Abstract
It has been widely accepted that DNA can adopt other biologically relevant structures beside the Watson-Crick double helix. One recent important example is the guanine-quadruplex (G-quadruplex) structure formed by guanine tracts found in the MYC (or c-myc) promoter region, which regulates the transcription of the MYC oncogene. Stabilization of this G-quadruplex by ligands, such as the cationic porphyrin TMPyP4, decreases the transcriptional level of MYC. Here, we report the first structure of a DNA fragment containing five guanine tracts from this region. An unusual G-quadruplex fold, which was derived from NMR restraints using unambiguous model-independent resonance assignment approaches, involves a core of three stacked guanine tetrads formed by four parallel guanine tracts with all anti guanines and a snapback 3′-end syn guanine. We have determined the structure of the complex formed between this G-quadruplex and TMPγP4. This structural information, combined with details of small-molecule interaction, provides a platform for the design of anticancer drugs targeting multi-guanine-tract sequences that are found in the MYC and other oncogenic promoters, as well as in telomeres.
Human c-MYC is a transcription factor that is central to regulation of cell growth, proliferation, differentiation and apoptosis1–4. The MYC (also known as c-myc) gene that encodes this protein is tightly regulated in normal cells and its aberrant overexpression is associated with the progression of many cancers5. An important element in the MYC promoter region, termed nuclease-hypersensitivity element III1 (NHE III1), controls up to 90% of total MYC transcription6–9. Previous studies have suggested that this element, composed of a pyrimidine-rich and a purine-rich strand, may form other structures beyond the canonical B-DNA Watson-Crick duplex6,8,10–12. In particular, the 27-nucleotide (nt) purine-rich strand (Pu27) of this element, which contains six guanine tracts (Table 1), forms multiple G-quadruplex structures12–15 built from the stacking of G·G·G·G tetrads16. Furthermore, the G-quadruplex structure(s) involving four central guanine tracts of Pu27 is biologically relevant, as its destabilization (by guanine-to-adenine mutations) and its stabilization (by ligands, such as TMPyP4) increases and decreases the MYC transcriptional level, respectively12,17. Recently, sequences consisting of these four guanine tracts13–15 have been reported to form a so-called ‘propeller-type’ parallel-stranded G-quadruplex18.
Table 1.
Investigated guanine-rich sequences in the MYC promoter NHE III1a
| Name | Sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Loop 1 | Loop 2 | Loop 3 | Loop 4 | ||||||||
| Pu27 | TGGGGA | GGG | T | GGGG | A | GGG | T | GGGG | AA | GG | |
| Pu24 | TGA | GGG | T | GGGG | A | GGG | T | GGGG | AA | GG | |
| Pu24I | TGA | GGG | T | GGIG | A | GGG | T | GGGG | AA | GG | |
| myc-2345b | TGA | GGG | T | GGGG | A | GGG | T | GGGG | AA | ||
| Pu24T | TGA | GGG | T | GGTG | A | GGG | T | GGGG | AA | GG | |
| Pu24I-T12 | TGA | GGG | T | GGIG | T | GGG | T | GGGG | AA | GG | |
| Pu24Δ-T12 | TGA | GGG | T | GG - - | T | GGG | T | GGGG | AA | GG | |
| Pu24I-C21 | TGA | GGG | T | GGIG | A | GGG | T | GGGG | CA | GG | |
| Pu22I | - - A | GGG | T | GGIG | A | GGG | T | GGGG | AA | GG | |
| Pu25I | TGA | GGG | T | GGIG | A | GGG | T | GGGG | AA | GG | T |
| Pu24-A9 | TGA | GGG | T | GAGG | A | GGG | T | GGGG | AA | GG | |
| Pu24-A14 | TGA | GGG | T | GGGG | A | GAG | T | GGGG | AA | GG | |
Modifications in sequences derived from Pu24 are underlined and in boldface.
This sequence was studied in ref. 13.
Here we present the structure of a 24-nt five-guanine-tract sequence from the guanine-rich strand of the MYC NHE III1 in K+ solution. The structure represents a unique intramolecular parallel-stranded foldback G-quadruplex, in which a guanine from the 3′ tail is plugged back into the core through guanine-tetrad (G-tetrad) formation. This is the first intramolecular G-quadruplex structure of a five-guanine-tract sequence, in contrast to previously reported intramolecular G-quadruplexes of different four-guanine-tract sequences18–21. Mutations in the sequence allowed us to understand the importance of different structural motifs in this unique quadruplex folding topology. Several structural elements were revealed as potential binding sites for small-molecule ligands. Our study of the interaction of this G-quadruplex with four different ligands indicated that they all stack on the top of the G-tetrad core, the most favorable binding site. However, different patterns of NMR spectrum perturbations reflected differences in exact position, stability and kinetics of the binding events. In particular, we found that TMPyP4 binds to this G-quadruplex in slow exchange; the structure of the G-quadruplex–TMPyP4 complex revealed how stacking and electrostatic interactions contribute to the stability of the complex.
