Arresting flexibility to prevent RNA from undergoing functionally important conformational transitions is an established strategy for developing RNA-targeting therapeutics.[1] To understand the role of RNA flexibility in adaptive recognition[2] is also important for the rational design of small molecules that bind their RNA target with high affinity and specificity.[3] Yet few studies have quantitatively examined how RNA-binding therapeutics affect the flexibility of their RNA targets. As a result, little is known about the RNA–ligand interactions that are important for arresting different types of RNA functional flexibility. Here, we provide direct evidence by using NMR residual dipolar couplings (RDCs)[4] that electrostatic interactions play a primary role in dictating the degree to which small molecules arrest global motions in RNA.
The transactivation response element (TAR) RNA from HIV-1 (Figure 1a) is a primary RNA target for developing anti-HIV therapeutics[5] and its molecular flexibility is implicated in its function. Upon binding to its cognate target, the transactivator protein (Tat), TAR undergoes large conformational changes that involve reorientation of two helical domains from a bent to a coaxially aligned state.[6] By using RDCs, we previously reported evidence for significant interdomain flexibility in free TAR which may play a direct role in mediating the conformational changes in TAR that accompany Tat recognition.[7a,b] We also demonstrated that Mg2+ or the ligand mimic of Tat, argininamide, can bind TAR and completely arrest these interdomain motions.[7c,d]
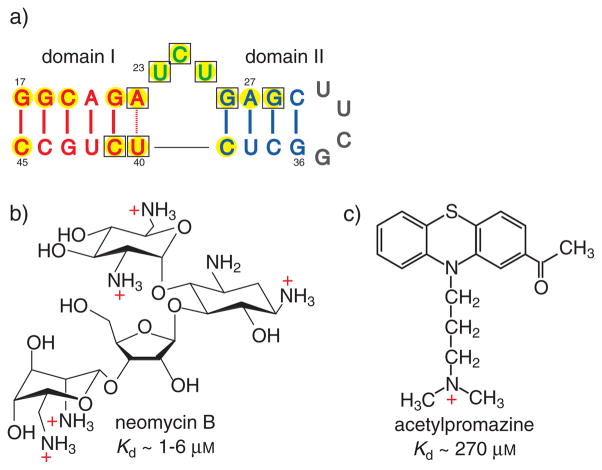
Figure 1.
a) Secondary structure of TAR (transactivation response element) RNA in which the wild-type loop is replaced by a UUCG counterpart. The highlighted residues undergo Δδ>0.1 ppm changes in 1H NMR chemical shifts upon binding to neomycin B (yellow) and acetylpromazine (□). b,c) Structures and TAR-dissociation constants (Kd) for neomycin B (b; NeoB)[9, 11] and acetylpromazine (c; AcP).[14]
The RDC methodology and TAR provide a unique opportunity to examine RNA–ligand interactions that are particularly important for arresting a general class of RNA motions, which involve global reorientation of helical domains. Numerous studies on RNA–aminoglycoside recognition have demonstrated that electrostatic interactions between cationic groups on the ligand and regions of high negative-charge density in the RNA can contribute significantly to aminoglycoside-binding affinities and specificities.[8] As global motions in RNA originate from local mobility in the negatively charged backbone and as functional groups from many residues can be involved in forming pockets of high negative-charge density in RNA, we reasoned that similar electrostatic interactions are also likely to be important for arresting RNA global motions.
To examine this hypothesis, we employed RDC NMR methodology to characterize the conformational dynamics of TAR when it is bound to two molecules that have different electrostatic charges: the aminoglycoside neomycin B (NeoB),[9] which bears five positively charged ammonium groups, and the small organic molecule acetylpromazine (AcP),[10] which carries a single positive charge (Figure 1b,c). Both NeoB and AcP bind TAR and inhibit its interaction with Tat.[9,10] A previously reported NOE-based NMR spectroscopic study of the structure of the TAR–NeoB complex[11] showed that NeoB binds TAR in the minor groove and stabilizes a coaxially stacked conformation similar to that observed for TAR bound to Tat peptides and Arg (arginine).[6a,b,d] In contrast, the NMR-determined structure of the TAR–AcP complex indicates that AcP induces minor changes in the conformation of TAR and that it binds a cavity at the interdomain interface.[12] Both NMR studies did not report on the global dynamics of TAR in the ligand-bound state.
