Abstract
Objective
To determine the contribution of rare variants as genetic modifiers of the expressivity, penetrance, and severity of systemic sclerosis (SSc).
Methods
We performed whole-exome sequencing of 78 European American systemic sclerosis patients, including 35 patients without pulmonary arterial hypertension (SSc-PAH−) and 43 patients with PAH (SSc-PAH+). Association testing of case-control probability for rare variants was performed using the aSKAT-O method with small sample adjustment by comparing all SSc patients with a reference population of 3,179 controls from the ESP 5,500 exome dataset. Replication genotyping was performed in an independent sample of 3,263 patients (415 SSc and 2,848 controls). We conducted expression profiling of mRNA from 61 SSc patients (19 SSc-PAH− and 42 SSc-PAH+) and 41 corresponding controls.
Results
The ATP8B4 gene was associated with a significant increase in the risk of SSc (P = 3.18 × 10−7). Among the 64 ATP8B4 variants tested, a single missense variant, c.1308C>G (F436L, rs55687265), provided the most compelling evidence for association (P = 9.35 × 10−10; OR = 6.11), which was confirmed in the replication cohort (P = 0.012; OR = 1.86) and meta-analysis (P = 1.92 x 10−7; OR = 2.5). Genes involved in E3 ubiquitin-protein ligase complex (ASB10) and cyclic nucleotide gated channelopathies (CNGB3) as well as HLA-DRB5 and HSPB2 (aka heat shock protein 27) provided additional evidence for association (P < 10−5). Differential ATP8B4 expression was observed among the SSc patients compared to the controls (P = 0.0005).
Conclusion
ATP8B4 may represent a putative genetic risk factor for SSc and pulmonary vascular complications.
Systemic sclerosis (SSc) is a rare multisystem autoimmune disease estimated to have a prevalence of 240/million in the US population (based on a predominantly European American population) (1). SSc is characterized by small vessel vasculopathy, autoantibody production, excessive collagen deposition in the skin and internal organs, and abnormalities of the immune system. Whether presenting in the limited or diffuse form, the survival rate of SSc is negatively impacted by demographic characteristics such as older age of onset and male gender and by internal organ involvement of the lung and kidney (2). In fact, the most common cause of death in SSc is pulmonary disease, manifesting as either interstitial lung disease (ILD) or pulmonary arterial hypertension (PAH) (3). PAH affects approximately 10% of patients with SSc and these patients (SSc-PAH+) typically have a poorer response to PAH-specific medications and worse prognosis compared to patients with idiopathic PAH (IPAH) (4). Thus, SSc-PAH− and SSc-PAH+ subtypes (or SSc patients with other common complications that are known to worsen outcomes such as ILD) are at opposite severity extremes within the spectrum of SSc diseases.
Genetic factors may play an essential role in SSc etiology. Multiple reports from candidate gene association and replication studies, together with genome-wide association studies (GWAS) in SSc have suggested genes in the major histocompatibility complex (MHC) region, STAT4, IRF5, BLK, BANK1, TNFSF4, CD247 and TNIP1 are SSc susceptibility genes (5–7). Additional loci in IL12RB2 (8), CSK (9) and PPARG (10) were confirmed through GWAS follow-up. However, the contribution of rare variants (minor allele frequency (MAF) of less than 1–5%) to SSc susceptibility remains largely unexplored. Further, no genes directly involved in vascular or fibrotic pathways, the two critical mechanisms contributing to SSc pathogenesis, have been reproducibly identified. Whole-exome sequencing (WES) is a powerful and effective strategy for the discovery of rare coding variants that may be deleterious and relevant to disease risk (11). We hypothesized that rare variants of large effect influencing susceptibility to SSc are present at a much higher frequency in affected individuals than in the general population. Furthermore, multiple rare variants in the same gene could each have similar consequences on protein function and thus collectively identify an important SSc-associated gene. As part of National Heart, Lung, and Blood Institute (NHLBI)-supported Exome Sequencing Project (ESP), we compared rare variants in WES data from 78 SSc affected individuals to those found in 3,172 exomes from unaffected controls using the ESP 5,500 exome dataset.
To our knowledge, this is the first report of WES on SSc, a complex autoimmune disorder, with a novel gene identified for which we have demonstrated replication in an independent cohort and determined the mRNA expression levels in peripheral blood mononuclear cells (PBMCs) from patients.
PATIENTS AND METHODS
Study population
Our study included 493 European American patients with SSc followed at the Johns Hopkins Scleroderma Center and/or referred to the Johns Hopkins Pulmonary Hypertension Program for evaluation of pulmonary hypertension and performance of right heart catheterization (RHC) between October 2006 and June 2010. The Johns Hopkins University Institutional Review Board approved the conduct of this study.
