Abstract
Mono-, di- and trimethylated states of particular histone lysine residues are selectively found in different regions of chromatin, thereby implying specialized biological functions for these marks ranging from heterochromatin formation to X-chromosome inactivation and transcriptional regulation1–3. A major challenge in chromatin biology has centred on efforts to define the connection between specific methylation states and distinct biological readouts impacting on function4. For example, histone H3 trimethylated at lysine 4 (H3K4me3) is associated with transcription start sites of active genes5–7, but the molecular ‘effectors’ involved in specific recognition of H3K4me3 tails remain poorly understood. Here we demonstrate the molecular basis for specific recognition of H3(1–15)K4me3 (residues 1–15 of histone H3 trimethylated at K4) by a plant homeodomain (PHD) finger of human BPTF (bromodomain and PHD domain transcription factor), the largest subunit of the ATP-dependent chromatin-remodelling complex, NURF (nucleosome remodelling factor). We report on crystallographic and NMR structures of the bromodomain-proximal PHD finger of BPTF in free and H3(1–15)K4me3-bound states. H3(1–15)K4me3 interacts through anti-parallel β-sheet formation on the surface of the PHD finger, with the long side chains of arginine 2 (R2) and K4me3 fitting snugly in adjacent preformed surface pockets, and bracketing an invariant tryptophan. The observed stapling role by non-adjacent R2 and K4me3 provides a molecular explanation for H3K4me3 site specificity. Binding studies establish that the BPTF PHD finger exhibits a modest preference for K4me3- over K4me2-containing H3 peptides, and discriminates against monomethylated and unmodified counterparts. Furthermore, we identified key specificity-determining residues from binding studies of H3(1–15)K4me3 with PHD finger point mutants. Our findings call attention to the PHD finger as a previously uncharacterized chromatin-binding module found in a large number of chromatin-associated proteins.
The packaging of DNA within chromosomes, the orderly replication and distribution of chromosomes, the maintenance of genomic integrity, and the regulated expression of genes depend upon nucleosomal histone proteins8–11. Introduction of covalent histone modifications, ATP-dependent chromatin remodelling, and utilization of histone variants are three major mechanisms by which variation can be introduced into the chromatin fibre. Histones methylated at lysine residues constitute a key epigenetic indexing system, demarcating transcriptionally active from inactive chromatin domains in eukaryotic genomes1–3. In particular, H3K4me3 has been linked to transcriptional activation in a range of eukaryotic species5–7. Recently, the chromo-domain protein CHD1 (refs 12–14) and WDR5, a WD40 repeat protein15, have been shown to target H3 methylated at K4.
Because the trimethylated, rather than the dimethylated, form of K4 in H3 marks the transcription start site of eukaryotic genes, efforts have been made to identify and characterize a H3K4me3 reader. An accompanying paper16 reports on the discovery of trimethyl-specific H3K4me3 effectors: human BPTF and its orthologue, Drosophila NURF301, the largest subunit of NURF, the chromatin-remodelling complex that has been shown to facilitate transcriptional activation17–19.
Human BPTF and Drosophila NURF301 contain a bromodomain-proximal PHD finger (Fig. 1a and sequence alignment in Supplementary Fig. S1) involved in complex formation with H3 peptides trimethylated at K4 (ref. 16). NMR-based structures20–23 of PHD domains have previously defined an interleaved, compact zinc finger scaffold containing a conserved Cys4HisCys3 segment coordinated to two Zn2+ ions. We observe well-resolved 1H,15N-heteronuclear single quantum coherence (HSQC) spectra for the backbone amide region of 15N-labelled human BPTF PHD finger in both the free state (Supplementary Fig. S2a) and in complex with H3(1–15)K4me3 peptide (Supplementary Fig. S2b). Several amide protons undergo shifts (up to 0.5 parts per million, p.p.m.) on complex formation (Supplementary Figs S3a and S4), including three tyrosines (Y10, Y17 and Y23) and one tryptophan (W32) (Fig. 1b, left panel). The chemical shift dependence of the amide resonance of Y17 as a function of added H3(1–15) peptides containing different K4 methylation states is plotted in Fig. 1b, right panel. These data establish that the BPTF PHD finger binds most tightly to H3(1–15)K4me3, somewhat weaker to H3(1–15)K4me2, and with the binding affinity decreasing further for the monomethylated and unmethylated states. Dissociation constants (Kd) and binding enthalpies (ΔH) have been estimated from calorimetric measurements with Kd ≈ 2.7μM and ΔH ≈ − 13.1 kcal mol−1 for the H3K4me3 complex (Fig. 1c, left panel and Supplementary Fig. S5) and Kd ≈ 5.0μM and ΔH ≈ − 12.1 kcal mol−1 for the H3K4me2 complex (Fig. 1c, right panel and Supplementary Fig. S5). Binding has also been monitored using surface plasmon resonance with binding decreasing in the order K4me3 > K4me2 > K4me1 (Fig. 1d).
