Abstract
♦ Background:
Peritonitis is a major complication of peritoneal dialysis (PD) and is associated with significant morbidity and mortality. Early empirical antibiotic therapy is recommended, with the choice of agents guided by local resistance patterns. As routine use of specific antimicrobial agents can drive resistance, regular assessment of causative organisms and their susceptibility to empirical therapy is essential.
♦ Methods:
We conducted a retrospective review of all PD peritonitis cases and positive PD fluid cultures obtained over a 5-year period in Western Australia following the introduction of a statewide protocol for the initial management of PD peritonitis with intraperitoneal vancomycin and gentamicin.
♦ Results:
The incidence of PD peritonitis decreased from 1 in 16 patient months (0.75/year at risk) to 1 in 29 patient months (0.41/year at risk) over the 5 years. There were 1,319 culture-positive samples and 1,069 unique isolates identified. Gram-positive bacteria accounted for 69.9% of positive cultures, with vancomycin resistance averaging 2% over the study period. Gram-negative bacteria accounted for 25.4% of positive cultures, with gentamicin resistance identified in an average of 8% of organisms. No increase in antimicrobial resistance to vancomycin or gentamicin occurred over the 5 years and there was no change in the proportion of gram-positive (69.9%), gram-negative (25.4%) or fungal (4.4%) organisms causing PD peritonitis.
♦ Conclusions:
Over time, the peritonitis rates have dramatically improved although the profile of causative organisms remains similar. Empirical treatment of PD peritonitis with intraperitoneal vancomycin and gentamicin remains efficacious, with high levels of susceptibility and no evidence that the introduction of this statewide empirical PD peritonitis treatment protocol is driving resistance to these agents.
Keywords: Peritonitis, empirical therapy, vancomycin, gentamicin, resistance patterns
Peritoneal dialysis (PD) is the dialysis modality for approximately 20% of Australians with end-stage kidney disease (1). A major complication of PD is the development of peritonitis, which is associated with significant morbidity and costs, and may contribute to the deaths of up to 27% of patients on PD (2). Even in patients who recover from peritonitis, infections can result in shortened modality survival or modality failure (3). Current guidelines recommend PD peritonitis should occur less than once in every 18 patient months on treatment (0.67 episodes per year at risk) (4).
One reason for the failure of antimicrobial therapy in PD peritonitis is antimicrobial resistance (5). Knowledge of common causative organisms and local antimicrobial susceptibility patterns is therefore essential when deciding upon appropriate therapy. In 2007, a state-wide protocol for the management of PD peritonitis was introduced in Western Australia, with initial empirical treatment consisting of intraperitoneal vancomycin and gentamicin. To determine whether this protocol might influence antimicrobial resistance or the pattern of organisms causing peritonitis (6), we performed a retrospective analysis of all cases of PD peritonitis that occurred during the 5 years following its introduction.
Patients and Methods
We conducted a retrospective review of culture and antimicrobial susceptibility data from all samples of PD fluid from patients with suspected peritonitis between July 2008 and July 2013, following introduction of the statewide peritonitis treatment protocol. Only adult patients were included in the review, and all positive culture results were included in the analysis of antibiotic resistance. Patients presenting with suspected peritonitis received empirical treatment with intraperitoneal gentamicin and vancomycin according to the following protocol; patients weighing 60 kg or less received 2 g of vancomycin and 140 mg of gentamicin, while patients > 60 kg received 2.5 g of vancomycin and 200 mg gentamicin. Prophylaxis with intranasal application of mupirocin was continued throughout this time and patients were reviewed 1-week post-initial presentation for repeat dosing of vancomycin (if required), and assessment of PD technique.
With the exception of some remote communities in Western Australia, which are geographically closer to metropolitan centers in other Australian states, dialysate samples from all regional centers as well as the 3 main tertiary hospitals in Western Australia and the 2 home dialysis training centers were processed through a single provider, PathWest Laboratory Medicine WA, with patient data recorded through the Western Australian Home Dialysis Program and reported nationally to the Australia and New Zealand Dialysis and Transplant Registry (ANZDATA). For the purpose of the study, peritonitis was defined as the presence of 2 of the following: 1) cloudy dialysate fluid and/or abdominal pain and/or fever; 2) dialysate white cell count > 100/μL, with > 50% neutrophils; 3) positive dialysate microbiological culture.