Each SSc patient satisfied 1 of these criteria (12): (1) American College of Rheumatology (ACR) criteria for SSc (13); (2) 3 or more of the 5 features of the CREST syndrome (calcinosis, Raynaud’s phenomenon (RP), esophageal dysmotility, sclerodactyly, telangiectasias) (14, 15); or (3) the combination of definite RP, abnormal nailfold capillaries, and a SSc-specific autoantibody (16). The onset of SSc was defined by the first non-Raynaud’s phenomenon manifestation. (2) The SSc only patients in our cohort were free of PAH when the samples were collected from patients at diagnosis, and remained free of PAH in follow-up observation. PAH was defined as a mean pulmonary artery pressure (mPAP) of >25mmHg, pulmonary artery wedge pressure (PAWP) ≤15mmHg, and pulmonary vascular resistance (PVR)≥ 3 wood units by RHC, in the absence of other known causes of pulmonary hypertension (17). Data for the following were available for all patients in both the discovery and replication cohorts: age, gender, race, severity and duration of illness; data related to clinical, hemodynamic (right atrial pressure (RAP), mPAP, cardiac index (CI) and PVR), and echocardiographic parameters (e.g., TAPSE measurement) were available for the SSc-PAH+ patients. Hemodynamic measurements obtained closest to the date of blood collection were utilized for genomic analyses.
A subset of unrelated European American individuals with SSc was selected for the discovery cohort as part of the Exome Sequencing Project; these included SSc-PAH− and SSc-PAH+ patients (n = 78) matched on age (at blood draw), gender and subtype of SSc (limited (lcSSc) and diffuse (dcSSc) cutaneous involvement) (14). Briefly, we prioritized on (1) SSc-PAH+ patients for whom data on subtype of SSc and age of onset at PAH were available; (2) individuals with male gender, dcSSc or both (Table 1). 3,179 unaffected European American controls in the discovery cohort were drawn from the ESP 5,500 exome dataset. For unaffected controls in the replication cohort, we selected a subset of 2,848 European American subjects from Lung Health Study (18) who participated in exome array genotyping but were not part of the ESP project (Supplementary Appendix).
Table 1.
Patient Characteristics for the Exome Discovery and Replication Cohorts*
| Exome Discovery | Replication | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| SSc-PAH− | SSc-PAH+ | All SSc | SSc-PAH− | SSc-PAH+ | All SSc | |
| Demographics | ||||||
| No. of Subjects | 35 | 43 | 78 | 323 | 92 | 415 |
| Age, years (mean ± SD) | 69.6±5.4 | 61.9±11.8 | 65.3±10.2 | 56.2±12.3; | 62.3±10.3 | 57.5±12.1* |
| Female, N (%) | 28 (80.0%) | 37 (86.0%) | 65 (83.3%) | 276 (85.4%) | 79 (85.9%) | 355 (85.5%) |
| Age of onset at SSc | ||||||
| Years (mean± SD); N | 54.4±10. 8; 35 | 47.5±13. 5; 43 | 51.0±12.7; 78 | 43.1±12.5; 318 | 49.9±13. 5; 89 | 44.6±13.0 *; 407 |
| Older adult Onset (≥40 yrs), N (%) | 33/35 (94.3%) | 30/43 (69.8%) | 63/78 (80.8%) | 194/318 (61.0%) | 71/89 (79.8%) | 265/407 (65.1%)* |
| Type of SSc | ||||||
| Diffuse/limited, N (%); Total | 6/ 29 (17.1%); 35 | 5/ 29 (12.2%); 41 | 11/ 58 (14.4%); 76 | 105/211 (33.2%); 311 | 8/50 (11.1%); 72 | 113/261 (29.5%); 383 |
All numeric measurements are displayed as mean ± SD; actual number of subjects (N) available for each variable was also shown;
P < 2.2 x 10−16 from t-tests (for Age, Age of onset at SSc and Older adult onset) comparing the ‘All SSc’ group in the replication to the exome discovery cohort.
Whole-exome sequencing
DNA was extracted from whole blood samples collected from patients at the time of diagnosis. Details on quality control of sample DNA, library production and exome capture, clustering and sequencing, read mapping and variant analysis, have been described previously (19). Standard quality control (QC) approaches appropriate for exome data were used to assess both individual samples and variants (Supplementary Appendix). Originally, 80 subjects were included in the exome sequencing; 2 outliers as indicated by the different ‘heterozygous to homozygous ratio’ were discarded after applying quality control filters. Three ATP8B4 sites were removed due to low quality (read depth <10).
Genotyping and quantification of ATP8B4 mRNA expression in PBMCs from patients
Genomic DNA was extracted from peripheral blood samples. Validation genotyping for rs55687265 in ATP8B4 was performed in an independent replication cohort of 415 European American patients with SSc-PAH+ (n = 92) and SSc-PAH− (n = 323) by using TaqMan® Allelic discrimination Assays on the 7900HT Sequence Detection System (Applied Biosystems). Positive controls representing 3 genotype clusters were run on every plate to ensure accurate clustering and allele calling. We replicated 10% of the samples for quality control. Additional genotyping for controls in the replication cohort was performed as part of the Illumina HumanExome-12v1_A Beadchip array (Supplementary Appendix).