Figure 1. NMR, calorimetry and surface plasmon resonance-based studies of the binding of the BPTF PHD finger by H3(1–15)K4 peptides as a function of methylation state.
a, Sequences of the PHD finger of human BPTF and histone H3(1–20). b, Left panel: superposition of 1H,15N-HSQC NMR spectra showing BPTF PHD finger amide resonances in the absence (black) and presence (red) of 5 equivalents of H3(1–15)K4me3 peptide. Right panel: plots of chemical shift changes of the Y17 amide resonance of the BPTF PHD domain as a function of peptide concentration for different methylation states of H3K4. c, Isothermal titration calorimetry enthalpy plots for the binding of the BPTF PHD finger by H3(1–15)K4me3 (left panel) and H3(1–15)K4me2 (right panel) peptides. d, Surface plasmon resonance-based binding curves for the BPTF PHD finger with biotin-labelled H3(1–20) peptides as a function of K4 methylation state.
We have solved the 2.0 Å crystal structures (Supplementary Table S1) of a PHD finger-linker-bromodomain fragment of BPTF (sequence in Fig. 2a) in the free state (Fig. 2b) and in complex with H3(1–15)K4me3 peptide (Fig. 2c). The free (Fig. 2b) and bound (Fig. 2c) PHD finger-linker-bromodomain structures exhibit similar overall scaffolds (root mean square deviation (r.m.s.d.) of 0.76 Å for 165 Cα atoms), with an α-helical linker connecting the PHD finger and bromodomains. We have computed 1,030 Å2 of buried surface area on complex formation.
Figure 2. Crystal structures of human BPTF PHD finger-linker-bromodomain in the free state and bound to H3(1–15)K4me3.
a, Domain architecture of BPTF bromodomain and proximal PHD finger. b, Ribbon representation of the crystal structure of the BPTF PHD finger-linker-bromodomain in the free state. Two bound Zn ions within the PHD fold are shown as balls. c, Crystal structure of the H3(1–15)K4me3-bound complex, with bound peptide shown in a space-filling representation. d, Structure of the PHD finger complex with the 2Fo − Fc omit electron density (0.8j level) highlighted for the bound H3(1–15)K4me3 peptide. e, Positioning of the H3(1–15)K4me3 peptide (space-filling representation) on the surface of the PHD finger portion (electrostatic surface representation with red as negatively charged and blue as positively charged surface) of the complex. f, Different view emphasizing the positioning of R2 and K4me3 in adjacent channels.
We can readily and accurately trace the first six residues, A1-R2-T3-K4me3-Q5-T6, of the H3(1–15)K4me3 peptide in the complex (Fig. 2d and Supplementary Fig. S7a). This peptide segment is positioned on the surface of the BPTF finger (Figs 2e and 2f) through formation of an anti-parallel β-sheet with the K21 to D27 segment (Fig. 3a), stabilized by three direct hydrogen bonds between backbone amide and carbonyl groups involving the R2-T3-K4me3 segment. Notably, R2 and K4me3 are positioned in adjacent surface channels (Fig. 2f) separated by W32 (Fig. 2d, f), with this alignment facilitated by a hydrogen bond between the side chains of T3 and Q5 on the opposite side of the peptide backbone. The NMR complexation shifts are localized to the H3(1–15)K4me3-peptide-binding face of the PHD domain (Supplementary Fig. S3b).
Figure 3. Details of the intermolecular contacts in the H3(1–15)K4me3 peptide–BPTF PHD finger complex and comparison with its H3(1–15)K4me2-bound counterpart.
a, Intermolecular backbone interactions between the A1–T6 segment of bound H3(1–15)K4me3 peptide and the PHD finger in the complex. b, Intermolecular hydrogen-bonding interactions involving the guanidinium group of R2 in the complex. c, Superposition of free (coloured green) and H3(1–15)K4me3-bound complex (coloured yellow) of the BPTF PHD finger. d, e, Positioning of the trimethylated lysine of the H3(1–15)K4me3 peptide (d) and the dimethylated lysine of the H3(1–15)K4me2 peptide (e) within a four-aromatic-amino-acid cage of the BPTF PHD finger. Two bridging water molecules link the NH of K4me2 to the carboxylate of D6, as indicated by dashed lines.