Microbiological Methods
Culture and antimicrobial susceptibility testing was performed at PathWest Laboratory Medicine WA according to International Society for Peritoneal Dialysis (ISPD) recommended methods (4). Briefly, 2 x 20 mL volumes of peritoneal dialysate were aseptically removed from the spent dialysate bag and centrifuged, and the sediment was inoculated onto a range of media for aerobic and anaerobic incubation. Remaining sediments were inoculated into Bactec aerobic and anaerobic blood culture vials for a minimum of 5 days' incubation. Prior to 2012, isolates were identified by a combination of Vitek automated systems (bioMérieux, Inc., Durham, NC, USA), API (bioMérieux, Inc., Durham, NC, USA), and phenotypic tests along with fatty acid analysis. Incongruous fatty acid and phenotypic results were clarified by 16S rDNA polymerase chain reaction (PCR). From 2012 onwards, matrix-assisted laser desorption/ionization-time of flight mass spectrometry was used as the primary means of identification.
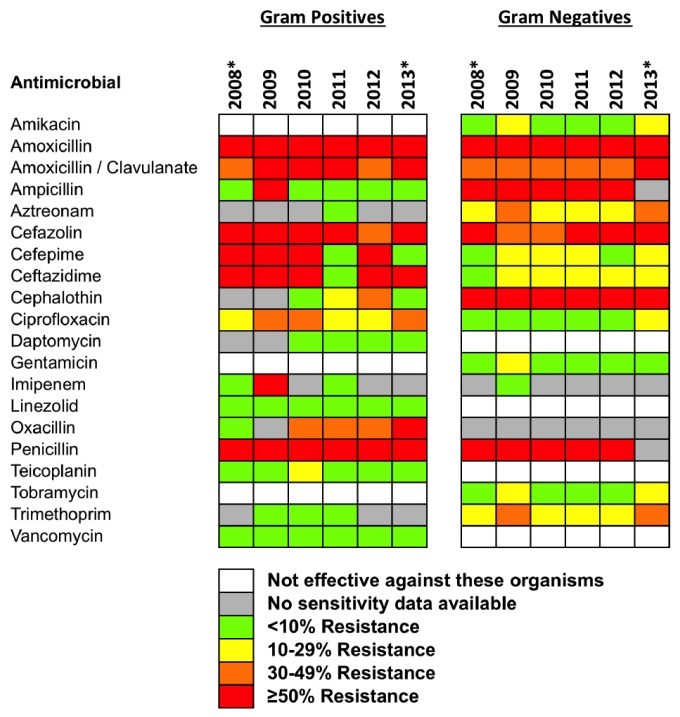
The susceptibility of staphylococci and enterococci was tested on the automated Biomerieux Vitek 2 system. A combination of oxacillin and cefoxitin was used to determine methicillin resistance on the Vitek 2. Oxacillin-resistant S. aureus and coagulase-negative staphylococci (CoNS) were considered resistant to other beta-lactam agents (with the exception of the cephalosporins with anti-methicillin-resistant S. aureus [MRSA] activity), and carbapenems, as convincing clinical data that document clinical efficacy for those agents has not been presented (7). Ambiguous results were repeated using disc diffusion or e-test methods and PCR for the mec/nuc gene. Where vancomycin resistance was detected, vanA and vanB PCR was performed. Streptococci, gram-positive bacilli and slow-growing nonfermentative gram-negative bacilli were tested by disc diffusion and e-tests. Other gram-negative bacilli were tested by agar dilution, except for swarming bacteria that were tested using the Vitek 2. Interpretation of the susceptibility results was based on guidelines and breakpoints from the Clinical and Laboratory Standards Institute (7). Isolates were classified as multi-drug resistant if they were resistant to at least 1 antimicrobial agent in 3 or more of the categories listed in Supplementary Table 1 (8). Resistance to antimicrobial agents was classified as grey (no resistance data available), green (< 10% of isolates displaying resistance), yellow (10 – 29% resistance), orange (30 – 49% resistance) and red (≥ 50% resistance) (Figure 1).
Figure 1 —
Rates of resistance to antimicrobial agents listed in the 2010 ISPD guidelines for intraperitoneal administration to PD patients. Resistance data are shown by calendar year for gram-positive and gram-negative organisms. *Denotes partial calendar years. ISPD = International Society for Peritoneal Dialysis; PD = peritoneal dialysis.
Statistical Analyses
All samples coded by the laboratory as PD fluid that yielded positive cultures were included in the analyses. Analyses were performed in Microsoft Excel 2011 and GraphPad Prism Version 6 (GraphPad Software, Inc., CA, USA).