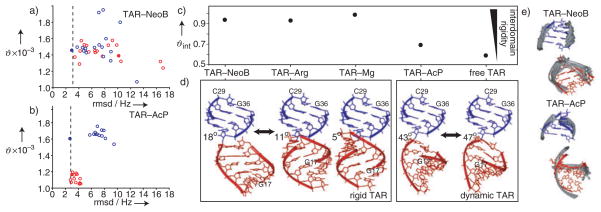
Shown in Figure 2 are the RDC-derived best-fit generalized degrees of order (ϑ)[13] determined for each domain in the two TAR complexes. The generalized degree of order describes the degree of alignment experienced by each domain which should be identical if the domains are held rigid relative to one another.[13] Results are shown when different input domain structures were used in the order tensor calculations, including idealized A-form helices generated by using Insight II (Molecular Simulations Inc.) and the previous NOE-based NMR spectroscopically determined structures of TAR–NeoB (pdb 1QD3)[11] and TAR–AcP (pdb 1LVJ).[12] Also shown are the values of the root-mean-square deviation (rmsd) between measured and back-calculated RDC values which provide a measure of agreement between a domain structure and measured RDC values.
Figure 2.
The global structural dynamics of TAR–NeoB and TAR–AcP. The best-fit generalized degree of order (ϑ) for domains I and II in a) TAR–NeoB and b) TAR–AcP as a function of the root-mean-square deviation (rmsd) between measured residual dipolar couplings (RDCs) and values back-calculated by using the best-fit order tensor (red unfilled circle: domain I NMR models; red filled circle: domainI idealized; blue unfilled circle: domain II NMR models; blue filled circle: domain II idealized). The experimental uncertainty (rmsd) in RDCs (see Supporting Information) is indicated by a dashed vertical line. c) The internal generalized degree of order, ϑint = ϑdomain I/ϑdomain II, determined for TAR–NeoB and TAR–AcP and a comparison with previous values determined for free TAR,[7a] TAR–Mg,[7c] and TAR–Arg.[7d] d) The best-fit relative domain orientation in TAR–NeoB and TAR–AcP determined by superimposing order tensor frames and a comparison with free TAR,[7a] TAR—Mg,[7c] and TAR–Arg.[7d] Domain II (blue) is superimposed and the best-fit interhelical angle is shown next to each structure. The residues for the bulge are not shown. The estimated uncertainty in the interhelical angle is less than ±6°. e) Comparison of the RDC-derived interdomain alignment (red and blue) with structures as determined by NOE-based NMR spectroscopy (in gray) of TAR–NeoB (pdb1QD3) and TAR–AcP (pdb1LVJ).
In the structure determined from NMR spectroscopic studies, NeoB induces local distortions in domain I.[11] Accordingly, most NMR models for domain I in TAR–NeoB yield rmsd values that are lower than the idealized A-form helix (Figure 2a). More importantly, the ϑ value determined for the two domains in TAR–NeoB are similar, particularly for domains that yield the lowest rmsd values (Figure 2a). The best-fit ϑint value (ϑdomainI/ϑdomainII),[13] which provides a measure of interdomain motional amplitudes, is close to unity (ϑint = 0.94) and thus indicates that NeoB arrests interdomain motions in TAR (Figure 2c). Remarkably, this is not the case for TAR bound to AcP. As previously observed for free TAR,[7a] the ϑ value for domain I is significantly smaller than that for domain II (Figure 2b), and the best-fit ϑint value (0.69) for TAR–AcP is only slightly higher than that previously reported for free TAR (0.59; Figure 2c). The low ϑint value for TAR–AcP cannot be attributed to errors in the input domain structures because the best-fit domains yield rmsd values that are equal to or smaller than the experimental uncertainty in the RDC values (see Figure 2b and Supporting Information) and because low ϑint values are observed independent of both input domain structure (Figure 2b) and RDCs (see Supporting Information). The low ϑint value for TAR–AcP could arise from fast exchange between free and AcP-bound TAR states. On the basis of the concentrations of TAR (≈1 mM) and AcP (≈2 mM) used in the RDC measurements and the binding constant (Ka) of Ka ≈270 μm,[14] approximately 81% of TAR is computed to be in the bound state. If a population-weighted average of free and bound ϑint values is assumed (although, strictly, averages over RDCs should be considered), then a similar population-corrected ϑint value for TAR–AcP of 0.71 is obtained. In agreement with this analysis, an ϑint value of 0.71 was determined for molar ratios of about 1:5 for TAR/AcP in which the bound state is approximately 94% populated (data not shown). Rather, the low ϑint value determined for TAR–AcP argues that unlike NeoB, AcP only marginally arrests global motions in TAR.