The guanidinium group of R2 is directed towards G25 and is anchored in place through a number of direct and water-mediated hydrogen bonds involving its Nε and Nη protons and electrostatic interactions with D27 in the complex (Fig. 3b). The trimethyl group of K4me3 is positioned within a cage of four aromatic amino acids (Fig. 3c), of which Y23 forms the base and Y10, Y17 and W32 form three walls. The trimethyl ammonium group is stabilized by van der Waals and cation–π interactions, similar to earlier results in the H3K9me3–HP1 chromodomain complex24–26 (Supplementary Fig. S8a, b). There seem to be small conformational changes in the BPTF PHD finger on complex formation with H3(1–15)K4me3 peptide (stereo view in Fig. 3c).
The structure of the complex also provides a likely explanation as to why the PHD finger of BPTF binds specifically to H3K4me3 but not to H3K9me3 or H3K27me3. The arrangement of W32-separated binding channels (Fig. 2d, f) requires that R2 and K4me3 be separated by one residue, as occurs for R2-T3-K4me3 but not for R8-K9me3 (Supplementary Fig. S8c, d) or R26-K27me3. We also note that the amino terminus of the peptide is anchored in position through a pair of hydrogen bonds from the NH2 backbone to adjacent backbone carbonyls at the I48–D49 step (Fig. 3a).
We have also solved the crystal structure of the PHD finger-linker-bromodomain fragment of BPTF in complex with H3(1–15)K4me2 peptide at 1.9 Å resolution (Supplementary Table S1). There is very good superposition (r.m.s.d. = 0.12 Å for 176 Cα atoms) of the crystal structures of the H3(1–15)K4me3-bound and H3(1–15)K4me2-bound complexes (stereo view in Supplementary Fig. S9). The dimethylammonium group of K4me2 of this complex (Fig. 3e) is positioned within the aromatic-amino-acid-lined cage of the PHD finger in an orientation similar to that observed for the trimethyl-ammonium group of K4me3 in its complex (Fig. 3d). Importantly, the proton on the dimethylammonium group of K4me2 is linked through two bridging water molecules to the carboxylate of D6 (Fig. 3e; electron densities of bridging waters are shown in Supplementary Fig. S7b). The modest reduction in calorimetrically measured binding affinity and enthalpy from the K4me3 complex to the K4me2 complex (Fig. 1c) might reflect the loss of π-stacking following replacement of a methyl group by a proton.
We have also solved the NMR structure of the BPTF PHD finger in the free state (superpositioned structures in Fig. 4a; statistics in Supplementary Table S2) and when bound to H3(1–15)K4me3 peptide (superpositioned structures in Fig. 4b and Supplementary Fig. S10; statistics are in Supplementary Table S2). There is excellent correspondence between the recognition features observed in the crystalline and solution states with r.m.s.d. values (51 Cα backbone atoms) of 1.35 Å for the free BPTF PHD finger and 1.42 Å for the complex with bound H3(1–15)K4me3.
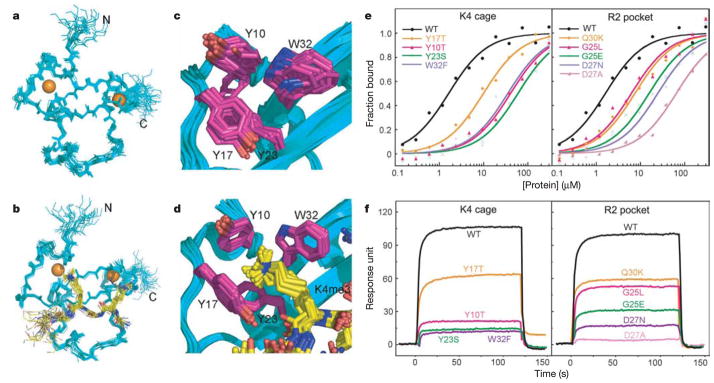
Figure 4. NMR-based structures of free and H3(1–15)K4me3-bound BPTF PHD finger, together with binding studies of PHD finger mutants with H3(1–15)K4me3 peptide.
a, b, Backbone superposition of 20 NMR-based structures of the free BPTF PHD finger (residues 8–58) (a) and the H3(1–15)K4me3-bound BPTF PHD finger in solution (b). c, d, Superposition of structures centred about a cluster of aromatic residues (Y10, Y17, Y23 and W32) in the free BPTF PHD finger (c) and when bound to H3(1–15)K4me3 (d). e, f, Fluorescence polarization-based binding curves for fluorescein-labelled H3(1–15)K4me3 peptide (e) and surface plasmon resonance-based binding curves for biotin-labelled H3(1–20)K4me3 peptide (f) with BPTF PHD finger mutants that cluster about either the K4-binding aromatic-amino-acid-lined cage (left panels) or the R2-binding pocket (right panels).