We utilized existing data from high-throughput expression profiling using Illumina Sentrix Human BeadChips (HT12_v3) with mRNA from PBMCs of SSc-PAH− (n = 19) and SSc-PAH+ (n = 42) patients and corresponding controls (n = 41). Control subjects were healthy individuals who had no known cardiovascular, pulmonary and kidney disease. Details on isolation of PBMCs, purification of RNA, the microarray experiment and analysis have been described previously (20). For comparison of gene expression between specified pairs of groups, fold changes, P-values and Benjamini-Hochberg false discovery rates (BH-FDRs) (21) were obtained using custom software written in IDL (Interactive Data Language; Exelis Visual Information Solutions, Boulder, CO) (Supplementary Appendix) (20). For the validation analysis using quantitative RT-PCR (qRT-PCR), we selected 24 samples (12 samples from the 61 cases and 12 from the controls) to represent the entire range of microarray expression values in the full dataset, and the correlation between ATP8B4 mRNA levels as detected by microarray and by RT-PCR was assessed (Supplementary Appendix).
Statistical analysis
For the exome sequencing discovery cohort, we obtained P values for association by comparing all 78 patients with SSc to the reference population of 3,179 controls using the Sequence Kernel Association Test with optimal kernel weighting and the small sample adjustment (aSKAT-O) (22). Testing was done collapsing by genes, and the model was adjusted for ancestry, using scores for four principal components (PC1, PC2, PC3 and PC16) from the principal-component decomposition of the exome data (Supplementary Appendix). The significance level was defined as 3.2 x 10−6 (P value of 0.05 divided by 15,625 genes tested). Analysis was restricted to the functional variants that are missense, nonsense, and splicing sites. Only genes with at least four functional variants with MAF<0.05 were considered. We used a flexible beta weight that is the default for aSKAT-O, to upweight the influence of rarer variants. For both the discovery and replication cohorts, single variant association testing between variant rs55687265 and disease status was done using the Pearson Chi-square test with Yates continuity correction, and we also performed a Fisher meta-analysis combining 2 Fisher’s exact tests weighted by sample size with a Mantel-Haenszel odds ratio calculated.
RESULTS
Patient characteristics
In the discovery cohort of 78 exomes from SSc patients (Table 1), affected individuals were predominantly women (83.3%) with a mean age of 65.3 (standard deviation (SD), ±10.2) years at sample collection and the mean age at SSc onset was 50.8 (SD, ±12.7) years compared to 58.8% women and a mean age of 55.6 (SD, ±13.6) years in the European American unaffected controls. Although distributed among all 4 classes of the New York Heart Association functional classification (NYHA FC) of pulmonary hypertension (23), patients with moderate to severe PAH (Class III and IV) accounted for 50% of the SSc-PAH+ group (Table S1 in the Supplementary Appendix).
ATP8B4 deleterious variant is associated with risk of SSc
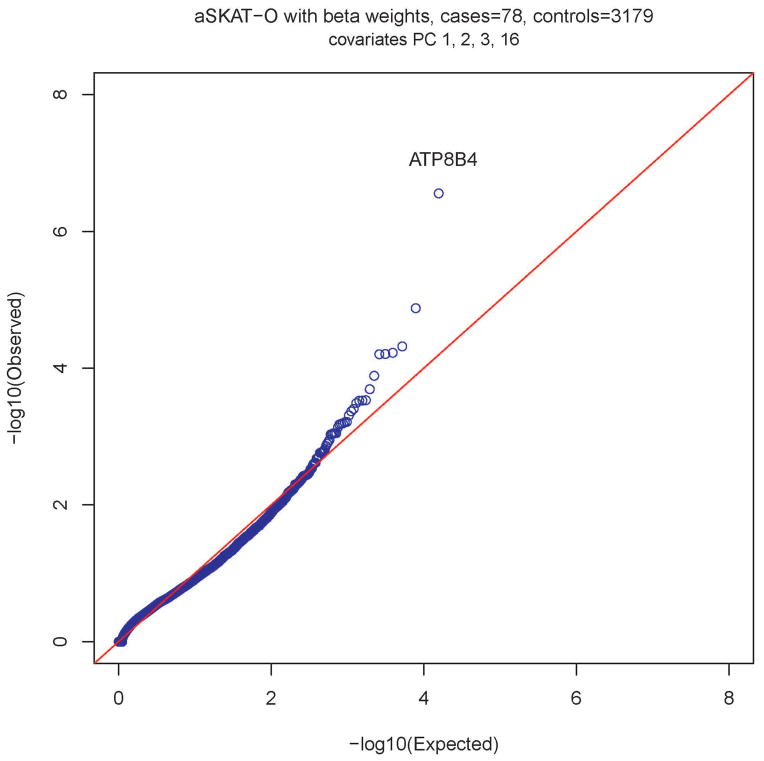
Testing for association between rare functional variants and risk of SSc was performed. The top 20 genes identified using aSKAT-O with P value <10−3 are presented in Table 2. After Bonferroni adjustment, a single gene, ATP8B4 on chromosome 15q21.2, was significantly associated with risk of development of SSc (P(beta weight) = 2.77×10−7; Bonferroni adjusted P value = 0.0043 (P-value of beta weighted analysis multiplied by 15,625 genes tested; Fig. 1). This result remained unchanged when the analysis was restricted to females only in both the discovery cohort (n = 65) and the control population (n = 1682) (Fig. S3 in the Supplementary Appendix).
Table 2.
