ is expressed at much lower levels than nNOSµ but is highly localized to the Golgi apparatus in close juxtaposition with soluble guanylate cyclase and protein kinase G, suggesting that activation of nNOSβ is important for an NO-cGMP signaling pathway at the Golgi (12). Mice lacking both nNOSµ and nNOSβ have smaller muscles with reduced specific force and high susceptibility to fatigue (12).
Restricted spatial NO signaling is likely determined by localization of NOS. Although a highly diffusible molecule, NO action is limited to areas near its site of synthesis by the high concentration of scavengers (e.g. myoglobin and glutathione). Although some nNOSµ is present in the cytosol, most of the enzyme is restricted to the sarcolemma in normal muscle. During muscle atrophy, nNOSµ becomes mislocalized moving from the sarcolemma to the cytosol where the NO produced can activate genes that induce muscle atrophy pathways (13). In Duchenne muscular dystrophy (DMD), a severe, progressive and fatal muscle disease caused by mutations in dystrophin, nNOSµ is also lost from the sarcolemma (14). This mislocalization results in a lack of sarcolemmal NO production that prevents the override of sympathetic vasoconstriction resulting in functional ischemia of the muscle (15). Experiments in mice show that loss of sarcolemmal nNOSµ not only leads to functional ischemia but also makes the muscle more susceptible to fatigue (16) and damage from eccentric contractions (17).
The mdx mouse serves as a dystrophin-deficient mouse model of DMD. It shares many of the key characteristics of DMD including utrophin upregulation, functional ischemia, increased fatigue and damage from eccentric contractions even though the continuous degeneration/regeneration cycles are not observed in adult mdx limb muscles. Studies published over the past decade or more have suggested that transgenic overexpression of nNOSα in mdx muscle improves the dystrophic pathology, even when not targeted to the sarcolemma (18,19,20). However, a major limitation of these studies was the lack of any measurements of muscle function, which is ultimately the most relevant determinant of mobility and longevity in DMD patients. Given the important regulatory role of nNOSµ on normal skeletal muscle contractility (21), we aimed to test whether sarcolemmal nNOSµ targeting was required for improved function of dystrophic muscles.
Here, we report that a novel form of nNOSμ, targeted to the sarcolemma by addition of a palmitoylation sequence, is remarkably effective at improving functional aspects of skeletal muscle including resistance to eccentric contraction-induced injury and fatigue. In contrast, expression of similar amounts of non-targeted nNOSμ, which remains in the cytosol, shows no improvement. Interestingly, we also demonstrate that targeted nNOSμ effectively upregulates sarcolemmal expression of utrophin and other members of the dystrophin protein complex (DPC). Together, our results demonstrate that targeted nNOSμ is a promising therapeutic approach for DMD and other neuromuscular diseases that exhibit reduced or absent sarcolemmal nNOSμ.
Results
Generation of transgenic mice
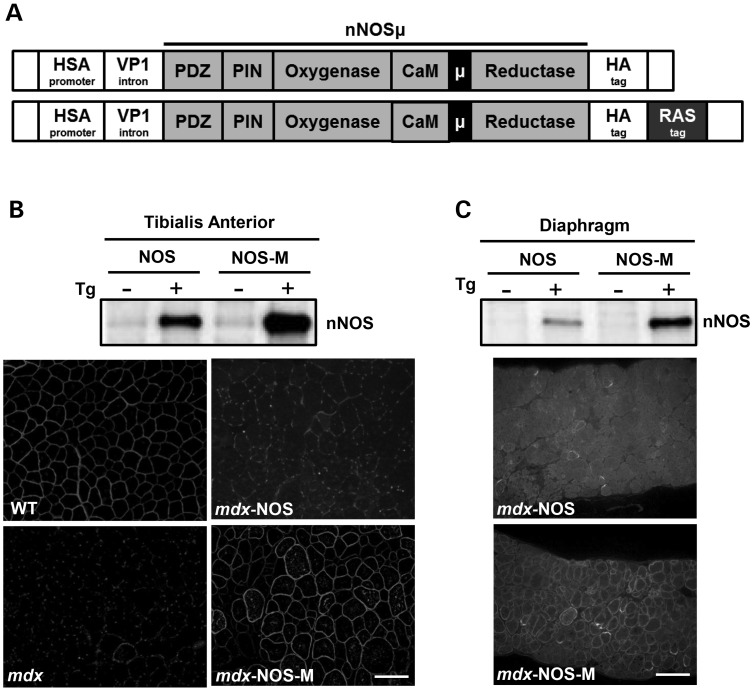
To compare the effects of general nNOSμ with sarcolemma-targeted nNOSμ expression, we generated two lines of transgenic mice. Both used the human skeletal actin (HSA) promoter to drive transcription (Fig. 1A). This promoter is only active in skeletal muscle and is turned on early in embryonic muscle development and maintained throughout mature skeletal muscle (22). A hemagglutinin tag (HA) was placed at the C-terminal end of nNOSμ in both lines. To one transgene we added a C-terminal palmitoylation signal sequence from the K-Ras oncogene (RAS) to target nNOSμ to the sarcolemma. Hereafter, this construct will be referred to as NOS-M, whereas NOS denotes the non-targeted, nNOSμ-HA construct. We obtained three founder mice expressing NOS and two founders expressing NOS-M. Each founder line was bred onto the mdx background (C57Bl/10ScSnJ) for at least three generations before use in experiments with non-transgenic littermates as controls.