We have generated and purified point mutations of amino acids lining the K4me3-binding aromatic cage (Y10, Y17, Y23 and W32) and adjacent R2-binding channel (G25, D27 and Q30) of the BPTF PHD finger and estimated their impact on H3(1–15)K4me3 binding affinity by fluorescence polarization (Fig. 4e) and surface plasmon resonance (Fig. 4f) measurements. Fluorescence polarization measurements establish that aromatic cage mutants have an impact on binding affinity (Fig. 4e), with values dropping from Kd = 1.6 ± 0.1μM for wild-type to 10.0 ± 0.8μM for Y17T, 34.4 ± 1.1μM for W32F, 40.3 ± 4.5μM for Y10T and 59.7 ± 8.3μM for Y23S mutants. Thus, it seems that the aromatic cage residues, especially tryptophan at position 32, are critical for recognition. Y17, which is the least affected, varies the most among PHD finger homologues (Supplementary Fig. S11a). Corresponding reductions in binding affinity are also observed for R2-binding channel mutants, with values of 7.5 ± 0.4μM for Q30K, 6.2 ± 0.3μM for G25L, 15.8 ± 1.1μM for G25E, 24.6 ± 0.6μM for D27N and 59.2 ± 3.4μM for D27A mutants. Positions G25 and Q30 are also known to vary among PHD finger homologues (Supplementary Fig. S11a), whereas the more pronounced effect of D27 mutants suggests that this residue contributes critical electrostatic and hydrogen-bonding contacts within the R2 pocket. Surface plasmon resonance measurements yield a similar overall trend in reduced binding affinity (Fig. 4f).
The linker connecting the PHD and bromodomains of BPTF (Supplementary Fig. S12a) adopts an α-helical conformation between residues 54–65 in both the free (Fig. 2b) and H3(1–15)K4me3 peptide-bound (Fig. 2c) states. This solvent-exposed α-helix projects a hydrophilic surface with its N terminus fixed due to coordination involving the second zinc (stereo view in Supplementary Fig. S12b). Residues 66–71 adopt an extended conformation that forms extensive hydrogen bonds with elements of the bromodomain, with further stabilization associated with burial of L69, a conserved hydrophobic residue (Supplementary Fig. S12b).
Acetylated lysines in histone tails form complexes with bromo-domains, an extensive family of protein modules (Supplementary Fig. S13a) found in histone acetyltransferases27,28. Structural studies of acetylated lysine-containing histone peptides in complex with bromodomains have enriched our understanding of the molecular basis of ligand selectivity29,30. The BPTF bromodomain superimposes well with the reported structure of yeast GCN5 bromodomain bound to H4K16ac peptide30 (r.m.s.d. = 0.88 Å for 100 Cα atoms) (Supplementary Fig. S13b). The extent to which paired PHD finger-bromodomain in BPTF recognizes combinatorially methyl and acetyl marks in one or multiple tails, in keeping with the histone code hypothesis, remains to be determined.
Our work has provided the structural basis for substrate recognition by a tri- and dimethylated lysine-specific binding module: the PHD finger of BPTF/NURF301. The current working model, supported by the functional data in an accompanying paper16, is that PHD finger-mediated binding of BPTF/NURF301 to H3K4me3 tails stabilizes the NURF complex on chromatin, resulting in NURF-catalysed nucleosome sliding and activation of HOX genes. As PHD fingers appear in a wide range of chromatin-associated proteins31, we anticipate that our complementary structural and functional studies will pave the way for new initiatives on these modification-binding modules and shed light on their role in a wide variety of proteins associated with chromatin, its remodelling and transcriptional regulation.
METHODS
Protein and peptide preparation
The preparation and purification of the BPTF PHD finger and dual PHD finger-bromodomain, as well as isotope labelling with 15N and 13C,15N of the PHD finger for NMR studies and selenomethionine labelling of the dual PHD finger-bromodomain for X-ray studies, are described in detail in Supplementary Methods. Unmodified and methylated K4-modified H3 peptides and their carboxy-terminal fluorescein-and biotin-labelled counterparts were prepared and purified at our Institutional Core Facilities and their methylation states confirmed by mass spectroscopy.