Results
The peritonitis rate fell from 1 in 16 patient months (0.75/year at risk) in 2008 to 1 in 29 patient months (0.41/year at risk) in 2013. Over the 5 years, there were 1,319 culture-positive PD fluid samples. The culture-negative rate averaged 16.8% (min. 13.7%, max. 19.6%) and the number of patients on PD (numbers as of 31st December each year) and patient months on treatment (data given for full calendar years only) remained relatively constant over the study period (2008: 218 patients, 2009: 220 patients and 2,751 patient-months, 2010: 223 patients and 2,723 patient months, 2011: 220 patients and 2,663 patient months, 2012: 220 patients and 2,580 patient months, 2013: 225 patients). Approximately 65% of patients were managed on continuous ambulatory peritoneal dialysis (CAPD) and 35% on automated peritoneal dialysis (APD) during the study period. Of the positive samples, 1,247 were from adults, of which 1,069 were unique (excluding samples from the same patient with identical culture and antimicrobial susceptibility profiles within a 7-day period; where susceptibility profiles differed between cultures of the same organism, these organisms were considered individually in analyses of resistance). There were 13 cases of refractory peritonitis, 28 cases of relapsing peritonitis, 23 cases of recurrent peritonitis and 41 cases of repeat peritonitis according to ISPD guideline definitions (4). The refractory cases were caused by: Candida tropicalis (2 cases), Candida glabrata (1 case), Candida albicans (1 case), Cryptococcus neoformans (1 case), vancomycin-resistant enterococci (VRE) (1 case), Stenotrophomonas maltophila (1 case), Enterococcus faecalis (1 case), Escherichia coli (1 case), and Burkholderia cepacia (1 case). In the remaining 3 cases, the organisms isolated were sensitive to either vancomycin or gentamicin, but clearance took > 5 days.
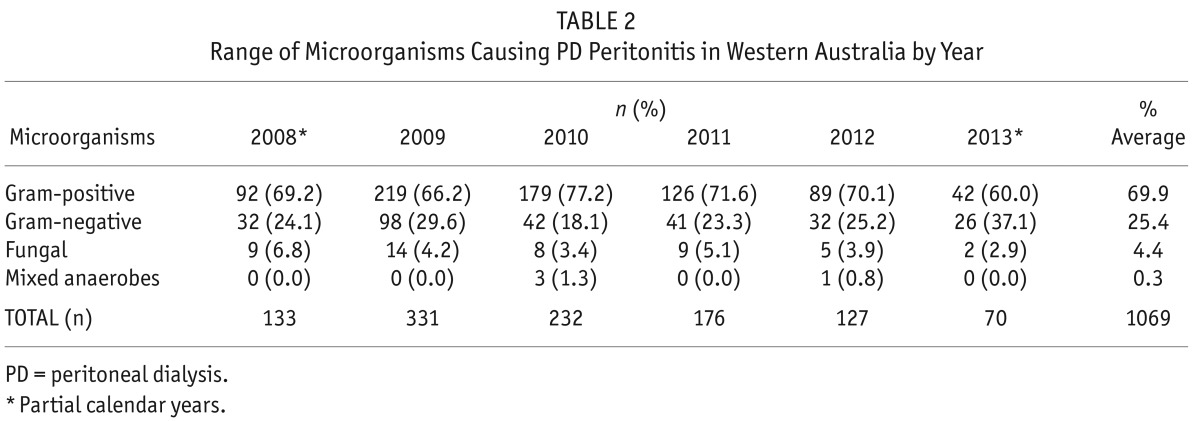
Gram-negative species accounted for 25.4% of all positive cultures (n = 271) and 47 fungal pathogens (4.4%) were isolated. Table 1 shows the breakdown of microorganisms cultured between July 2008 and July 2013. Coagulase-negative staphylococci were the predominant gram-positive species, comprising 42% of the isolates, while Klebsiella sp., E. coli and Pseudomonas sp. were the most common gram-negative microorganisms (approximately 5% each). The range of organisms cultured by year is shown in Table 2, with partial year data available for 2008 and 2013. The number of cases of PD peritonitis decreased from 331 in 2009 to 127 in 2012.
TABLE 1.