Figure 1.
Design of transgenes and expression of nNOSµ in muscles. (A) Schematic representation of the two nNOSµ transgenes with the regular NOS (top) and the modified, NOS-M version (lower) showing the addition of the palmitoylation (RAS) tag (see text for details). (B) TA and (C) diaphragm representative western blots and immunostaining showing nNOSµ from NOS and NOS-M transgenic mice. Note the strong sarcolemmal staining in NOS-M but not NOS muscles. Scale bar = 100 µm.
Both NOS and NOS-M were expressed well in skeletal muscle (Fig. 1B, C). Western blots of tissue homogenates showed high expression of the nNOS transgene products in both tibialis anterior (TA) and diaphragm muscles. The relative amounts of NOS and NOS-M were similar. Immunofluorescence studies of these two muscles show that transgenically expressed NOS is only faintly labeled at the sarcolemma whereas NOS-M shows robust sarcolemmal labeling (Fig. 1B, C).
Analysis of muscle function
TA in situ
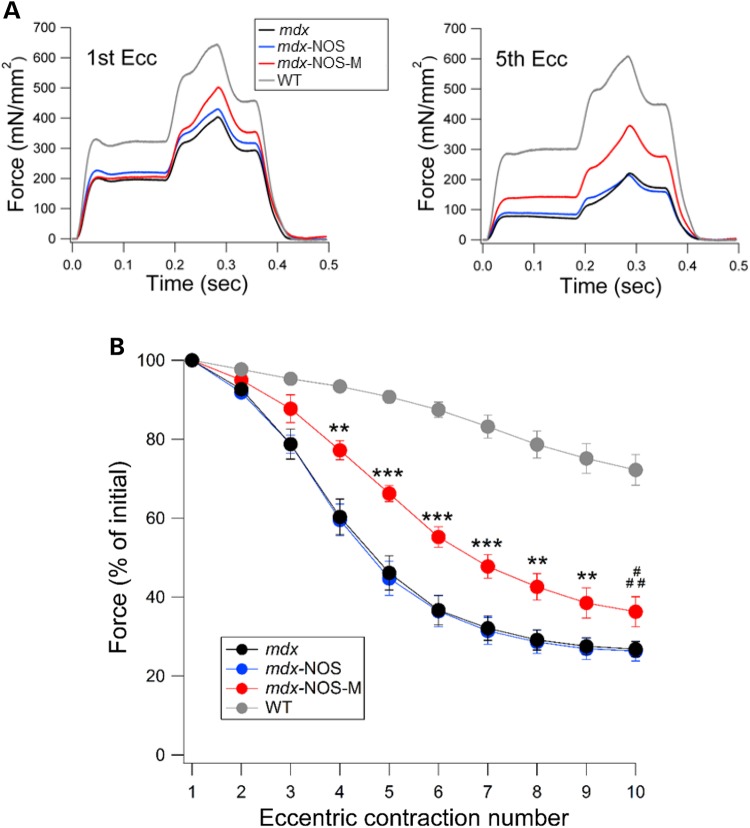
Dystrophic muscles are particularly vulnerable to damage from eccentric contractions (23). We therefore tested the performance of skeletal muscle from WT, mdx, and both transgenic lines (bred onto the mdx background) during eccentric contractions. These experiments were performed in situ with the knee immobilized and the distal tendon of the TA muscle attached to a force transducer (24). A major advantage of this preparation is that in vivo vascular function is maintained, thus preserving proper oxygenation. Maintenance of in vivo oxygen levels is necessary when studying functions involving NO (21). Isometric force was measured during sequential contractions initiated by direct stimulation of the sciatic nerve (Fig. 2). The dramatic reduction of force in the mdx mice is evident as early as the third contraction. After five contractions, the force generated by the mdx and mdx-NOS muscle fell to ∼40% of the initial force whereas for the mdx-NOS-M muscle, force only dropped to ∼60% of its initial value (note WT drops to ∼90% of initial force). From eccentric contractions 4–10, the force produced by mdx-NOS-M was significantly greater than mdx and mdx-NOS (see Fig. 2). Thus, sarcolemmal expression of nNOS provides considerable protection against force loss in dystrophic muscle, particularly during the early eccentric contractions, where most of the force loss occurs in regular mdx mice.
Figure 2.