The top 20 genes identified using aSKAT-O method (sorted by p-value from beta weighted analysis) comparing 78 SSc patients in the exome discovery cohort to the 3,179 controls
| Chromosome | Gene | adj.p.beta* | adj.p.logistic† |
|---|---|---|---|
| 15 | ATP8B4 | 2.77 x 10−7 | 1.32 x 10−7 |
| 7 | ASB10 | 1.32 x 10−5 | 1.18 x 10−5 |
| 8 | CNGB3 | 4.79 x 10−5 | 0.015 |
| 11 | HSPB2 | 5.93 x 10−5 | 5.59 x 10−5 |
| 6 | HLA-DRB5 | 6.18 x 10−5 | 6.18 x 10−5 |
| 16 | FAM195A | 6.24 x 10−5 | 6.17 x 10−5 |
| 10 | ADD3 | 0.00012886 | 0.00010348 |
| 14 | SNX6 | 0.0002026 | 0.00020171 |
| 17 | KRTAP17-1 | 0.00029338 | 0.00029338 |
| 10 | GDF2 | 0.00029692 | 0.00029162 |
| 5 | CTNND2 | 0.0002986 | 0.00028746 |
| 11 | OR8B2 | 0.00032285 | 1.92E-06 |
| 16 | LYRM1 | 0.00039352 | 0.00039535 |
| 17 | TOM1L2 | 0.00042753 | 0.00040262 |
| 13 | ZIC5 | 0.00048856 | 0.00046994 |
| 11 | APIP | 0.00060887 | 0.00061461 |
| 20 | SDC4 | 0.00061663 | 0.00061266 |
| 17 | NGFR | 0.00063978 | 0.00042046 |
| 13 | CCDC70 | 0.00065038 | 0.00061089 |
| 20 | FKBP1A-SDCBP2 | 0.00066472 | 0.00066472 |
P-values from the beta weighted aSKAT-O analyses;
P-values from the logistic weighted aSKAT-O analyses.
Figure 1.
Quantile-quantile plot of P values for a rare variant test of association between each gene and presence of SSc phenotype. A total of 15,625 genes with at least four rare variants were tested for associations with risk of SSc. The most significant association (P = 2.77×10−7) was with ATP8B4, with a collapsed score based on one single missense variant at c.1308C>G (rs55687265). The x axis represents −log10 expected P values, and the y axis represents −log10 observed P values.
Among the 64 rare functional variants (missense, nonsense or at/near a splice site) tested for ATP8B4, a single missense variant, F436L (rs55687265, chr15: 50226359) provided the most compelling evidence for association (P = 9.35 × 10−10; OR = 6.11) with an MAF of 8% among SSc individuals as compared to MAF = 1.4% in the control population. Among the 3,179 controls (3,172 with non-missing calls), 89 were heterozygous for the non-reference allele (C) at rs55687265. In contrast, among the 78 cases (75 with non-missing calls), there was 1 homozygous and 10 heterozygous. Computational protein prediction program, Polymorphism Phenotyping v2 (PolyPhen-2), predicted rs55687265 as a damaging variant with a score of 0.941 (under the HumDiv model) on a scale of 0 to 1 (0 is benign) (24). Notably, repeating the aSKAT-O analysis after removing variant rs55687265 eliminated the association with ATP8B4 (P(beta weight) = 0.5), suggesting that rs55687265 was the main deleterious variant responsible for the association signal observed for ATP8B4. Indeed, rs55687265 had the highest MAF among the 64 rare variants tested for ATP8B4. Two additional intronic variants (rs17494791 and rs187777730) were in linkage disequilibrium (LD) with variant rs55687265 (r2 threshold of 0.8 in Europeans was used), according to HaploReg v3 (http://www.broadinstitute.org/mammals/haploreg/haploreg_v3.php). Of note, SNP rs55687265, but not the other two SNPs, overlaps with an enhancer in fetal heart, suggesting the potential role of rs55687265 influencing differential ATP8B4 expression. However, neither of these variants was captured on the latest GWAS arrays (e.g., The Human Omni 2.5 BeadChip) or appeared on the GWAS Catalog (http://www.genome.gov/gwastudies/), a curated resource of SNP-trait associations.
Results from sub-group analysis remained significant when we compared the SSc-PAH− or SSc-PAH+ group to the controls separately (P = 8.98 × 10−6 and 1.41 × 10−4, respectively; Table S2 in the Supplementary Appendix).
Validation in an independent cohort
The association with variant rs55687265 was validated in an independent cohort of 415 European American SSc patients (323 SSc-PAH−, 92 SSc-PAH+; Table 1) and 2,848 control subjects (female gender: 37.1%; mean age = 48.6 years, SD = 6.8). SSc-PAH+ patients in the replication cohort had comparable characteristics compared to the discovery group, and there was no significant difference for hemodynamic measurements among the SSc-PAH+ patients (Table S1 in the Supplementary Appendix). While in the replication cohort SSc-PAH− patients were also predominantly female (85.5%) but younger, there were fewer (61%) older adult onset SSc (first SSc symptom after age 40 yrs, see Supplementary Appendix) compared to the discovery cohort (94.3%). Significant differences (P < 2.2 x 10−16) were observed for age, age of onset at SSc and older adult onset when the combined SSc group in the replication cohort was compared to the exome discovery cohort (Table 1). Among the replication set, the frequency of carriers for the non-reference allele at rs55687265 among 415 SSc patients (with 21 carriers including 2 homozygous, MAF = 2.77%) was significantly higher (P = 0.012, OR = 1.86, 95% CI: 1.134–3.025) than that among the control group of 2,848 subjects (with 84 carriers including 2 homozygous, MAF = 1.51%). A meta-analysis of 493 cases (with an MAF of 3.57%) and 6,027 controls (MAF = 1.45%) combining the discovery and replication cohorts provided enhanced signal for the association with susceptibility of SSc (P = 1.9 x 10−7, OR = 2.5, 95% CI: 1.714–3.65).