NMR assignments, complex formation and structure determination
NMR experiments and data processing that result in backbone and side-chain resonance assignments of 13C,15N-labelled BPTF PHD finger in the absence and presence of H3(1–15)K4me3 peptide, and structure determination computations for the PHD finger in free and H3(1–15)K4me3 peptide-bound states (statistics provided in Supplementary Table S2) are described in detail in Supplementary Information.
The NMR titration protocols for monitoring complex formation between the BPTF PHD finger and H3 K4 peptides as a function of methylation state are described in detail in Supplementary Information.
Crystallization, data collection and structure determination
Crystallization conditions, MAD data set collection of the free Se-Met-labelled BPTF PHD finger-chromodomain, as well as its complex with H3(1–15)K4me3 and H3(1–15)K4me2 peptides, and processing of data sets are described in detail in Supplementary Information.
The free form crystal belongs to space group P21212. MAD phasing and initial automatic model building were performed with the program Solve/Resolve, followed by structure refinement and model rebuilding using the programs CNS and O, respectively. The model of free protein was determined at 2.0 Å against the Se-MAD peak data set with final R work/R free of 21.7/23.4. The H3(1–15)K4me3 and H3(1–15)K4me2 complex crystals belong to space group P21 with three molecules in one asymmetric unit. The structure of the H3(1–15)K4me3 complex was solved by molecular replacement using Molrep in the CCP4 software package with the free protein as a search model. The model was refined to R work/R free 19.8/22.7 at 2.0 Å by CNS. The structure of the H3(1–15)K4me2 complex was solved by difference Fourier synthesis using the CNS program with the H3(1–15)K4me3 complex structure as the starting model. The model was refined to R work/R free of 20.0/22.0 at 1.9 Å by CNS. Detailed refinement statistics are summarized in Supplementary Table S1.
Additional protocols
The protocols for fluorescence polarization, surface plasmon resonance and isothermal titration calorimetry measurements of the energetics of complex formation between H3K4me3 peptide as a function of methylation state and PHD mutants are described in detail in Supplementary Information.
Supplementary Material
Acknowledgments
D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust, and the Dewitt Wallace and Maloris Foundations. C.D.A. is supported by an NIH MERIT award and funds from Rockefeller University. S.I. holds a Ruth Kirschstein NIH postdoctoral fellowship and J.W. holds a Damon-Runyon CRF Fellowship. We thank the Peptide Core Facilities at Sloan-Kettering (S. S. Yi at Microchemistry and Proteomics) and Rockefeller University for the synthesis and purification of K4-methylated H3 peptides. We would like to thank the staff at beam line X25 at the Brookhaven National Laboratory and beam lines 23ID-D and 24ID-C of the Advanced Photon Source at the Argonne National Laboratory, supported by the US Department of Energy, for assistance with data collection. D.J.P. is a member of the New York Structural Biology Center, supported in part by funds from the NIH.
Footnotes
Author Contributions H.L. is responsible for the X-ray and surface plasmon resonance studies, S.I. for the NMR studies, W.W. for the calorimetric studies, B.D. for fluorescence polarization studies and J.W. for identification and functional characterization of the BPTF PHD finger as a H3K4me3 reader. D.J.P. and C.D.A. supervised the structural and functional (see companion paper) aspects of the project, respectively, and take overall responsibility for their joint research. All authors discussed the results and commented on the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Coordinates of the X-ray structures of the BPTF PHD finger-linker-bromodomain in the free state and when bound to H3(1–15)K4me3 and H3(1–15)K4me2 peptides have been deposited in the RCSB Protein Data Bank under accession codes 2F6N, 2F6J and 2FSA, respectively. Coordinates of the NMR structures of the BPTF PHD finger in the free state and H3(1–15)K4me3 peptide-bound state have been deposited in the RCSB Protein Data Bank under accession codes 2FUI and 2FUU, respectively.
The authors declare no competing financial interests.
References
- 1.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 4.Fischle W, Wang Y, Allis D. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 5.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 6.Santos-Rosa H, et al. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 7.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 12.Pray-Grant MG, et al. Chd1 chromodomain links histone H3 methylation with SAGA-SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 13.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 15.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006 May 21; doi: 10.1038/nature04815. advance online publication. [DOI] [PubMed] [Google Scholar]
- 17.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 18.Xiao H, et al. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 19.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 20.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 21.Capili AD, Schultz DC, Rauscher FJ, III, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottomley MJ, et al. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1) J Biol Chem. 2005;280:11505–11512. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 23.Kwan AH, et al. Engineering a protein scaffold from a PHD finger. Structure. 2003;11:803–813. doi: 10.1016/s0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 26.Min J, Zhang Y, Xu RM. Structural basis for specific binding of polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 28.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–469. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 30.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.