NOS-M reduces force loss from eccentric contractions in mdx TA muscle. (A) Representative traces showing force production during the first and fifth eccentric contractions for each group of mice. (B) Pooled data showing the isometric force production over 10 eccentric contractions, expressed as a percent of first contraction. **P < 0.01 mdx-NOS-M versus mdx and mdx-NOS, ***P < 0.001 mdx-NOS-M versus mdx and mdx-NOS, #P < 0.05 mdx-NOS-M versus mdx, ##P < 0.01 mdx-NOS-M versus mdx-NOS.
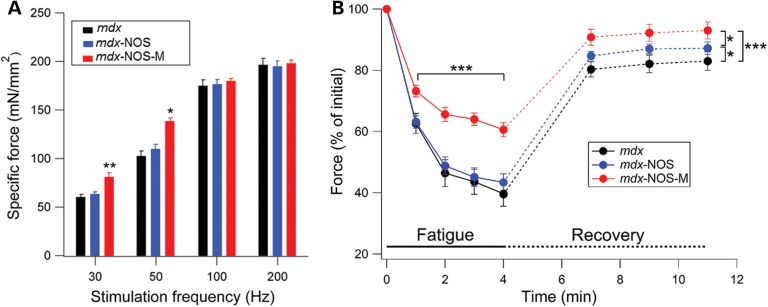
We then determined the effect of sarcolemmal nNOSμ restoration on TA skeletal muscle-specific force (force normalized to cross-sectional area). The specific force of TA muscle was significantly increased in NOS-M mice at low (30 and 50 Hz) stimulation frequencies but not at higher frequencies compared with mdx-NOS and normal mdx mice (Fig. 3A). In addition to eccentric contractions, muscle fatigue is another major cause of muscle weakness in dystrophic muscles (25). Muscle force was substantially higher (20%) for mdx-NOS-M muscles compared with those of mdx-NOS and regular mdx mice over 4 min of fatiguing contractions (Fig. 3B). In addition, recovery from fatigue was significantly greater for mdx-NOS-M muscle than the other two groups of mdx mice, whereas mdx-NOS mice had a small but significantly greater recovery than regular mdx mice (see Fig. 3B).
Figure 3.
NOS-M enhances low-frequency force, protects against muscle fatigue and improves force recovery in mdx TA muscle. (A) Specific force values at various stimulation frequencies for each group. *P < 0.05 mdx-NOS-M versus mdx and mdx-NOS, **P < 0.01 mdx-NOS-M versus mdx and mdx-NOS. (B) Mean force levels measured each minute during 4 min of fatigue (sold lines) and during fatigue recovery (dotted lines). For fatigue, ***P < 0.001 mdx-NOS-M versus mdx and mdx-NOS. For fatigue recovery, *P < 0.05, ***P < 0.001.
Diaphragm ex vivo
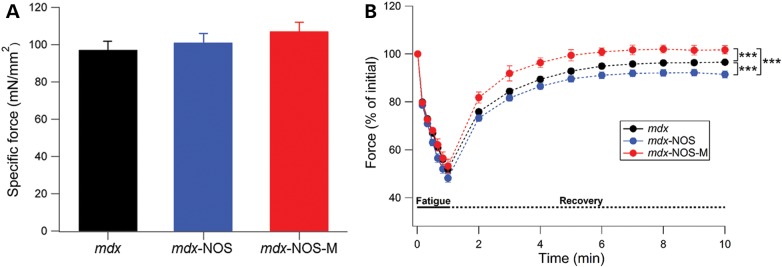
We also measured diaphragm function using small muscle strips ex vivo (26). Here, specific force at 120 Hz stimulation was not improved in mdx muscles by either the NOS-M or NOS transgene (Fig. 4A). Diaphragm fatigue over 1 min of repetitive stimulation was also not different between the groups (Fig. 4B). However, force recovery after fatigue was significantly greater for the mdx-NOS-M muscles compared with both mdx-NOS and regular mdx, whereas mdx-NOS was significantly lower than regular mdx mice (see Fig. 4B).
Figure 4.
NOS-M improves fatigue recovery in mdx diaphragm muscle. (A) Specific force for diaphragm muscles during 120-Hz stimulation. There were no significant differences between any groups. (B) Mean force levels during 1 min of fatigue (sold lines) and for 10 min of fatigue recovery (dotted lines). For fatigue recovery, ***P < 0.001.
From these physiological studies, we conclude that nNOSµ targeted to the sarcolemma markedly improves the contractile function of mdx muscle during eccentric contractions and muscle fatigue. In fact, the magnitude of these improvements is similar to those attained by treatments that upregulate utrophin and current dystrophin gene therapy approaches (27,28,29). In contrast, untargeted nNOSμ shows no improvement in protection against force loss from eccentric contractions or fatigue in TA, and while these muscles show a small improvement in TA fatigue recovery compared with regular mdx muscle, fatigue recovery is worse than mdx mice in diaphragm muscles.