Similarly, we performed sub-group analysis to test whether the variant was differentially associated with disease subtypes. Interestingly we found older adult onset patients (61%) drove associations for the comparison between SSc-PAH− patients and the controls, in the replication cohort (P = 3.29 × 10−3). In contrast, older adult onset patients were 79.8% among the SSc-PAH+ patients in the replication cohort and the comparison between SSc-PAH+ patients and the controls were slightly more significant (P = 7.93 × 10−4; Table S2 in the Supplementary Appendix).
Notably, carriers were significantly more likely to have older adult onset (≥40 yrs) for SSc (P = 0.005) (Fig. S5 in the Supplementary Appendix). Moreover, we recently reported that late-age onset patients with SSc (≥65 yrs) in this study population had a higher incidence of PAH at 5 years compared to those with early-age onset disease (12). Thus, our finding suggests that the rs55687265 variant may represent a genetic risk factor for greater severity as measured by PAH and thus poorer outcomes of SSc.
Over-expression of ATP8B4 in PBMCs of patients with scleroderma
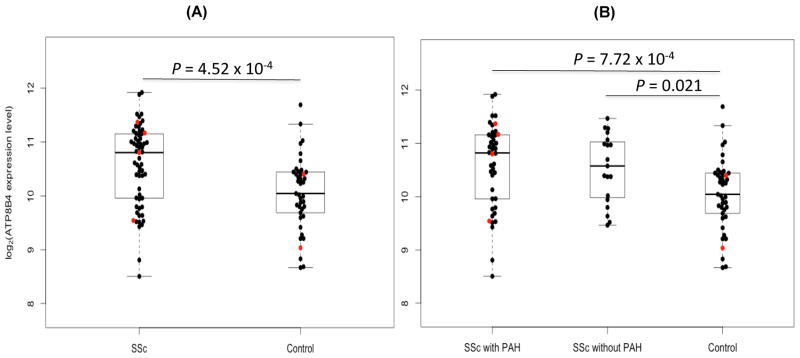
A summary of demographic and clinical descriptions for patients enrolled in the genomic analysis was presented previously (Table 1 and Table S3 in reference #20). We observed significant over-expression of ATP8B4 among 61 SSc patients (19 SSc-PAH− and 42 SSc-PAH+) compared to 41 controls (P = 4.52 × 10−4, BH-FDR = 0.003, fold change = 1.43) (Fig. 2A). We also performed a sub-group analysis comparing the expression levels of ATP8B4 in the SSc-PAH− and SSc-PAH+ groups to controls. The same trend of differential expression of ATP8B4 remained when we examined SSc-PAH− and SSc-PAH+ patients separately (P = 0.021 and 7.72 × 10−4, respectively; Fig. 2B). The direct comparisons between SSc-PAH− and SSc-PAH+ patients did not yield any significant differences, neither at the genetic nor genomic level suggesting that the SSc-PAH+ subtype represents the extreme of SSc severity rather than being a distinct independent phenotype as compared to the SSc-PAH− subtype.
Figure 2.
Overexpression of ATP8B4 mRNA in peripheral blood mononuclear cell of scleroderma patients. Box and whisker plots of ATP8B4 (ILMN_1783956) expression values: (A) comparing the combined SSc-PAH− (n=19) and SSc-PAH+ (n=42) patients to corresponding controls (n=41) (P = 4.52 × 10−4), and (B) comparing either the SSc-PAH− or SSc-PAH+ group to the controls separately (P = 0.021 and 7.72 × 10−4, respectively). Boxes represent the interquartile range (IQR) and the horizontal lines are the medians. Whiskers represent the closest values within 1.5 times the IQR. The sample values for the 4 cases and 2 controls known to have the GC genotype are colored in red.
We further validated ATP8B4 over-expression in the combined SSc case group compared to controls using qRT-PCR, in selected samples of 24 (12 samples from each group). The ATP8B4 mRNA levels detected by microarray and RT-PCR were correlated (Fig. S4 in the Supplementary Appendix, Pearson correlation was 0.765, P = 1.3×10−5). However, we were unable to definitively determine the effect of the variant rs55687265 genotype on the expression level of ATP8B4 due to limited sample size. Both the ATP8B4 expression level and variant genotype were available for 56 patients (either SSc-PAH− or SSc-PAH+) and 32 controls. Among the cases, 4 were carriers (all 4 of which were SSc-PAH+) who had the higher mean ATP8B4 expression (presented as log base 2 of scaled expression values: 10.72 ± 0.82, 95%CI: 9.42–12.03) compared to the 2 carriers among the controls (9.72 ± 0.97, 95%CI: 1.05–18.39). Cases who were non-carriers (n = 52) had significantly higher ATP8B4 expression (10.53 ± 0.78, 95%CI: 10.31–10.75) compared to the controls (n = 30, 10.12 ± 0.66, 95%CI: 9.88–10.37) that were also non-carriers (P = 0.015). Therefore, the tendency of ATP8B4 over-expression in cases versus controls was preserved, albeit with a small number of subjects having the variant genotype. Thus, over-expression of ATP8B4 gene alone correlated with SSc status and is, therefore, a potential biomarker for SSc.