Microorganisms Isolated from Adult PD Fluid Samples Across Western Australia: July 2008 – July 2013
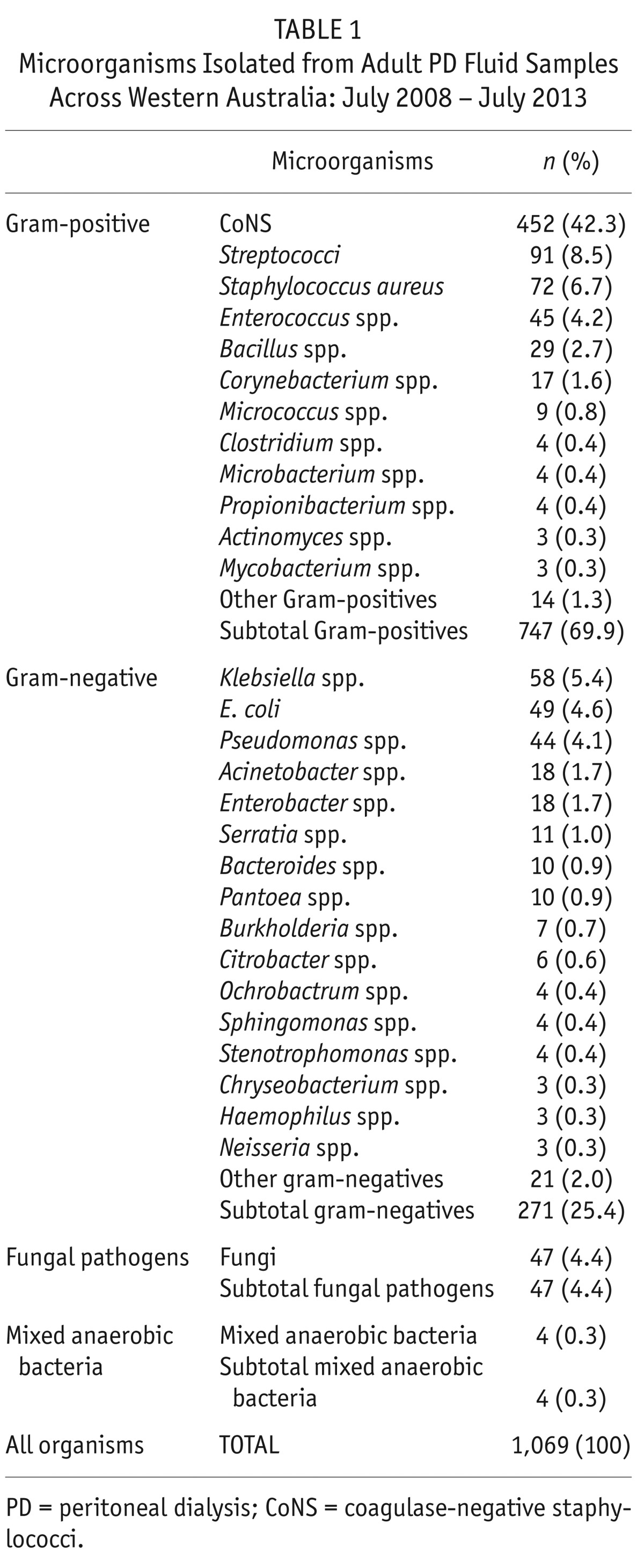
TABLE 2.
Range of Microorganisms Causing PD Peritonitis in Western Australia by Year
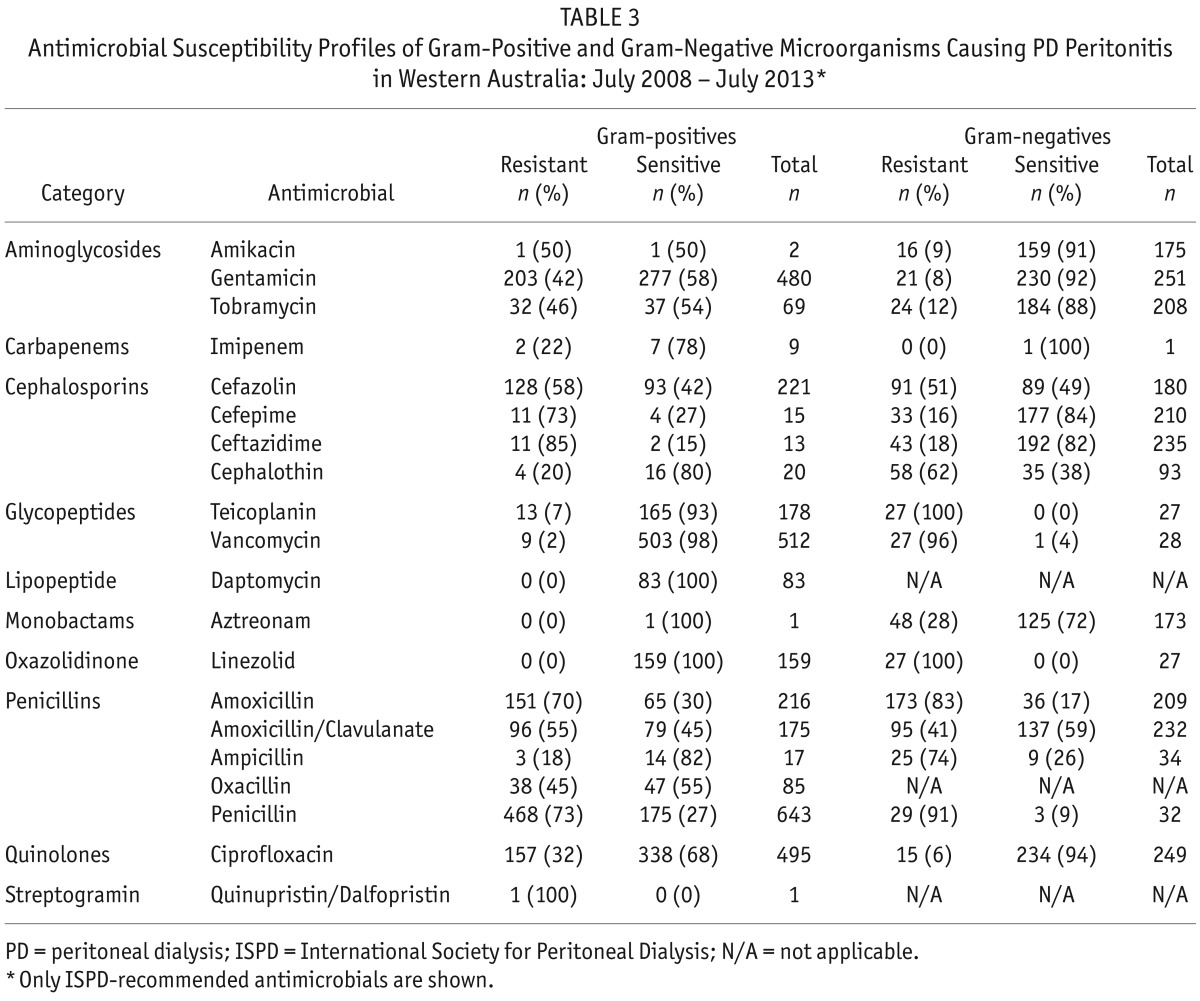
Antimicrobial susceptibility results for the ISPD-recommended intraperitoneal agents grouped into gram-positive and gram-negative isolates are shown in Table 3. Data for 2 ISPD-recommended antifungals were available: fluconazole (n = 13 isolates; n = 1 [8%] resistant) and amphotericin B (n = 9 isolates; all were sensitive). Detailed antimicrobial susceptibility results on all antimicrobial agents tested are shown in Supplementary Table 1.
TABLE 3.
Antimicrobial Susceptibility Profiles of Gram-Positive and Gram-Negative Microorganisms Causing PD Peritonitis in Western Australia: July 2008 – July 2013*
The overall resistance profiles for the ISPD-recommended antimicrobial agents, grouped by gram-positive or gram-negative microorganisms and sorted by calendar year, are shown in Figure 1. Vancomycin resistance was detected in 9 isolates of gram-positive bacteria (Enterococcus faecium [n = 3], Enterococcus gallinarum [n = 2], Lactobacillus species [n = 1], Leuconostoc species [n = 1], Staphylococcus epidermidis [n = 1] and Enterococcus faecalis [n = 1]). The resistance of E. gallinarum (n = 2), Lactobacillus sp. (n = 1) and Leuconostoc sp. (n = 1) to vancomycin is consistent with their known intrinsic resistance to this antibiotic (9,10). Only 1 of the 452 isolates of CoNS was resistant to vancomycin indicating that CoNS generally remain sensitive to vancomycin (the minimum inhibitory concentration [MIC] breakpoints for CoNS to vancomycin were: susceptible ≤ 4 μg/mL, intermediate 8-16 μg/mL and resistant ≥ 32 μg/mL) (11,12).
The majority of the gentamicin-resistant gram-negative bacteria (16/21) were non-fermenting bacilli amongst which antibiotic resistance, including resistance to aminoglycosides, is common (13) (Burkholderia cepacia complex [n = 7], Chryseobacterium indologenes [n = 1], Escherichia coli [n = 4], Ochrobactrum anthropi [n = 3], Pseudomonas mendocina [n = 1], Raoultella ornithinolytica [n = 1], Sphingobacterium species [n = 1], Stenotrophomonas maltophilia [n = 3]).