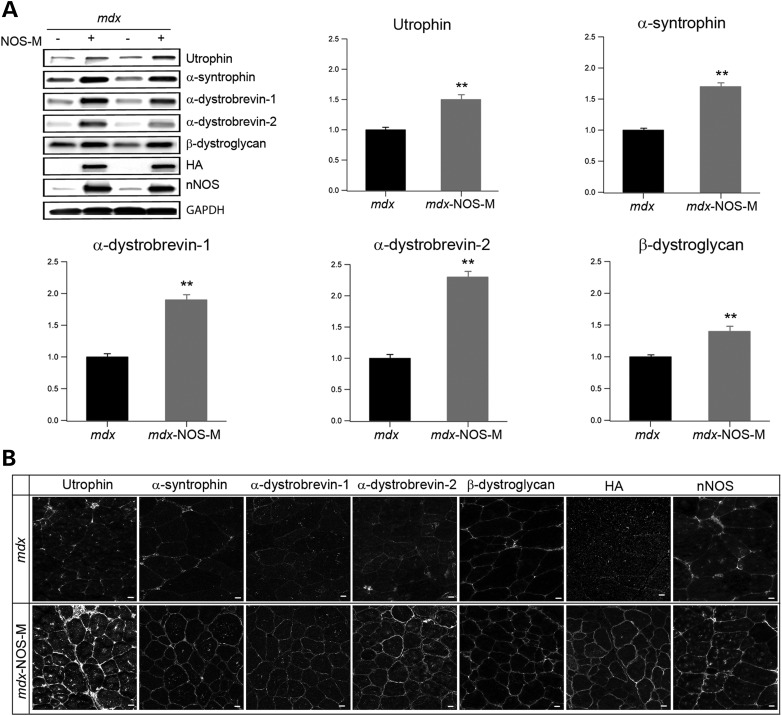
NOS-M targets utrophin and dystrophin-associated proteins to the sarcolemma
Transgenic expression and upregulation of utrophin improves the dystrophic pathology in the mdx mouse (27,30). As increased NO bioavailability upregulates utrophin levels in mdx muscle (31,32), we tested the hypothesis that NOS-M improves mdx contractile function by increasing sarcolemmal utrophin levels. We found that NOS-M markedly increases expression (Fig. 5A) and sarcolemmal localization (Fig. 5B) of utrophin in mdx TA muscles. In addition, several other key proteins of the DPC including β-dystroglycan, α-dystrobrevin-1 and -2 and α-syntrophin are also upregulated and localized at the sarcolemma. Therefore, our results indicate that proper localization of nNOSµ on the sarcolemma is required for the observed increase in utrophin and proteins of the DPC. Whether these restorations are mediated by NOS-M scaffolding, by increased NO bioavailability, or both, requires further study. The pathway mediating the upregulation of utrophin and DPC proteins increase does not appear to involve increased transcription/mRNA stabilization. We found no significant difference in mRNA levels of utrophin or α-dystrobrevin when comparing muscle RNA isolated from mdx and mdx-NOS-M mice ( Supplementary Material, Fig. S1).
Figure 5.
NOS-M increases the expression and sarcolemmal localization of utrophin and DPC proteins. (A) Representative western blots (top left) and pooled data showing the levels of utrophin and DPC proteins from TA muscles of mdx and mdx-NOS-M mice. Data were pooled from 5–10 mice/group. The western blots also show the levels of nNOSµ and the HA tag associated with the transgene. **P < 0.01. (B) Representative immunostaining images showing the increased sarcolemmal localization of utrophin and DPC proteins in mdx-NOS-M muscle. Scale bar = 10 µm.
Discussion
We have developed a novel approach to restore nNOSµ at the sarcolemmal membrane in skeletal muscle. Introduction of a K-RAS palmitoylation signal sequence at the carboxy terminus of nNOSµ causes the modified enzyme (NOS-M) to be localized on the surface membrane, as expected for proteins that are palmitoylated. The rationale for this approach is based on the hypothesis that NO bioavailability is determined in large part by the localization of its site of synthesis. In contrast to unmodified nNOSµ (NOS), NOS-M improves two important aspects of contractile function in mdx muscle: resistance to eccentric contraction-induced force loss and improved resistance to muscle fatigue. Eccentric damage and fatigue are important causes of muscle weakness in skeletal muscle of mdx mice and DMD boys.