RESULTS
Favoring a possible biologically relevant G-quadruplex
The imino proton NMR spectrum of the six-guanine-tract (27-nt) MYC promoter sequence Pu27 in K+ solution (Fig. 1a) indicated the presence of multiple G-quadruplex forms. Earlier data derived from chemical probing12 suggested that the first guanine tract is not involved in biologically relevant G-quadruplex formation. The 24-nt sequence Pu24 lacking this guanine tract (Table 1) gave a much better resolved NMR spectrum (Fig. 1a). The number and intensity of major peaks for Pu24 indicated the presence of a major conformation. A single guanine-to-inosine substitution at position 10 of Pu24, resulting in the Pu24I sequence (Table 1), further improved the quality of NMR spectra (Fig. 1a). The pronounced similarity between the NMR spectra (and assignments) of Pu24 and Pu24I (Fig. 1a) are indicative of these sequences adopting the same major G-quadruplex fold. Notably, similar NMR spectra were also observed for Pu24I in solution containing 160 mM K+, 10 mM Na+, 1 mM Ca2+ and 13 mM Mg2+ (data not shown), suggesting formation of the same G-quadruplex fold at physiological condition.
Figure 1.
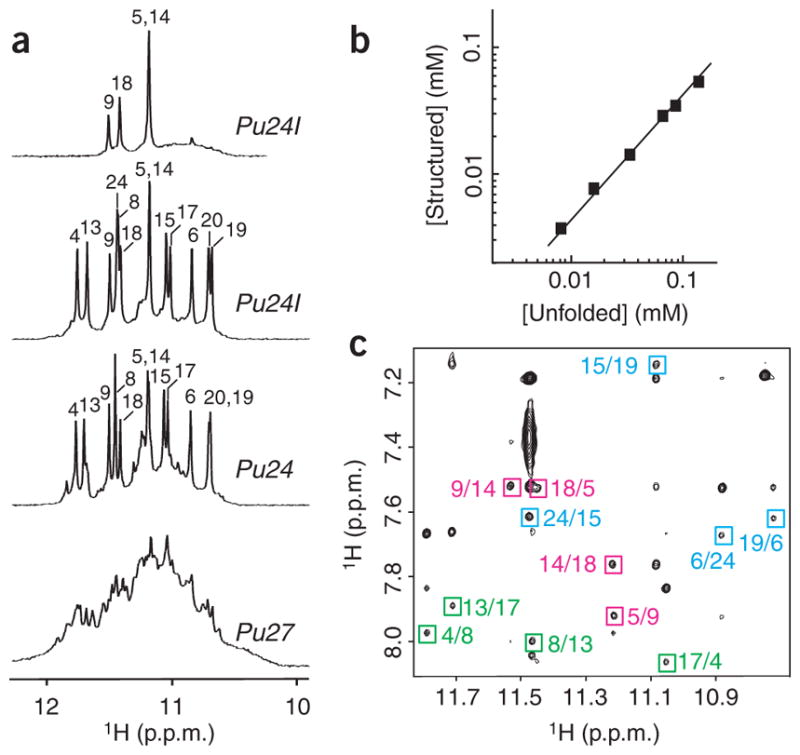
NMR study of the MYC promoter guanine-rich sequences. (a) The 600 MHz imino proton spectra of Pu27 (bottom), Pu24 (lower middle) and Pu24I in H2O (upper middle) and of Pu24I after 4 h in D2O (top), with unambiguous resonance assignments for both Pu24 and Pu24I listed over the spectra. (b) Determination of stoichiometry of Pu24I by NMR (see Methods). Line of slope 1 is drawn through the data points. (c) NOESY spectrum (mixing time, 200 ms) of Pu24I. Characteristic imino-H8 cross peaks that identify three G-tetrads (colored cyan, magenta and green) are framed and labeled with the number of imino protons in the first position and that of H8 in the second position.
Folding topology of Pu24I quadruplex
NMR titration of the concentration-dependent equilibrium between the structured and unfolded forms of Pu24I (Fig. 1b) indicated the formation of a monomeric structure, and hence an intramolecular monomeric G-quadruplex. To elucidate a novel DNA fold by NMR, unambiguous and model-independent resonance assignments are required. To this end, we used site-specific low-enrichment labeling and natural-abundance through-bond correlation approaches22,23 (Supplementary Fig. 1 online).
On the basis of the characteristic imino-H8 connectivity patterns observed in NOESY spectra of Pu24I (Fig. 1c), we established the formation of a core of three G-tetrads: (G4·G8·G13·G17), (G5·G9·G14·G18) and (G6·G24·G15·G19) (Supplementary Fig. 2 online). Observation of a strong intraresidue H8-H1′ NOE for G24 and weak H8-H1′ NOEs for other guanines (data not shown) indicated a syn glycosidic conformation for G24 and an anti conformation for other guanines. This is consistent with the G-tetrad core containing G4–G6, G8–G9, G13–G15 and G17–G19 stretches oriented to one direction and the 3′ tail oriented to the opposite direction, plugging G24 back into the core through (G6·G24·G15·G19) tetrad formation (Supplementary Fig. 2). Connecting sequential residues with loops revealed an unusual intramolecular parallel-stranded foldback G-quadruplex (Fig. 2). There are three double-chain-reversal loops connecting adjacent parallel strands: two (loops 1 and 3) are single-residue (T7 and T16) loops bridging three G-tetrad layers; another (loop 2) contains three residues (I10-G11-A12) and bridges two G-tetrad layers. There is a diagonal loop (loop 4) formed by the G20-A21-A22-G23 segment, connecting two opposite corners of the bottom G-tetrad. In this diagonal loop, the formation of G20·(A22-G23) triad (Fig. 2) is consistent with the observation of a sharp imino proton for G20 (Fig. 1a) and the identification of NOEs between A22(H8) and G23(H8) and between A22(H8) and G20(NH2) (data not shown).