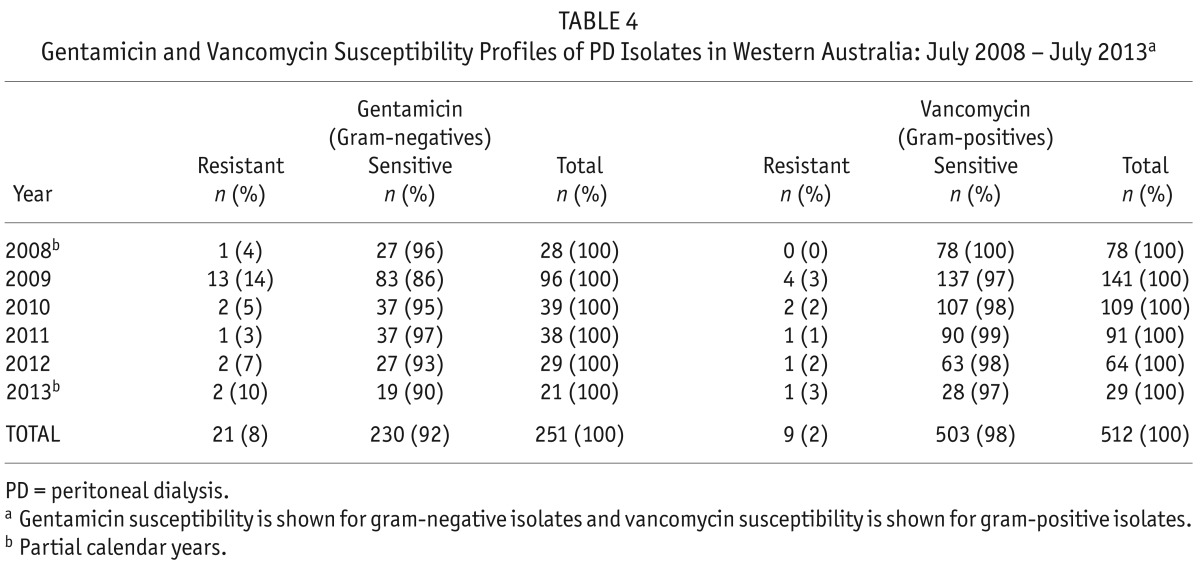
We asked next whether the profile of organisms causing PD peritonitis or their resistance profiles changed over the 5-year period following the introduction of the intraperitoneal vancomycin/gentamicin empirical treatment regimen. Despite the significant reduction in the peritonitis rate during the study, there was no change in the types of organisms causing peritonitis, or the proportion of gram-positive vs gram-negative bacteria. Resistance to gentamicin and vancomycin, the empirical treatments for PD peritonitis in Western Australia, remained low across the 5-year period studied (Table 4). Gentamicin resistance was present in 8% of gram-negative isolates (range: 3 – 14%) and vancomycin resistance was present in 2% of gram-positive isolates (range: 0 – 3%). There were no trends toward an increase in antimicrobial resistance to these agents over time, although an increase in resistance to oxacillin was noted.
TABLE 4.
Gentamicin and Vancomycin Susceptibility Profiles of PD Isolates in Western Australia: July 2008 – July 2013a
Multi-drug resistance (MDR) is defined as an isolate with resistance to at least 1 agent in 3 or more categories of antimicrobial agents (8). Multi-drug resistance was present in 395 (37%) of the positive cultures. Of the MDR isolates, 71% were gram-positive and 29% were gram-negative. Methicillin resistance occurred in 18% of Staphylococcus species tested (3% of S. aureus isolates and 20% of CoNS) (p = 0.0001, Fisher's exact test).
Discussion
An appreciation of local antimicrobial susceptibility patterns is a necessary first step toward optimizing outcomes for patients with peritonitis and devising appropriate empirical prescribing strategies (4,14,15). Following implementation of a statewide agreement for home therapies that began in 2007, a uniform protocol for the management of PD peritonitis was developed, providing an opportunity to assess the effects of this program on the profile of organisms causing PD peritonitis and their antimicrobial resistance profiles. We report 5-year data following the introduction of intraperitoneal gentamicin (high dose) and vancomycin empirical therapy for PD peritonitis, and demonstrate that this regimen has not promoted resistance to these agents, and provides outcomes comparable to those reported from other centers, in terms of mortality and technique survival (1,4).