Our results conflict with results published by Tidball and colleagues, in which they claim that improvement of pathological features of mdx muscle by transgenic expression of nNOS does not require sarcolemmal localization (18,19,20). Several differences in approach and analyses may explain the conflicting results. First, the Tidball group used nNOSα, the brain form of nNOS, rather than the muscle-specific form, nNOSµ. The function of the µ insert is not known but its location between the oxygenase and reductase catalytic regions and adjacent to the CaM binding site suggests an important role in regulating NO production. Second, the transgenic expression levels of nNOSα in their experiments were exceptionally high, especially in the ‘supernatant fraction’. High levels of untargeted, cytosolic NOS may have deleterious effects caused, for example, by inappropriate non-physiological nitrosylation of proteins. Also, translocation of nNOSµ from the sarcolemma to the cytosol is a key factor in muscle atrophy during mechanical unloading (13). Finally, no increase in utrophin localization on the sarcolemma was observed in mdx muscle from transgenic muscle expressing nNOSα (20). In fact, total utrophin levels were reduced, compared with mdx muscle. Although some pathological features were improved by nNOSα expression in mdx muscle (reduced inflammation and reduction of infiltrating macrophages) early in the disease process, no measurements of contractile function were conducted.
Upregulation of utrophin, a homolog of dystrophin, has received major attention as a possible therapeutic target, based on initial studies showing that muscle-specific expression of utrophin is highly effective in reducing the pathological deficits caused by the loss of dystrophin. One advantage of utrophin expression, compared with genetic treatments with micro-dystrophin constructs, is the greatly reduced likelihood of eliciting an immune response. A potential disadvantage is that the utrophin complex fails to restore nNOSµ to the sarcolemma (33), perhaps because of differences in the repeat 16–17 region that is essential for nNOSµ association with the DPC (22). In addition to increasing sarcolemmal utrophin levels, NOS-M has the added benefit of restoring nNOSµ to its proper location. NOS-M does not appear to affect transcription or transcript stabilization of utrophin but may increase utrophin levels via S-nitrosylation-mediated inhibition of calpain (34), or by serving as a scaffold that stabilizes utrophin and associated proteins in a sarcolemmal complex. Regardless of the mechanism by which utrophin levels are increased, the observation that NOS-M increases levels of utrophin (and associated proteins) and partially restores normal function in dystrophic muscle makes it a potential therapeutic candidate. As NOS-M is much smaller than dystrophin/utrophin, it may serve as a more amenable protein for use in DMD-directed gene therapy.
Although our studies were focused on the mdx mouse model of DMD, NOS-M may also have therapeutic value in other muscle diseases in which nNOSµ expression and localization are perturbed. These include forms of limb-girdle muscular dystrophy (primarily the sarcoglycanopathies) (35), amyotrophic lateral sclerosis (36), myasthenia gravis (37), disuse atrophy (13,38), sarcopenia (39) and muscle mitochondrial diseases (40). Development of methods for viral delivery of NOS-M to skeletal muscle will be needed to test the therapeutic effectiveness in these devastating human muscle diseases.
Materials and Methods
nNOSμ cloning and transgenic mice generation
nNOSμ cDNA was derived from total skeletal muscle RNA using reverse transcription–PCR and the presence of the µ insert confirmed by sequencing. We used PCR to add a single heme-agglutinin (HA) tag epitope to the C-terminal of nNOSμ or the HA tag preceded by the palmitylation signal sequence from the K-ras oncogene (amino acid sequence: KDGKKKKKKSKTKCVIM) (41). These two nNOSμ constructs were cloned into the NotI/PacI sites of pBSX-HSAvpA vector (42,43) (a gift from Drs. Jeff Chamberlain and Stephen Hauschka, University of Washington). The pBSX-HSAvpA vector contains the HSA promoter that provides skeletal muscle-specific expression of the nNOSμ transgenes. Linearized constructs were injected into oocytes of a cross of the C57BL/6 and C3H mouse strains (Transgenic Resources Program, University of Washington). Lines of mice expressing the transgenes were bred onto WT and mdx, both on the C57BL/10ScSn background, for a minimum of three generations. All experimental procedures performed on mice were approved by the Institutional Animal Care and Use Committee at the University of Washington.
In situ analysis of skeletal muscle contractile function
Anesthetized mice were positioned on a 37°C heated platform, the distal tendon of the TA muscle was surgically isolated, the knee joint was restrained with a surgical needle and the distal tendon attached to the lever arm of a servomotor (Aurora Scientific, ON, Canada). The TA muscle was activated by stimulation of the sciatic nerve using two needle electrodes. The muscle was adjusted to an optimum length (Lo) to produce the maximum tetanic force. While held at Lo, the TA was stimulated every 2 min at increasing frequencies (10 to 200 Hz) to generate force frequency curves, to obtain the maximal tetanic force (Po). Then, both Lo and TA mass were recorded and used to normalize to the physiological cross-sectional area ([Lo × density]/mass) and calculate specific tetanic force. For each mouse, the left TA was used for testing resistance to exercise-induced fatigue, whereas the right was used to test susceptibility to contraction-induced injury. To test resistance to exercise-induced fatigue, TA muscles were subjected to a series of repeated isometric stimulations (200 Hz) at 2-s intervals for 4 min. Recovery from fatigue was assessed by recording force output at 3, 5 and 7 min after the completion of the fatigue period.