Figure 2.
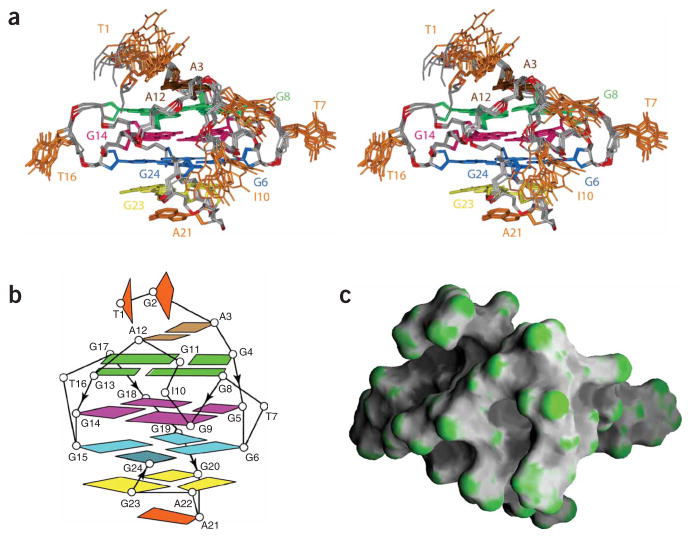
Structure of the Pu24I quadruplex. (a) Stereo view of eight superposed refined structures of the Pu24I quadruplex. The (G6·G24·G15·G19) tetrad is colored cyan, the (G5·G9·G14·G18) tetrad is colored magenta, the (G4·G8·G13·G17) tetrad is colored green, the G20· (A22-G23) triad is colored yellow and the A3·A12 pair is colored brown, with the remaining bases in orange. The backbone is colored white, phosphorus atoms are colored red, and the exocyclic backbone oxygens are omitted in the interest of clarity. (b) Schematic structure of the Pu24I quadruplex. (c) Surface view of a representative refined structure of the Pu24I quadruplex.
The imino proton spectrum of Pu24I after 4 h in D2O at 25 °C (Fig. 1a) showed that the four most protected peaks belonged to guanine imino protons of the central G-tetrad (G5, G9, G14 and G18), supporting the folding topology (Fig. 2b). Their exchange times, which were measured in real time at 25 °C by NMR, are 8 h for G9 and 5 d for G5, G14 and G18.
Solution structure of Pu24I quadruplex
The structure of the Pu24I quadruplex (Fig. 2) was calculated on the basis of NMR restraints (Supplementary Table 1 online). There are three G-tetrads (colored cyan, magenta and green) in the core of the Pu24I quadruplex, which are sandwiched between the G20· (A22-G23) triad (yellow) at the bottom and the A3·A12 pair (brown) at the top (Fig. 2a,b). The bottom triad is further capped with the well-defined stacking of A21; the top terminates with two less well defined 5′-terminal bases T1 and G2 among the ensemble of refined structures. The tetrad, triad, pair and base planes are continuously stacked in the Pu24I quadruplex (Supplementary Fig. 3 online). Two G-tetrads (green and magenta) adopt anti·anti·anti·anti alignments and one G-tetrad (cyan) adopts an anti·syn·anti·anti alignment.
The 3′-terminal tail folds back and G24 is plugged into the G-tetrad core by participating in G6(anti)·G24(syn)·G15(anti)·G19(anti) tetrad formation (Fig. 2b). Without the last two 3′-end guanines, the four-guanine-tract sequence myc-2345 (Table 1) forms a propeller-type parallel-stranded G-quadruplex involving G10 in the G-tetrad core13. In contrast, in the Pu24I quadruplex, I10 loops out from the core, being displaced by G24. This is a unique feature of a quadruplex architecture, and its origin may be related to the stability of a G·(A-G) triad–containing diagonal loop. This displacement of residue 10 is more favorable in Pu24I (with the G10I substitution) than in Pu24, because a G·I·G·G tetrad lacking one hydrogen bond is less stable than a G·G·G·G tetrad. This explains why Pu24I gives a cleaner NMR spectrum than Pu24. However, the similarity between the major forms of Pu24 and Pu24I (Fig. 1) indicated that G10 would also be displaced by G24 in the Pu24 sequence.