DISCUSSION
In this study, we have identified a novel gene and its variants as susceptibility loci for development of SSc and for vascular complications within SSc. Using WES and highly phenotyped subjects, we describe a variant in phospholipid transporter gene ATP8B4 that is strongly associated with the risk of development of SSc, with or without PAH, a devastating complication with high morbidity and mortality. Further, we have replicated these findings in a large, well-matched cohort, thereby strengthening the validity of these findings. Additionally, gene expression profiling data demonstrating over-expression of the ATP8B4 gene in both the study and replication cohort in both those with and without gene variants suggests a possible role in disease pathogenesis.
SSc is commonly complicated by PAH, a leading cause of mortality in the SSc spectrum of diseases. The main results from prior studies using both candidate gene and GWAS approaches have been the identification of genes belonging to immune pathways, most of the time also associated with some other autoimmune genes (“shared auto-immunity”). Many of these studies have focused on risk of development of SSc and not specifically on the risk of complications of SSc such as PAH. Interestingly, one study examining genetic associations with PAH in SSc also identified an immune gene TNFAIP3 (25), suggesting a possible role for immune system perturbations in the development of pulmonary vascular complications in SSc. As evidenced by recent efforts of WES in successful identification of mutations in caveolin-1 (CAV1) (26) and KCNK3 (the gene encoding potassium channel subfamily K, member 3) in familial and IPAH (27), it is highly suspected that rare variants with strong effects remain to be discovered for SSc and its associated pulmonary vascular complications. In the current study, to strengthen the possibility of identification of genetic susceptibility factors for vascular complications, we included both subtypes of SSc (SSc-PAH− and SSc-PAH+) in a small, well-phenotyped cohort in the exome discovery phase, with targeted follow-up in an independent larger case-control cohort. Indeed, the identification of a novel phospholipid transporter gene ATP8B4 and variants as susceptibility loci for SSc and pulmonary vascular complications (i.e., PAH) in our data suggest that this is a productive approach for identifying missing heritability in complex traits. Moreover, we identified rare coding-region variants in novel risk loci for SSc rather than those known loci (e.g., STAT4 and IRF5) revealed by GWAS studies, supporting the notion that rare variants contribute to less than 3% of the heritability explained by common variants at known risk loci for autoimmune diseases (28).
ATP8B4, or probable phospholipid-transporting ATPase IM, is thought to participate in ATP biosynthesis and phospholipid transport via a variety of potential mechanisms. ATP8B4 was recently identified as a risk factor for Alzheimer disease via a GWAS study (29); ATP8B4 variants have also been associated with blood lipid phenotypes in the Framingham Heart Study (30), and risk of stroke in the STAMPEED study (dbGaP Accession: pha002887.1). However, the function and mechanisms of ATP8B4 remain largely unexplored given that there are only 10 related articles published to date. Human Class I ATPase (ATP8A1, ATP8B1, ATP8B2, ATP8B3, ATP8B4) is also called “P4-type ATPase”, which functions as ATP dependent aminophospholipid translocase and catalyzes phospholipid transport from the outer to the inner leaflet of membrane bilayers. While no high-resolution structure for a P4-ATPase has been determined, the presence of well-conserved motifs found in all P-type ATPases suggests that they adopt the four-domain structure observed with the Ca++, Na+/K+ and H+ ATPases that have been crystallized: the nucleotide-binding (N), phosphorylation (P), actuator (A) and membrane (M) domains (31). All of these pumps are predicted to have 10 transmembrane segments (TMs) composing the membrane domain. The A domain folds from cytosolic sequences preceding TM1 and in the loop between TM2 and TM3, while the P and N domains are formed from the cytosolic loop between TM4 and TM5. For cation transporters, binding of ATP to the N domain and the subsequent phosphorylation of an Asp residue in the P domain induces dramatic conformational transitions described as E1 to E2~P in the Post-Albers catalytic cycle (32–34). The deleterious ATP8B4 variant F436L is located at amino acid position 436 which is between TM4 and TM5 where the P and N domains are formed from the cytosolic loop, thus, possibly affecting the E1 to E2~P conformational transitions. Because the molecular mechanisms in which P4-type ATPases relate to severe inherited human diseases remain largely unexplored, the study of these proteins presents a challenge. Mutations in ATP8B1 cause progressive familial intrahepatic cholestasis type 1 (FIC1) or Byler disease (35), and are associated with increased risk for pneumonia with markedly elevated amounts of cardiolipin (a major phospholipid of the mitochondrial inner membrane and a key for apoptosis (36)) in lung fluid (37). Defects in cardiolipin may also play an important role in the rapid development of right ventricular dysfunction and right heart failure in persistent pulmonary hypertension of the newborn (38). Given the structural and functional similarities between ATP8B1 and ATP8B4, we speculate that defects in ATP8B4 may have important roles in regulating phospholipids in SSc associated diseases as well as other organs in which it is a candidate for genetic and acquired diseases.