Resistance to eccentric contraction (ECC)-induced injury
TA muscles were subjected to a series of 10 lengthening (eccentric) contractions. Muscles were maximally stimulated (200 Hz) for 175 ms at Lo to achieve maximal isometric tension, and then stretched while contracting by 20% of Lo at a velocity of two fiber lengths per second. The total stimulation time for each contraction was 350 ms. Each eccentric contraction was separated by 1 min. The maximum isometric force measured before the stretch was recorded for each contraction and expressed as a percent of the first contraction.
Ex vivo analysis of diaphragm muscle function
Diaphragm muscle strips, 2- to 4-mm wide were used for ex vivo function (26). Briefly, strips were dissected in physiological buffer, bubbled with 95% O2 and 5% CO2 (pH 7.4). The central tendon of the strip was attached to the lever of a servomotor (Aurora Scientific), and the rib bone was attached to a stainless steel hook. Muscle stimulation was provided by two platinum electrodes, attached to the inside walls of the chamber, which are connected to a stimulator (Aurora Scientific). A length-force curve was measured using 120 Hz stimulation to establish the optimum length (Lo), and specific force was later determined by dividing force by cross-sectional area. The muscle strip was then subjected to a fatigue protocol. Here, the muscle was stimulated at 120 Hz, every second for 60 s. Recovery of force following fatigue was measured at 1-min intervals up to 10 min.
Antibodies
Our antibodies to utrophin, α-syntrophin and α-dystrobrevin have been described previously (44). We used the following commercial antibodies. Rabbit polyclonal antibodies: glyceraldehyde-3-phosphate dehydrogenase (Millipore), nNOS (Invitrogen) and HA (Sigma). Mouse monoclonal antibodies: pan dystrobrevin (BD Biosciences) and β-dystroglycan (Novacastra). Goat polyclonal anti HA (Abcam).
Western blotting
Muscle extracts in 1% Triton X-100 were prepared as described previously (44). Protein concentrations were determined using the bicinchoninic acid reagent (Pierce), and equal amounts of muscle protein were used for SDS–PAGE. Proteins were transferred to a PVDF membrane (Millipore) and the primary antibodies detected using HRP-conjugated secondary antibodies (Jackson Immunoresearch) and ECL substrate (Pierce) with a Protein Simple Fluorochem M imager. Band intensity was determined using Image J software and plotted with the average mdx value set to 1. N-values (where N refers to an individual mouse) were as follows: for utrophin–mdx N = 9, mdx-NOS-M N = 10, for syntrophin dystrobrevin and dystroglycan mdx N = 5, mdx-NOS-M N = 9.
Immunohistochemistry
Muscles from 8- to 9-week-old mice were dissected, embedded in Tissue Tek OCT, frozen in liquid nitrogen-cooled isopentane. Cryostat sections (10 μm thick) were incubated with primary antibody at 4°C overnight then with Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. Images were obtained using a Zeiss 510 Meta confocal microscope at the W.M. Keck Center for Advanced Studies in Neural Signaling at the University of Washington.
Data analysis
In all experiments, transgenic mice were compared with non-transgenic littermates. As different lines were obtained, and results from different lines for the same transgene performed very similarly, we used pooled data. Thus, C57Bl10 and mdx non-transgenic data correspond to negative littermates of all transgenic lines. Data from cytosolic NOS transgenic mice correspond to the average of three different lines. All values are expressed as the mean ± SEM, and statistical significance was set at P < 0.05. For two experimental groups, a Student's t-test was used for statistical analysis. For more than two groups, two-way ANOVA was used. Where an ANOVA was significant, an LSD post hoc test was used to evaluate differences between independent variables (Data Desk, Ithaca, New York).
Supplementary Material
Funding
This work supported by funding from a predoctoral fellowship from CONICYT and MECESUP to D.L.R., a Cardiovascular Training grant T32HL007828 from NHLBI to M.J.K., the Raymond and Beverly Sackler Foundation, and National Institutes of Health grant R01 AR056221. The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Supplementary Material
Acknowledgements
The authors thank Dr. Kenneth Bible for his assistance with many aspects of this work and Dr. Justin Percival for his advice, discussions and instructional assistance.
Conflict of Interest statement: None declared.