The G20-A21-A22-G23 segment diagonally spans the bottom of the quadruplex and connects opposite corners G19 and G24, providing the foldback position for G24 (Fig. 2b). This GAAG sequence adopts a new diagonal loop stabilized by G· (A-G) triad formation24. The G20·(A22-G23) triad is sandwiched between the (G6·G24·G15·G19) tetrad and the single-residue turn A21, with very good stacking. In this triad, G20 is in the central position and hydrogen bonded with both A22 and G23 (Supplementary Fig. 4 online). The G· (A-G) triad seen here can be compared with a G· (C-A) triad25, a T· (A-A) triad26 and an A·(T-A) triad27, which were observed previously in the context of G-quadruplexes. It can be seen as part of the recently reported pentads28,29, hexads30 and heptads31.
Two single-residue (T7 and T16) loops, which bridge three G-tetrad layers, adopt similar alignments, with the T7 and T16 bases pointing outward from the quadruplex scaffold. The three-residue I10-G11-A12 fragment forms a double-chain-reversal loop that bridges two G-tetrad layers (Supplementary Fig. 4). Within these three residues, two residues, I10 and G11, loop out and are not very well defined. In contrast, A12 is well defined and seems to form a mismatch pair with A3 (Fig. 2a), although no hydrogen-bond constraints were imposed during the structure calculation.
The four grooves in the Pu24I quadruplex differ by the orientation of strands that define them and by the presence of loops. As shown in a surface view (Fig. 2c), the shapes of the grooves are quite different and could serve as potential targets for small-molecule ligands.
Analysis of modified Pu24 sequences
The importance of different structural motifs in the quadruplex fold of Pu24 and Pu24I was analyzed through various modifications in the sequence (Table 1).
Modifications in loops
Here we did not create mutations in loops 1 and 3, because previous studies have already indicated that a single residue forms the most stable double-chain-reversal loop bridging three G-tetrad layers13,32 and that the structure of such a loop does not depend much on the nature of the base in it (thymine or adenine)29.
As discussed above, the G10I substitution in loop 2 favors the looping out of this base, resulting in cleaner NMR spectra for Pu24I than for Pu24. This was also true for the G10T substitution in the Pu24T sequence: the NMR spectrum of Pu24T was almost identical to that of Pu24I except for the peaks of residue 10 (Supplementary Fig. 5 online). The A3·A12 pair was observed in Pu24I, but is likely not to be essential for the general fold, because A12 could be replaced by a thymine (in the Pu24I-T12 sequence), and the three residues 10-11-12 could be replaced by a single thymine (in the Pu24Δ-T12 sequence), without affecting the overall fold (data not shown).
In loop 4, the structure of Pu24I predicts that A21, forming a single-base turn, can be replaced by other bases without altering the folding of the loop and the general topology. This prediction was confirmed in the A21C mutant Pu24I-C21 (Supplementary Fig. 5).
Modifications in terminal residues: 5′ end can be truncated; 3′ end can be extended
In the ensemble of structures calculated for Pu24I (Fig. 2a), the two 5′-end residues T1 and G2 are not well defined, suggesting that these residues are flexible and not required for the fold. Indeed, the NMR spectrum of the Pu22I sequence lacking these residues (Table 1) was very similar to that of Pu24I (Supplementary Fig. 5), indicating that Pu22I adopts the same fold.
The foldback conformation of the 3′-end residue G24 in Pu24I raised the question of whether this foldback feature is unique for the very end residue or can also occur with an internal residue. The proton spectrum of the Pu25I sequence (Table 1), which was extended to the 3′ direction by a thymine, was similar to that of Pu24I (Supplementary Fig. 5). This was confirmed for several unambiguous site-specific resonance assignments in Pu25I. The observation of the sharp imino proton of G24 confirmed the involvement of this base in G-tetrad formation. Similarities between the NOESY spectra of Pu25I and Pu24I (data not shown) indicated that these sequences adopt the same overall G-quadruplex topology. The structure of Pu25I, which was calculated on the basis of the Pu24I quadruplex structure and additional restraints involving T25 in Pu25I, is shown in Supplementary Figure 6 online. In this structure, T25(3′-OH) is pointed outward from the quadruplex core, suggesting the possibility of further extension of the 3′ end. This finding indicates that the folding topology presented here may exist in the context of longer sequences and represent a G-quadruplex pseudoknot.
Modifications in guanine tracts that increase MYC transcriptional activity
Two specific guanine-to-adenine mutations have been reported to greatly increase the transcriptional level of MYC12,17. These mutations are in positions 9 and 14 of our five-guanine-tract sequences, corresponding to A9 of Pu24 and A14 of Pu24, respectively (Pu24-A9 and Pu24-A14; Table 1). NMR spectra of these sequences were very different from that of Pu24 (Supplementary Fig. 5), indicating that these mutations alter the folding topology.