Peritoneal dialysis peritonitis is predominantly caused by gram-positive bacteria (60 – 70%), with a minority of gram-negative organisms (20 – 25%), and rarely, fungal, anaerobic and mixed infections (3,4). Outcomes of PD peritonitis are strongly linked to the causative organism, with a higher rate of transfer to hemodialysis from gram-negative bacteria, fungi, or mycobacteria (3). Recent data from Spain, collected on all PD peritonitis patients between 2004 and 2009, revealed 64.6% gram-positive (primarily CoNS, and Streptococcus sp.) and 20.5% gram-negative microorganisms (principally E. coli), while fungal and anaerobic pathogens were rare (16). In the USA and Canada, gram-positive microorganisms accounted for 62% and 61% of PD peritonitis, respectively, and gram-negative microorganisms were responsible for 20.5% and 23.6%, respectively (17). The most frequently isolated gram-positive organisms in the USA and Canada were CoNS, S. aureus, Streptococcus spp., Enterococcus spp. and Corynebacterium spp. Frequently isolated gram-negative bacteria included E. coli, Pseudomonas spp., Klebsiella spp., Enterobacter spp., Acinetobacter spp. and Citrobacter spp. The proportions of gram-positive (69.9%), gram-negative (25.4%), and fungal (4.4%) pathogens in our patient populations are consistent with those seen elsewhere in the world, and remained relatively constant throughout the study period (4).
In keeping with data from other Australian centers (1), the incidence of PD peritonitis in Western Australia has been declining over the past few years, with peritonitis rates falling from 1 in 16 patient months (0.75/year at risk) in 2008 to 1 in 29 patient months (0.41/year at risk) in 2013. Data from the ANZDATA registry has shown a trend toward decreased gram-negative and non-MRSA S. aureus infections, and an increase in culture-negative rates over the past few years (1). However, this trend was not seen in our cohort, with the relative proportions of gram-positive and gram-negative organisms remaining stable despite overall numbers of PD peritonitis cases declining (Table 2).
Retraining and a detailed review of hygiene practices are performed following each episode of peritonitis, which may explain the reduction in peritonitis cases; however this would be expected to result in a disproportionate decrease in gram-positive infections, which was not seen. One possible reason for the decrease in gram-negative infections over time is the increased use of biocompatible dialysate solutions, which have been linked to a reduction in non-pseudomonal gram-negative peritonitis (18).
Vancomycin is a glycopeptide antibiotic with activity against gram-positive organisms that is commonly employed for the empirical treatment of PD peritonitis due its ease of dosing and efficacy against methicillin-resistant bacteria. In contrast, gram-negative organisms, due to their different cell wall structure, are largely resistant to vancomycin. To provide adequate empirical coverage against gram-negative species, high dose intraperitoneal gentamicin was introduced in 2007, due to its bactericidal properties and concerns that resistance to alternative agents, such as the fluoroquinolones, can evolve rapidly (19,20). As trends in antimicrobial prescribing provide selection pressures that can shape resistance patterns, regular review to determine whether agents remain effective is important. Over the 5-year observation period of the present study, the resistance to vancomycin remained stable, and low, with only 2% of gram-positive isolates demonstrating resistance. Gentamicin resistance was present in 8% of gram-negative isolates; however, considering the high concentration of intraperitoneal gentamicin given in our protocol, in vivo resistance may have been even less. Of note, despite use of these high doses, no cases of gentamicin-related ototoxicity or nephrotoxicity were reported. The increase in resistance to oxacillin noted over the study period is consistent with data demonstrating the spread of oxacillin-resistant species in the community (21).
Although our study is limited by its single geographical location, a major strength of our data is that all peritonitis episodes were treated according to the same protocol and peritonitis data were reported centrally so that we can be confident of capturing almost all cases. The microorganisms causing peritonitis and the percentage of gram-positive vs gram-negative bacteria are broadly equivalent to those reported from other units, with gram-negative bacteria responsible for approximately a quarter of all cases of PD peritonitis and the most prevalent gram-negative isolates being Klebsiella sp., E. coli, and Pseudomonas spp. Outcomes from PD peritonitis may be improved the earlier that appropriate antimicrobials are given. Consequently, ensuring that empirical therapy provides optimal antimicrobial coverage without jeopardizing future therapeutic options is essential. Data from Western Australia indicate that intraperitoneal vancomycin and high-dose gentamicin remain suitable empiric therapies for PD peritonitis in our patient population. Despite changes in the peritonitis rates over time, empirical use of intraperitoneal vancomycin and gentamicin for patients presenting with PD peritonitis does not appear to have promoted resistance to these agents.
Disclosures
The authors declare that they have no conflicts of interest to disclose.
Acknowledgments
The authors would like to thank Ronan Murray, Paul Healy, Barbara Henderson, and Charmaine Tonkin from PathWest Laboratory Medicine WA for supplying the details of the laboratory methods in use for organism identification and susceptibility data, and Adriana Viola from Fresenius Medical Care for providing information on the number of patients on PD, and the incidence of PD peritonitis.