References
- 1.Thomas G.D., Sander M., Lau K.S., Huang P.L., Stull J.T., Victor R.G. (1998) Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc. Natl Acad. Sci. USA, 95, 15090–15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leary S.C., Shoubridge E.A. (2003) Mitochondrial biogenesis: which part of ‘NO’ do we understand? Bioessays, 25, 538–541. [DOI] [PubMed] [Google Scholar]
- 3.Kapur S., Bedard S., Marcotte B., Cote C.H., Marette A. (1997) Expression of nitric oxide synthase in skeletal muscle: a novel role for nitric oxide as a modulator of insulin action. Diabetes, 46, 1691–1700. [DOI] [PubMed] [Google Scholar]
- 4.Merry T.L., Steinberg G.R., Lynch G.S., McConell G.K. (2010) Skeletal muscle glucose uptake during contraction is regulated by nitric oxide and ROS independently of AMPK. Am. J. Physiol. Endocrinol. Metab., 298, E577–E585. [DOI] [PubMed] [Google Scholar]
- 5.Rando T.A. (2001) The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve, 24, 1575–1594. [DOI] [PubMed] [Google Scholar]
- 6.Lau K.S., Grange R.W., Isotani E., Sarelius I.H., Kamm K.E., Huang P.L., Stull J.T. (2000) nNOS and eNOS modulate cGMP formation and vascular response in contracting fast-twitch skeletal muscle. Physiol. Genomics, 2, 21–27. [DOI] [PubMed] [Google Scholar]
- 7.Xu L., Eu J.P., Meissner G., Stamler J.S. (1998) Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science, 279, 234–237. [DOI] [PubMed] [Google Scholar]
- 8.Silvagno F., Xia H., Bredt D.S. (1996) Neuronal nitric-oxide synthase-mu, an alternatively spliced isoform expressed in differentiated skeletal muscle. J. Biol. Chem., 271, 11204–11208. [DOI] [PubMed] [Google Scholar]
- 9.Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. (1995) Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell, 82, 743–752. [DOI] [PubMed] [Google Scholar]
- 10.Brenman J.E., Chao D.S., Gee S.H., McGee A.W., Craven S.E., Santillano D.R., Wu Z., Huang F., Xia H., Peters M.F. et al. (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell, 84, 757–767. [DOI] [PubMed] [Google Scholar]
- 11.Brenman J.E., Xia H., Chao D.S., Black S.M., Bredt D.S. (1997) Regulation of neuronal nitric oxide synthase through alternative transcripts. Dev. Neurosci., 19, 224–231. [DOI] [PubMed] [Google Scholar]
- 12.Percival J.M., Anderson K.N., Huang P., Adams M.E., Froehner S.C. (2010) Golgi and sarcolemmal neuronal NOS differentially regulate contraction-induced fatigue and vasoconstriction in exercising mouse skeletal muscle. J. Clin. Invest., 120, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki N., Motohashi N., Uezumi A., Fukada S., Yoshimura T., Itoyama Y., Aoki M., Miyagoe-Suzuki Y., Takeda S. (2007) NO production results in suspension-induced muscle atrophy through dislocation of neuronal NOS. J. Clin. Invest., 117, 2468–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grozdanovic Z., Gosztonyi G., Gossrau R. (1996) Nitric oxide synthase I (NOS-I) is deficient in the sarcolemma of striated muscle fibers in patients with Duchenne muscular dystrophy, suggesting an association with dystrophin. Acta Histochem., 98, 61–69. [DOI] [PubMed] [Google Scholar]
- 15.Sander M., Chavoshan B., Harris S.A., Iannaccone S.T., Stull J.T., Thomas G.D., Victor R.G. (2000) Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc. Natl Acad. Sci. USA, 97, 13818–13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi Y.M., Rader E.P., Crawford R.W., Iyengar N.K., Thedens D.R., Faulkner J.A., Parikh S.V., Weiss R.M., Chamberlain J.S., Moore S.A. et al. (2008) Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature, 456, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froehner S.C., Reed S.M., Anderson K.N., Huang P.L., Percival J.M. (2015) Loss of nNOS inhibits compensatory muscle hypertrophy and exacerbates inflammation and eccentric contraction-induced damage in mdx mice. Hum. Mol. Genet., 24, 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wehling M., Spencer M.J., Tidball J.G. (2001) A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell Biol., 155, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen H.X., Tidball J.G. (2003) Expression of a muscle-specific, nitric oxide synthase transgene prevents muscle membrane injury and reduces muscle inflammation during modified muscle use in mice. J. Physiol., 550, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tidball J.G., Wehling-Henricks M. (2004) Expression of a NOS transgene in dystrophin-deficient muscle reduces muscle membrane damage without increasing the expression of membrane-associated cytoskeletal proteins. Mol. Genet. Metab., 82, 312–320. [DOI] [PubMed] [Google Scholar]
- 21.Stamler J.S., Meissner G. (2001) Physiology of nitric oxide in skeletal muscle. Physiol. Rev., 81, 209–237. [DOI] [PubMed] [Google Scholar]
- 22.Lai Y., Zhao J., Yue Y., Duan D. (2013) alpha2 and alpha3 helices of dystrophin R16 and R17 frame a microdomain in the alpha1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl Acad. Sci. USA, 110, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen D.G., Zhang B.T., Whitehead N.P. (2010) Stretch-induced membrane damage in muscle: comparison of wild-type and mdx mice. Adv. Exp. Med. Biol., 682, 297–313. [DOI] [PubMed] [Google Scholar]