Interaction of Pu24I quadruplex with ligands
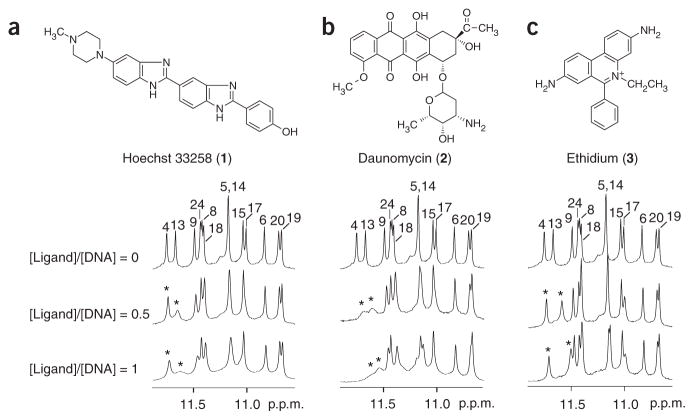
Because stabilization of G-quadruplexes in the MYC promoter can decrease MYC transcriptional level12, it is interesting to study the interaction and stabilization of the Pu24I quadruplex with different ligands. We used NMR to monitor the interaction of the Pu24I quadruplex with several small molecules that have been reported to bind G-quadruplexes: Hoechst 33258 (ref. 33; 1), daunomycin34 (2), ethidium35 (3) and TMPyP4 (ref. 12; 4) (Figs. 3 and 4). Modifications in these aromatic compounds can improve their affinity and specificity to G-quadruplexes34,36–38. Upon addition of ethidium, Hoechst 33258 and daunomycin to the solution of Pu24I, imino protons of the top G-tetrad’s residues (G4, G13, G8 and G17) were most perturbed (shifted upfield and broadened), consistent with the stacking of these ligands on the top G-tetrad. However, the patterns of these perturbations, which reflect the position, stability and kinetics of the binding events, differed between ligands. For instance, the perturbations of imino protons of G4 and G13 were different for the 1:1 ligand–DNA complexes: in the ethidium–Pu24I complex, the imino proton of G13 was shifted significantly upfield (~0.2 p.p.m.) but not much broadened (Fig. 3c), suggesting a well-defined stacking of ethidium over G13; in the Hoechst 33258–Pu24I complex, imino proton G13 was greatly broadened (Fig. 3a), suggesting multiple stacking configurations of Hoechst 33258 over G13; and in the daunomycin–Pu24I complex, both imino protons of G4 and G13 were shifted upfield and broadened (Fig. 3b), consistent with the previously observed stacking of three daunomycin molecules over a G-tetrad in the crystalline state34. Further addition of these ligands into the solution of Pu24I caused further broadening of imino protons (data not shown). In contrast to the three ligands above, TMPyP4 bound to Pu24I (under the same experimental conditions) in slow exchange regime (Fig. 4). At [TMPyP4]/[DNA] = 0.5, imino proton spectrum of Pu24I indicated the presence of both free and bound G-quadruplexes; at [TMPyP4]/[DNA] = 1, Pu24I existed mostly in the complex with TMPyP4.
Figure 3.
Interaction between the Pu24I quadruplex and different ligands as monitored by NMR. (a) Hoechst 33258. (b) Daunomycin. (c) Ethidium. Peaks are labeled with residue numbers. Peaks of G4 and G13 for samples containing ligands are labeled with stars. Pu24I concentration was 0.2 mM.
Figure 4.
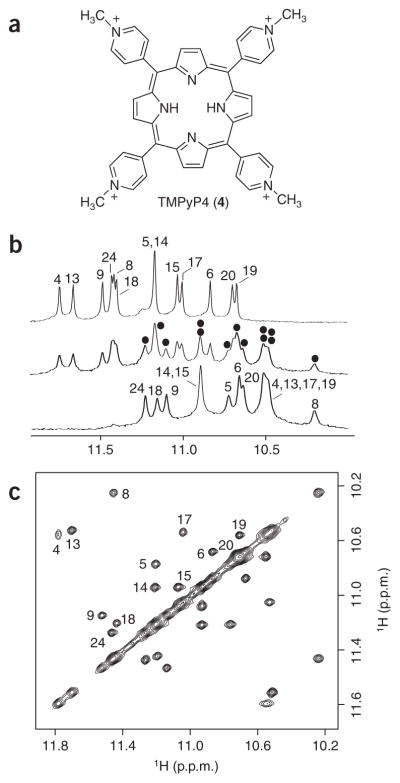
NMR study of the Pu24I–TMPyP4 complex. (a) The structure of the cationic porphyrin TMPyP4. (b) The interaction between the Pu24I quadruplex and TMPyP4 as monitored by imino proton NMR spectra. Top, middle and bottom spectra are for samples with the [TMPyP4]/[DNA] ratio being 0, 0.5, and 1, respectively. Peaks are labeled with residue numbers. At [TMPyP4]/[DNA] = 0.5, peaks for the Pu24I quadruplex–TMPyP4 complex are marked with black dots. (c) NOESY spectrum (mixing time, 75 ms) of Pu24I in the presence of 50% TMPyP4. Exchange cross-peaks between the free and bound Pu24I quadruplexes are labeled with residue numbers. Pu24I concentration was 0.2 mM for b and 0.4 mM for c.