In Figure 2d, we compare the average domain–domain orientations determined for TAR–NeoB and TAR–AcP by using RDCs with counterparts determined previously for free TAR[7a] and TAR bound to Mg2+[7c] and Arg.[7d] The more coaxially stacked TAR–NeoB conformation is similar to those of TAR–Arg and TAR–Mg, whereas TAR–AcP is very similar to free TAR. The RDC-derived global conformations for TAR–NeoB and TAR–AcP are also in excellent agreement with the corresponding results of NOE-based structural NMR studies (Figure 2e).[11,12]
Our results allude to a correlation between the average domain–domain orientation of TAR and dynamics with bent interdomain alignments being more globally flexible than their coaxially stacked counterparts (Figure 2c,d). Furthermore, all of the ligands that arrest global motions in TAR (NeoB, Arg, and Mg2+) are known to be involved in intermolecular electrostatic interactions. In the structure determined from NMR studies,[11] the five positively charged amino groups in NeoB are in close proximity to many backbone phosphate groups of TAR and other electronegative groups that belong to domains I and II and the bulge (U-C-U). The positively charged guanidinium group of Arg interacts with phosphate groups in domain I (A22) and the bulge (U23) as well as basic electronegative groups in domain II (G26).[6a,b,d] Recent studies indicate that two Arg ligands may be involved in electrostatic interactions with two regions of negative-charge density in TAR.[15] The crystal structure of TAR in the presence of Mg2+ and Ca2+ shows a network of inner- and outer-sphere interactions between three divalent ions and functional groups that span domains I (A22) and II (G26, A27, and G28) as well as the bulge (U23, C24, U25).[6e] In sharp contrast, the structure determined from NMR studies of TAR–AcP indicates that the RNA–ligand intermolecular contacts primarily involve stacking and hydrophobic interactions between the three-membered ring and base moieties in the bulge and neighboring residues.[12] There is also evidence that the aliphatic side chain that harbors the single positive charge is flexible and protrudes in and out of the TAR binding pocket.[14]
Our results can therefore be rationalized in part on the basis of electrostatic interactions between negatively charged pockets in TAR that can be formed by backbone phosphate groups as well as other sugar/base electronegative groups and cationic groups in the ligand. Such interactions can uniquely stabilize the negatively charged backbone that is responsible for activating global motions in TAR and/or allow simultaneous clamping interactions with electronegative groups that belong to residues that span the interdomain interface. In TAR, formation of such negatively charged pockets appears to require a degree of coaxial stacking which likely serves to bring backbone phosphates and other electronegative groups in the bulge and neighboring residues into spatial proximity.[15] In this manner, electrostatic interactions may simultaneously arrest global motions and stabilize coaxially aligned conformations of TAR.
To our knowledge, the results reported here for TAR–AcP represent the first example in which global motions are observed in an RNA complex. This illustrates how ligand binding in itself is not sufficient for arresting RNA global motions, even if the RNA-binding site is at a critical junction that intersperses helical domains. Our results also indicate that electrostatic interactions offer one approach for stabilizing the global alignment of RNA domains that are separated by flexible linkers. The RDC approach presented in this work provides insight into such dynamical features underlying RNA–small-molecule recognition that are critical to rational drug design and that are not available from static three-dimensional structures.
Experimental Section
Uniformly 15N/13C-labeled TAR was prepared by in vitro transcription. Samples for NMR studies contained TAR (≈1.0 mM), sodium phosphate (15 mM), sodium sulfate (25 mM), and EDTA (ethylenediaminetetraacetic acid, 0.1 mM) at pH 6.0–6.2. The samples of TAR–NeoB and TAR–AcP also contained NeoB (2 mM, MP Biomedical Inc.) or AcP (2 mM, Research Diagnostics), respectively. Identical samples that contained Pf1 phage (≈25 mgmL−1, ASLA Ltd) were also prepared for NMR studies.[16] NMR spectroscopy experiments were recorded at 600 MHz at 25°C. One-bond RDCs for C1′–H1′, C2–H2, C5–H5, C6–H6, C8–H8, N1–H1, and N3–H3 were measured twice by using experiments that yield splittings along either the direct (1H) and indirect (13C/15N) dimension, as previously described.[7d] Average RDC values were used whenever possible. Order tensors were computed using ORDERTEN SVD[17a] and REDCAT.[17b] A total of 20/18 and 21/19 RDCs were measured in domains I/II in TAR–NeoB and TAR–AcP, respectively. In all cases, RDCs from terminal residues G17 and C45 in domain I were omitted from analysis to avoid complications resulting from end-fraying effects. For NMR models of TAR–AcP, five RDC interactions from G18 and C44 had to be omitted owing to differences between the secondary structure of domain I in the two TAR constructs.