Despite the novelty of an exome sequencing approach in SSc, the use of phenotypically well-matched extremes and efficient statistical approaches for analysis of rare variants, there are limitations of our study. More generally, our results suggest that rare variants, as risk factors for SSc, deserve further attention using a larger sample size of SSc cases, i.e., genotyping or targeted sequencing of thousands of SSc patients in order to validate our findings. Furthermore, the mechanisms by which variants in ATP8B4 confer risk are unknown but clearly warrant further investigation. Expression modifiers are often located in promoters, distant promoters or even intronic regions. Notably, ATP8B4 gene expression is possibly regulated by HOXA9, a DNA-binding transcription factor that binds in a distal promoter of ATP8B4 and has been previously shown to be dysregulated in SSc (39). It is likely that genetic variants that act as “expression modifiers” affecting ATP8B4 gene expression in cis and in trans are yet to be identified. In contrast, damaging variants in ATP8B4 are likely to change the protein function such as the predicted E1 to E2-P conformational transition and subsequent phospholipid transport kinetic alteration. This may explain in part the weak correlation observed between the deleterious F436L variant and ATP8B4 overexpression in SSc patients. It is well-established that the plasma membrane exhibits an asymmetric distribution of lipids between the inner and outer leaflets of the lipid bilayer. Recent studies suggest that this asymmetric distribution changes locally and temporarily, accompanied by cellular events (40, 41). Future studies examining the function of the mutant ATP8B4 might provide insights into the function of the protein, such as evaluating the mutant effects on lipid uptake by measuring the fluorescence of nitrobenzoxadiazole (NBD)-labeled phosphatidylserine (PS), a negatively charged phospholipid component of cell and blood platelet membranes typically located in the inner leaflet of the membrane.
A limitation of the gene array studies performed in our study includes a focus on PBMCs, which limits our ability to interrogate the impact on single cell populations and which may have caused us to miss important differences in gene expression in relevant, specific subsets of cells. By analyzing global RNA expression within individual tissues or cells and treating the expression levels of genes as quantitative traits, variations in gene expression that are highly correlated with genetic variation can be identified as expression quantitative trait loci, or eQTLs. In future mechanistic studies, we anticipate utilizing whole transcriptome RNA sequencing (RNA-Seq) analysis for identifying allele-specific expression (ASE), and measuring gene expression variation derived from genetic variants as eQTLs. Single-cell RNA-seq analysis facilitated by single cell isolation methods such as fluorescence-activated cell sorting (FACS) or microfluidics, is also promising and provides the expression profile of individual cells, especially for profiling rare or heterogeneous populations of cells (42, 43). Thus, genome-scale studies using single-cell RNA-seq and in-depth functional characterization of the target genetic variants are warranted.
To evaluate the vascular phenotype of SSc patients, peripheral vasculopathy and digital ulcers must be taken into consideration in addition to PAH. Our inability to show tissue specificity (i.e., in the lung) of the expression of ATP8B4 gene and its protein products in our own data remains one of the limitations of our study. Correlations between genotype and tissue-specific gene expression levels will help identify regions of the genome that influence whether and how much a gene is indeed expressed. To explore the tissue-specific expression of the ATP8B4 gene, we searched the Genotype-Tissue Expression (GTEx) Project database with newly released RNA sequencing data from 1,641 samples across 43 human tissues from 175 individuals (http://www.gtexportal.org/home/eqtls/byGene?geneId=ATP8B4&tissueName=Lung). Although rs55687265 was not found in the GTEx database, significant SNP-gene eQTL association was observed in lung tissue samples (n=124) for 3 markers located directly upstream of the ATP8B4 gene: an intronic marker in gene SLC27A2 (rs112753726, P = 2.6 x 10−6); and two additional intronic markers in LD in gene HDC (rs55828351, P = 8.5 x 10−7; rs72737086, P = 1.1 x 10−6). This novel finding supports the lung as being the main target tissue where genetic variations in the genomic region of the ATP8B4 gene is acting in cis (locally) and thus influencing its expression. Our research suggests that further investigation of whether over-expression of the ATP8B4 gene by PBMCs can provide a non-invasive, cost-effective, easy to measure and sufficiently sensitive and specific molecular biomarker is warranted. Such studies would include deeper phenotyping of patient cohorts, comparison with other known biomarkers and characterization of signaling pathways that may support the role of ATP8B4 in SSc pathogenesis.