Structure and stability of Pu24I–TMPyP4 complex
The Pu24–TMPyP4 complex gave well-resolved NMR spectra suitable for structure determination (Fig. 4). At [TMPyP4]/[DNA] = 0.5, the Pu24I quadruplexes in the free and bound states interconverted in solution, so that the NOESY spectra showed exchange cross-peaks between protons of the two conformations (Fig. 4c). This enabled the unambiguous resonance assignments for the bound form on the basis of the peak assignments for the free form. Examples of such assignments for the imino protons are shown (Fig. 4). The largest chemical shift differences between protons of the free and bound forms are for the imino protons of G4, G8, G13 and G17 (Fig. 4) and for the aromatic and sugar protons of T1 and G2 (Supplementary Fig. 7 online), suggesting that TMPyP4 is stacked over the top G-tetrad. Analysis of the NOESY spectra of the Pu24–TMPyP4 complex revealed that the general fold of the Pu24I quadruplex remains unchanged.
We calculated the structure of the complex starting from the structure of the free Pu24I quadruplex by using the intermolecular NOEs between Pu24I and TMPyP4 (Supplementary Table 1) and intramolecular NOEs within Pu24I for the top part of the quadruplex (residues 1–4, 8, 13, 17 and 10–12). In the structure of the Pu24I–TMPyP4 complex, TMPyP4 is well defined and stacked on the top of the (G4·G8·G13·G17) tetrad, somewhat shifted toward the G4 corner (Fig. 5). The A3·A12 pair is not formed; A3 is located in the groove between G4 and G17; A12 is positioned between two pyridyl rings. Residues T1 and G2 are located above TMPyP4, consistent with their upfield-shifted resonances. The positive charges of the ligand are in close contact with negative phosphates of G2, A3, G5 and G12 (Fig. 5). A comparison between the structures of the Pu24I quadruplex and the Pu24I quadruplex–TMPyP4 complex is shown in Supplementary Figure 7.
Figure 5.
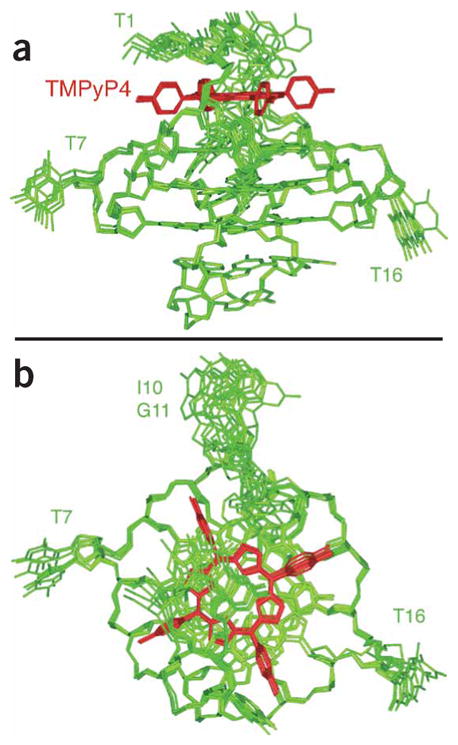
Six superposed refined structures of the Pu24I quadruplex–TMPyP4 complex. (a) Side view. (b) Top view. Pu24I quadruplex is colored green; TMPyP4 is colored red.
The imino proton spectrum of the Pu24I–TMPyP4 complex after dissolution in D2O again indicated that guanine imino protons of the central G-tetrad (G5, G9, G14 and G18) were most protected. Their exchange times at 25 °C (3 d for G9 and >40 d for G5, G14 and G18) are longer than those of the free Pu24I quadruplex by at least eightfold, consistent with the stabilization of the Pu24I quadruplex by TMPyP4.
DISCUSSION
We have established that the 24-nt five-guanine-tract Pu24 sequence from the MYC promoter folds into a distinct, parallel-stranded fold-back G-quadruplex topology in K+ solution. The identification of a parallel-stranded G-tetrad core associated with three double-chain-reversal loops highlights a general principle for intramolecular quadruplex folding in K+ solution13–15,18,29,31. However, our study also reveals several new structural motifs not seen previously. In particular, a guanine (G24) of the 3′ end is plugged back into the G-tetrad core by participating in G-tetrad formation and displacing another guanine (G10) of a continuous guanine tract into a loop. This configuration is provided by a stable diagonal loop, which contains a G· (A-G) triad stacking on and capping the G-tetrad core. These new folding features are direct consequences of the presence of five guanine tracts in the sequence, in contrast to four guanine tracts in sequences studied previously13–15,18–21.
The 27-nt Pu27 sequence (Table 1) is located in the center of a longer guanine-rich strand (~50 nt) of the MYC promoter NHE III1 (ref. 6). Containing many (six to ten) guanine tracts, these sequences are expected to adopt multiple G-quadruplex conformations using different guanine tracts and/or involving different strand orientation, syn/anti distribution and loop folding topologies. Indeed, this was observed in the NMR spectra of Pu27 (Fig. 1a). Shorter subsequences, such as Pu24 and myc-2345 (Table 1), adopt a major form and provide opportunities for structural characterization. An important question is how much (if any) of these structures is present in longer sequences and whether (and how) they are biologically relevant. It has been suggested that the formation of G-quadruplex(es), which involve four central guanine tracts of Pu27, suppresses the transcription of MYC12,17. Pu24 (five guanine tracts) and myc-2345 (four guanine tracts), which differ by two 3′-terminal guanines, both contain these guanine tracts, yet fold into different major forms. The role of the Pu24 quadruplex presented here in the full-length sequence context remains to be tested, because available biochemical data12 did not support its abundant presence in Pu27. Furthermore, even when a certain G-quadruplex structure does not exist naturally in sufficient abundance to control the transcription of MYC, it could be artificially stabilized by a ligand and, when generated in a pharmacological context, modulate the transcriptional level of MYC.