Supplementary Material
Footnotes
We thank Prof. H. Schwalbe, Dr. J. Wöhnert (University of Frankfurt), Dr. A. Phân (MSKCC), and Prof. O. Yaghi (University of Michigan) for stimulating discussions. H.M.A. was a visiting professor supported by SFB579 at the University of Frankfurt in the group of H. Schwalbe during part of this work. D.J.P. acknowledges funding from the NIH (CA46778).
Supporting information for this article (correlation plots for RDCs measured in two different experiments; measured and back-calculated RDCs; impact of choice of RDCs on order tensor analysis; table of measured RDCs) is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Stephen W. Pitt, Department of Pharmacology, Weil Medical College of Cornell University, New York, NY 10021 (USA)
Qi Zhang, Department of Chemistry and Biophysics Research Division, University of Michigan, Ann Arbor, MI 48109 (USA), Fax: (+1)734-647-4865.
Prof. Dinshaw J. Patel, Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021 (USA)
Prof. Hashim M. Al-Hashimi, Email: hashimi@umich.edu, Department of Chemistry and Biophysics Research Division, University of Michigan, Ann Arbor, MI 48109 (USA), Fax: (+1)734-647-4865
References
- 1.a) Porse BT, Leviev I, Mankin AS, Garrett RA. J Mol Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]; b) Pape T, Wintermeyer W, Rodnina MV. Nat Struct Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 2.a) Williamson JR. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]; b) Leulliot N, Varani G. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 3.a) Hermann T. Biochimie. 2002;84:869–875. doi: 10.1016/s0300-9084(02)01460-8. [DOI] [PubMed] [Google Scholar]; b) Vicens Q, Westhof E. Chem Bio Chem. 2003;4:1018–1023. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]
- 4.a) Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tjandra N, Bax A. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 5.a) Krebs A, Ludwig V, Boden O, Gobel MW. Chem Bio Chem. 2003;4:972–978. doi: 10.1002/cbic.200300652. [DOI] [PubMed] [Google Scholar]; b) Froeyen M, Herdewijn P. Curr Top Med Chem. 2002;2:1123–1145. doi: 10.2174/1568026023393200. [DOI] [PubMed] [Google Scholar]
- 6.a) Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]; b) Aboulela F, Karn J, Varani G. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]; c) Aboulela F, Karn J, Varani G. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Brodsky AS, Williamson JR. J Mol Biol. 1997;267:624–639. doi: 10.1006/jmbi.1996.0879. [DOI] [PubMed] [Google Scholar]; e) Ippolito JA, Steitz TA. Proc Natl Acad Sci USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]; b) Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. J Am Chem Soc. 2003;125:10530–10531. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]; c) Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. J Mol Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. J Mol Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Tor Y, Hermann T, Westhof E. Chem Biol. 1998;5:277–283. doi: 10.1016/s1074-5521(98)90286-1. [DOI] [PubMed] [Google Scholar]; b) Hermann T, Westhof E. J Mol Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]; c) Hermann T, Westhof E. J Med Chem. 1999;42:1250–1261. doi: 10.1021/jm981108g. [DOI] [PubMed] [Google Scholar]; d) Tor Y. Chem Bio Chem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 9.a) Wang S, Huber PW, Cui M, Czarnik AW, Mei HY. Biochemistry. 1998;37:5549–5557. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]; Mei HY, Galan AA, Halim NS, Mack DP, Moreland DW, Sanders KB, Truong HN, Czarnik AW. Bioorg Med Chem Lett. 1995;5:2755–2760. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 10.a) Filikov AV, Mohan V, Vickers TA, Griffey RH, Cook PD, Abagyan RA, James TL. J Comput-Aided Mol Des. 2000;14:593–610. doi: 10.1023/a:1008121029716. [DOI] [PubMed] [Google Scholar]; b) Lind KE, Du Z, Fujinaga K, Peterlin BM, James TL. Chem Biol. 2002;9:185–193. doi: 10.1016/s1074-5521(02)00106-0. [DOI] [PubMed] [Google Scholar]
- 11.Faber C, Sticht H, Schweimer K, Rösch P. J Biol Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Lind KE, James TL. Chem Biol. 2002;9:707–712. doi: 10.1016/s1074-5521(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 13.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. J Am Chem Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 14.Mayer M, James TL. J Am Chem Soc. 2004;126:4453–4460. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 15.Davis B, Afshar M, Varani G, Murchie AI, Karn J, Lentzen G, Drysdale M, Bower J, Potter AJ, Starkey ID, Swarbrick T, Aboulela F. J Mol Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 16.a) Hansen MR, Mueller L, Pardi A. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]; b) Clore GM, Starich MR, Gronenborn AM. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 17.a) Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]; b) Valafar H, Prestegard JH. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.