REFERENCES
- 1. Brown F, Gulyani A, McDonald S, Hurst K. Peritoneal dialysis. In: McDonald S, Clayton P, Hurst K, eds. ANZDATA Registry Report 2012. Adelaide: Australian and New Zealand Dialysis and Transplant Registry; 2012. [Google Scholar]
- 2. Boudville N, Kemp A, Clayton P, Lim W, Badve SV, Hawley CM, et al. Recent peritonitis associates with mortality among patients treated with peritoneal dialysis. J Am Soc Nephrol 2012; 23:1398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghali JR, Bannister KM, Brown FG, Rosman JB, Wiggins KJ, Johnson DW, et al. Microbiology and outcomes of peritonitis in Australian peritoneal dialysis patients. Perit Dial Int 2011; 31:651–62. [DOI] [PubMed] [Google Scholar]
- 4. Li PK-T, Szeto CC, Piraino B, Bernardini J, Figueiredo AE, Gupta A, et al. Peritoneal dialysis-related infections recommendations: 2010 update. Perit Dial Int 2010; 30:393–423. [DOI] [PubMed] [Google Scholar]
- 5. Camargo CH, da Cunha Mde L, Caramori JC, Mondelli AL, Montelli AC, Barretti P. Peritoneal dialysis-related peritonitis due to coagulase-negative Staphylococcus: a review of 115 cases in a Brazilian center. Clinical J Am Soc Nephrol 2014; 9:1074–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zelenitsky S, Barns L, Findlay I, Alfa M, Ariano R, Fine A, et al. Analysis of microbiological trends in peritoneal dialysis-related peritonitis from 1991 to 1998. Am J Kidney Dis 2000; 36:1009–13. [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 8. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18:268–81. [DOI] [PubMed] [Google Scholar]
- 9. Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, et al. Lactobacillus bacteremia, species identification, and antimicrobial susceptibility of 85 blood isolates. Clin Infect Dis 2006; 42:e35–44. [DOI] [PubMed] [Google Scholar]
- 10. Courvalin P. Resistance of enterococci to glycopeptides. Antimicrobial Agents and Chemotherapy. 1990; 34:2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srinivasan A, Dick JD, Perl TM. Vancomycin resistance in staphylococci. Clin Microbiol Rev 2002; 15:430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma XX, Wang EH, Liu Y, Luo EJ. Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): Emergence of teicoplanin-non-susceptible CoNS strains with inducible resistance to vancomycin. J Med Microbiol 2011; 60:1661–8. [DOI] [PubMed] [Google Scholar]
- 13. McGowan JE., Jr Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am J Med 2006; 119:S29–36. [DOI] [PubMed] [Google Scholar]
- 14. Barretti P, Pereira D, Brasil MA, de Lourdes Cunha M, Caramori JC, Montelli AC. Evolution of gram-negative bacilli susceptibility in peritoneal dialysis-related peritonitis in Brazil: a single center's experience over nine years. Perit Dial Int 2009; 29:230–3. [PubMed] [Google Scholar]
- 15. Kim DK, Yoo TH, Ryu DR, Xu ZG, Kim HJ, Choi KH, et al. Changes in causative organisms and their antimicrobial susceptibilities in CAPD peritonitis: a single center's experience over one decade. Perit Dial Int 2004; 24:424–32. [PubMed] [Google Scholar]
- 16. Quintanar Lartundo JA, Palomar R, Dominguez-Diez A, Salas C, Ruiz-Criado J, Rodrigo E, et al. Microbiological profile of peritoneal dialysis peritonitis and predictors of hospitalization. Adv Perit Dial 2011; 27:38–42. [PubMed] [Google Scholar]
- 17. Mujais S. Microbiology and outcomes of peritonitis in North America. Kidney Int Suppl 2006:S55–62. [DOI] [PubMed] [Google Scholar]
- 18. Johnson DW, Brown FG, Clarke M, Boudville N, Elias TJ, Foo MW, et al. The effects of biocompatible compared with standard peritoneal dialysis solutions on peritonitis microbiology, treatment, and outcomes: the balANZ trial. Perit Dial Int 2012; 32:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong SSY, Ho P-L, Yuen K-Y. Evolution of antibiotic resistance mechanisms and their relevance to dialysis-related infections. Perit Dial Int 2007; 27:S272–80. [PubMed] [Google Scholar]
- 20. Li XZ. Quinolone resistance in bacteria: emphasis on plasmid-mediated mechanisms. Int J Antimicrob Agents 2005; 25:453–63. [DOI] [PubMed] [Google Scholar]
- 21. Williamson DA, Coombs GW, Nimmo GR. Staphylococcus aureus ‘down under’: contemporary epidemiology of S. aureus in Australia, New Zealand and the South West Pacific. Clin Microbiol Infect 2014; 20:597–604. [DOI] [PubMed] [Google Scholar]