Stabilization of G-quadruplexes in the NHE by a ligand, such as TMPyP4, decreases the transcriptional level of MYC12. The G-quadruplex structure of the five-guanine-tract Pu24 sequence presented here may represent a potential anticancer target. The exceptional quality of the NMR spectra of Pu24I makes it an ideal system for NMR study of the interaction with various small molecules. Our study of the interaction between this G-quadruplex and several ligands brings insights into the common and most favorable binding site of Pu24I, the top end of the G-tetrad core. In particular, the structure of the Pu24I–TMPyP4 complex is especially valuable for the design of anticancer drugs. This structure emphasizes the stacking and the electrostatic contributions in stabilization of a G-quadruplex. In addition, this work also suggests several other structural elements (such as different grooves, different double-chain-reversal loops and a triad-containing diagonal loop) as potential binding sites for small-molecule ligands. Notably, this G-quadruplex can also be recognized by cellular proteins39, which may bind to the G-tetrad core or to different grooves and loops of the structure.
Finally, the new structural motifs and folding principles described here for Pu24 have direct relevance to ongoing efforts toward elucidating quadruplex structures formed by multi-guanine-tract DNA sequences in other oncogenic promoters and in telomeres, which are all currently attractive anticancer targets40–42, as well as by mRNA sequences that are targets of the Fragile X mental retardation protein43. One would envisage the existence of more foldback motifs with different loops in other sequence contexts.
METHODS
Sample preparation
Ethidium bromide, bisbenzimide (Hoechst 33258) tri-hydrochloride and daunomycin hydrochloride were purchased from Sigma-Aldrich. meso-Tetra(N-methyl-4-pyridyl)porphine (TMPyP4) tetrachloride was purchased from Frontier Scientific. Stock solutions (2–4 mM) were prepared by dissolving the powders in water. The unlabeled and the site-specific low-enrichment (2% 15N,13C-labeled for Pu24I and 2% 15N-labeled for Pu24 and Pu25I) oligonucleotides were synthesized and purified as described13,20. Unless otherwise stated, the strand concentration of the NMR samples was typically 0.5–2 mM; the solutions contained 70 mM of KCl and 20 mM of potassium phosphate (pH 7). The concentration of DNA and ligands was measured by NMR using the known concentration of dimethyl-2-silapentane-5-sulfonate (DSS) as internal reference.
NMR spectroscopy
Experiments were performed on 600 MHz Varian and 800 MHz Bruker spectrometers at 25 °C, unless otherwise specified. Resonances of the free quadruplexes were assigned unambiguously using site-specific low-enrichment labeling23 and through-bond correlations at natural abundance22. The resonances of the Pu24I–TMPyP4 complex were assigned through the peak assignments of the free Pu24I quadruplex by using exchange cross-peaks (in NOESY) between the free and TMPyP4-bound species obtained at [TMPyP4]/[Pu24I] = 0.5. Interproton distances were measured by using NOESY experiments.
Stoichiometry determination
The stoichiometry was determined on the basis of NMR titration of the concentration-dependent equilibrium at 60 °C between the structured and unfolded forms13,22. Measurements were carried out on samples in H2O based on signals of different imino and aromatic protons with a repetition delay of 5 s. The solutions contained 7 mM of KCl and 2 mM of potassium phosphate (pH 7).
Structure calculations
The structures of quadruplexes were calculated using the X-PLOR program44. NMR-restrained molecular dynamics (distance geometry and relaxation-matrix refinement) computations for Pu24I were performed essentially as described previously29. The structure of Pu25I was calculated from the structure of Pu24I by using additional distance restraints involving T25.
The structure of the Pu24I–TMPyP4 complex was calculated from the structure of Pu24I by using intermolecular distance restraints between TMPyP4 and Pu24I and intramolecular distance restraints for the top part of the Pu24I quadruplex. Residues 1, 2, 3 and 12 were lifted in order to manually place TMPyP4 on top of the (G4·G8·G13·G17) tetrad guided by TMPyP4-Pu24I intermolecular NOE restraints. Broken bonds between residues 11-12-13 and 3–4 were then restored, and the molecule was subjected to refinement with intra-DNA and intermolecular TMPyP4-DNA restraints. Because protons of TMPyP4 are not resolved (one single peak is observed for each type of proton), intermolecular restraints were treated as ambiguous with sum-averaging option for the refinement. The position of TMPyP4 within the complex was incrementally changed twice by a 15° rotation around the axis of the Pu24I G-tetrad core. In each of the three positions of TMPyP4 within the complex, angles between pyridyl rings and the porphyrin were initially set to ±60°, corresponding to previously reported crystal structure of the ligand45 and favorable van der Waals energy term. Next, the force constant for the torsion angle between pyridyl rings and the porphyrin was set to 0 (that is, free rotation was allowed); molecular dynamics and energy minimization was performed again for six complexes. The resulting torsion angle converged to values in the 65–82° range for different pyridyl rings of TMPyP4, and the ligand positions showed good convergence (see Results).