- 24.Percival J.M., Anderson K.N., Gregorevic P., Chamberlain J.S., Froehner S.C. (2008) Functional deficits in nNOSmu-deficient skeletal muscle: myopathy in nNOS knockout mice. PLoS One, 3, e3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen D.G. (2004) Skeletal muscle function: role of ionic changes in fatigue, damage and disease. Clin. Exp. Pharmacol. Physiol., 31, 485–493. [DOI] [PubMed] [Google Scholar]
- 26.Percival J.M., Whitehead N.P., Adams M.E., Adamo C.M., Beavo J.A., Froehner S.C. (2012) Sildenafil reduces respiratory muscle weakness and fibrosis in the mdx mouse model of Duchenne muscular dystrophy. J. Pathol., 228, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinsley J.M., Fairclough R.J., Storer R., Wilkes F.J., Potter A.C., Squire S.E., Powell D.S., Cozzoli A., Capogrosso R.F., Lambert A. et al. (2011) Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the mdx mouse. PLoS One, 6, e19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorwood C., Lozynska O., Suri N., Napper A.D., Diamond S.L., Khurana T.S. (2011) Drug discovery for Duchenne muscular dystrophy via utrophin promoter activation screening. PLoS One, 6, e26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amenta A.R., Yilmaz A., Bogdanovich S., McKechnie B.A., Abedi M., Khurana T.S., Fallon J.R. (2011) Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proc. Natl Acad. Sci. USA, 108, 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K. (1998) Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med., 4, 1441–1444. [DOI] [PubMed] [Google Scholar]
- 31.Chaubourt E., Fossier P., Baux G., Leprince C., Israel M., De La Porte S. (1999) Nitric oxide and l-arginine cause an accumulation of utrophin at the sarcolemma: a possible compensation for dystrophin loss in Duchenne muscular dystrophy. Neurobiol. Dis., 6, 499–507. [DOI] [PubMed] [Google Scholar]
- 32.Chaubourt E., Voisin V., Fossier P., Baux G., Israel M., de La Porte S. (2000) The NO way to increase muscular utrophin expression? C R Acad. Sci. III, 323, 735–740. [DOI] [PubMed] [Google Scholar]
- 33.Li D., Bareja A., Judge L., Yue Y., Lai Y., Fairclough R., Davies K.E., Chamberlain J.S., Duan D. (2010) Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J. Cell Sci., 123, 2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michetti M., Salamino F., Melloni E., Pontremoli S. (1995) Reversible inactivation of calpain isoforms by nitric oxide. Biochem. Biophys. Res. Commun., 207, 1009–1014. [DOI] [PubMed] [Google Scholar]
- 35.Crosbie R.H., Barresi R., Campbell K.P. (2002) Loss of sarcolemma nNOS in sarcoglycan-deficient muscle. FASEB J., 16, 1786–1791. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki N., Mizuno H., Warita H., Takeda S., Itoyama Y., Aoki M. (2010) Neuronal NOS is dislocated during muscle atrophy in amyotrophic lateral sclerosis. J. Neurol. Sci., 294, 95–101. [DOI] [PubMed] [Google Scholar]
- 37.Meinen S., Lin S., Ruegg M.A., Punga A.R. (2012) Fatigue and muscle atrophy in a mouse model of myasthenia gravis is paralleled by loss of sarcolemmal nNOS. PLoS One, 7, e44148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawler J.M., Kunst M., Hord J.M., Lee Y., Joshi K., Botchlett R.E., Ramirez A., Martinez D.A. (2014) EUK-134 ameliorates nNOSmu translocation and skeletal muscle fiber atrophy during short-term mechanical unloading. Am. J. Physiol. Regul. Integr. Comp. Physiol., 306, R470–R482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samengo G., Avik A., Fedor B., Whittaker D., Myung K.H., Wehling-Henricks M., Tidball J.G. (2012) Age-related loss of nitric oxide synthase in skeletal muscle causes reductions in calpain S-nitrosylation that increase myofibril degradation and sarcopenia. Aging Cell, 11, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finanger Hedderick E.L., Simmers J.L., Soleimani A., Andres-Mateos E., Marx R., Files D.C., King L., Crawford T.O., Corse A.M., Cohn R.D. (2011) Loss of sarcolemmal nNOS is common in acquired and inherited neuromuscular disorders. Neurology, 76, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahn S., Yamamoto F., Almoguera C., Winter E., Forrester K., Jordano J., Perucho M. (1987) The c-K-ras gene and human cancer (review). Anticancer Res., 7, 639–652. [PubMed] [Google Scholar]
- 42.Muscat G.E., Kedes L. (1987) Multiple 5′-flanking regions of the human alpha-skeletal actin gene synergistically modulate muscle-specific expression. Mol. Cell Biol., 7, 4089–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawford G.E., Faulkner J.A., Crosbie R.H., Campbell K.P., Froehner S.C., Chamberlain J.S. (2000) Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol., 150, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters M.F., Adams M.E., Froehner S.C. (1997) Differential association of syntrophin pairs with the dystrophin complex. J. Cell Biol., 138, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.