Supplementary Material
Acknowledgments
We thank Y. Modi for his participation at the early stage of this study. H.Y.G. is a student from the Bioinformatics Workshop at the City College of New York supported by a grant from Howard Hughes Medical Institute for undergraduate science education. D.J.P. is a member of the New York Structural Biology Center supported by US National Institutes of Health grant GM66354. This research was supported by US National Institutes of Health grant GM34504.
Footnotes
Accession codes. Protein Data Bank codes: The coordinates for the structures of Pu24I and the Pu24I–TMPyP4 complex have been deposited under accession codes 2A5P and 2A5R, respectively.
Note: Supplementary information is available on the Nature Chemical Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelengaris S, Rudolph B, Littlewood T. Action of Myc in vivo—proliferation and apoptosis. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 4.Jaattela M. Multiple cell death pathways as regulators of tumour initiation and progression. Oncogene. 2004;23:2746–2756. doi: 10.1038/sj.onc.1207513. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, deKernion JB, Verma IM, Cline MJ. Expression of cellular oncogenes in human malignancies. Science. 1984;224:256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 6.Siebenlist U, Hennighausen L, Battey J, Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 7.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 8.Davis TL, Firulli AB, Kinniburgh AJ. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc Natl Acad Sci USA. 1989;86:9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berberich SJ, Postel EH. PuF/NM23–H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 10.Boles TC, Hogan ME. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987;26:367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- 11.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seenisamy J, et al. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 15.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 16.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Edn Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 17.Grand CL, et al. Mutations in the G-quadruplex silencer element and their relationship to c-MYC overexpression, NM23 repression, and therapeutic rescue. Proc Natl Acad Sci USA. 2004;101:6140–6145. doi: 10.1073/pnas.0400460101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 19.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 22.Phan AT, Guéron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 23.Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J Am Chem Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 24.Kuryavyi VV, Jovin TM. Triangular complementarity of the triad-DNA duplex. In: Sarma RH, Sarma MH, editors. Biological Structure and Dynamics. Vol. 2. Adenine Press; Guilderland, New York, USA: 1996. pp. 91–103. [Google Scholar]
- 25.Kettani A, et al. A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework. J Mol Biol. 2000;301:129–146. doi: 10.1006/jmbi.2000.3932. [DOI] [PubMed] [Google Scholar]
- 26.Kuryavyi V, Kettani A, Wang W, Jones R, Patel DJ. A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads. J Mol Biol. 2000;295:455–469. doi: 10.1006/jmbi.1999.3345. [DOI] [PubMed] [Google Scholar]
- 27.Kettani A, Bouaziz S, Wang W, Jones RA, Patel DJ. Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nat Struct Biol. 1997;4:382–389. doi: 10.1038/nsb0597-382. [DOI] [PubMed] [Google Scholar]
- 28.Zhang N, et al. V-shaped scaffold: a new architectural motif identified in an A.(G.G.G.G) pentad-containing dimeric DNA quadruplex involving stacked G(anti)G(anti).G(anti).G(syn) tetrads. J Mol Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]
- 29.Phan AT, et al. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc Natl Acad Sci USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettani A, et al. A dimeric DNA interface stabilized by stacked A.(G.G.G.G). A hexads and coordinated monovalent cations. J Mol Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 31.Matsugami A, et al. An intramolecular quadruplex of (GGA)4 triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J Mol Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 32.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 33.Maiti S, Chaudhury NK, Chowdhury S. Hoechst 33258 binds to G-quadruplex in the promoter region of human c-myc. Biochem Biophys Res Commun. 2003;310:505–512. doi: 10.1016/j.bbrc.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 34.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J Am Chem Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 35.Guo Q, Lu M, Marky LA, Kallenbach NR. Interaction of the dye ethidium bromide with DNA containing guanine repeats. Biochemistry. 1992;31:2451–2455. doi: 10.1021/bi00124a002. [DOI] [PubMed] [Google Scholar]
- 36.Koeppel F, et al. Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res. 2001;29:1087–1096. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seenisamy J, et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita T, Uno T, Ishikawa Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg Med Chem. 2005;13:2423–2430. doi: 10.1016/j.bmc.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 39.Levens D, et al. DNA conformation, topology, and the regulation of c-myc expression. Curr Top Microbiol Immunol. 1997;224:33–46. doi: 10.1007/978-3-642-60801-8_3. [DOI] [PubMed] [Google Scholar]
- 40.Mergny JL, Héléne C. G-quadruplex DNA: a target for drug design. Nat Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 41.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 42.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 43.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 44.Brünger AT. X-PLOR: A System for X-ray Crystallography and NMR. Yale University Press; New Haven, Connecticut: 1992. [Google Scholar]
- 45.Bennett M, et al. A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling. Proc Natl Acad Sci USA. 2000;97:9476–9481. doi: 10.1073/pnas.160271897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.