It is noteworthy that evidence was also found for association of variants in several other genes with risk of SSc with P values less than 10−5 (Table 2). These included genes ASB10, CNGB3, MHC Class II gene HLA-DRB5 and HSPB2 (heat shock 27kDa protein 2, aka HSP27). Gene ASB10 (ankyrin repeat and SOCS box containing 10) is a component of the SCF-like ECS (Elongin-Cullin-SOCS-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins (44); it is also related to the pathway Class I MHC mediated antigen processing & presentation and Immune System. Gene CNGB3 (cyclic nucleotide gated channel beta 3) is involved in several pathways including Activation of cAMP-Dependent PKA, eNOS Signaling and Cellular Effects of Sildenafil (45, 46). An important paralog of this gene is KCNH6 (potassium voltage-gated channel, subfamily H (eag-related), member 6). It is well known that vascular, hormonal or neurological irregularities can alter production of nitric oxide by NOS and disturb the balance between synthesis and degradation of cGMP while sildenafil, a drug used to treat erectile dysfunction and PAH, can inhibit the action of PDE5 to increase cGMP levels and in turn enhance prolonged smooth muscle relaxation/vasodilation. Thus, our evidence suggests that ASB10-mediated ubiquitin degradation pathways and CNG channelopathies may play a role; functional and pharmacogenomics studies are needed to determine the clinical and genetic significance of these genes in the molecular pathogenesis of SSc and pulmonary vascular complications.
Utilizing optimized analytical approaches for rare variants and extensive studies of gene expression, our results provide compelling evidence that variants in ATP8B4 are associated with risk of SSc and pulmonary vascular complication. Dysfunction of ATP8B4 may also have implications for other neurodegenerative, cardiovascular, respiratory and autoimmune disorders depending on its targeted tissue specific expression and interaction with inflammation, angiogenesis and vascular remodeling as well as immune-effector pathways.
Supplementary Material
Acknowledgments
We thank the patients for participating in this study. The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Funding: Our work was supported in part by NHLBI HL102923 to KCB and LG; P50HL084946 to PMH; and R03HL114937 to LG, PMH and KCB. In addition, LG was supported in part by the Gilead Sciences Research Scholars Program in Pulmonary Arterial Hypertension. KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program and YK was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Barnes had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Barnes, Bamshad, Hassoun, Gao, Rich, Nickerson
Acquisition of data. Gao, Rafaels, Mathai, Hummers, Hassoun, Barnes, Bamshad, Nickerson, Cheadle, Vergara
Analysis and interpretation of data. Emond, Louie, Berger, Cheadle, Rafaels, Kim, Mathias, Taub, Ruczinski, Gao
References
- 1.Mayes MD, Lacey JV, Jr, Beebe-Dimmer J, Gillespie BW, Cooper B, Laing TJ, et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheum. 2003;48(8):2246–55. doi: 10.1002/art.11073. [DOI] [PubMed] [Google Scholar]
- 2.Barnes J, Mayes MD. Epidemiology of systemic sclerosis: incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr Opin Rheumatol. 2012;24(2):165–70. doi: 10.1097/BOR.0b013e32834ff2e8. [DOI] [PubMed] [Google Scholar]
- 3.Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972–2002. Ann Rheum Dis. 2007;66(7):940–4. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathai SC, Hassoun PM. Pulmonary arterial hypertension associated with systemic sclerosis. Expert Rev Respir Med. 2011;5(2):267–79. doi: 10.1586/ers.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorlova O, Martin JE, Rueda B, Koeleman BP, Ying J, Teruel M, et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS Genet. 2011;7(7):e1002178. doi: 10.1371/journal.pgen.1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42(5):426–9. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7(7):e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossini-Castillo L, Martin JE, Broen J, Gorlova O, Simeon CP, Beretta L, et al. A GWAS follow-up study reveals the association of the IL12RB2 gene with systemic sclerosis in Caucasian populations. Hum Mol Genet. 2012;21(4):926–33. doi: 10.1093/hmg/ddr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JE, Broen JC, Carmona FD, Teruel M, Simeon CP, Vonk MC, et al. Identification of CSK as a systemic sclerosis genetic risk factor through Genome Wide Association Study follow-up. Hum Mol Genet. 2012;21(12):2825–35. doi: 10.1093/hmg/dds099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Isac E, Bossini-Castillo L, Simeon CP, Egurbide MV, Alegre-Sancho JJ, Callejas JL, et al. A genome-wide association study follow-up suggests a possible role for PPARG in systemic sclerosis susceptibility. Arthritis Res Ther. 2014;16(1):R6. doi: 10.1186/ar4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manno RL, Wigley FM, Gelber AC, Hummers LK. Late-age onset systemic sclerosis. J Rheumatol. 2011;38(7):1317–25. doi: 10.3899/jrheum.100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preliminary criteria for the classification of systemic sclerosis (scleroderma) Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 1980;23(5):581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 14.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15(2):202–5. [PubMed] [Google Scholar]
- 15.Velayos EE, Masi AT, Stevens MB, Shulman LE. The ‘CREST’ syndrome. Comparison with systemic sclerosis (scleroderma) Arch Intern Med. 1979;139(11):1240–4. doi: 10.1001/archinte.139.11.1240. [DOI] [PubMed] [Google Scholar]
- 16.LeRoy EC, Medsger TA., Jr Criteria for the classification of early systemic sclerosis. J Rheumatol. 2001;28(7):1573–6. [PubMed] [Google Scholar]
- 17.Simonneau G, Gatzoulis MA, Adatia I, Celermajer D, Denton C, Ghofrani A, et al. Updated clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2013;62(25 Suppl):D34–41. doi: 10.1016/j.jacc.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Hansel NN, Ruczinski I, Rafaels N, Sin DD, Daley D, Malinina A, et al. Genome-wide study identifies two loci associated with lung function decline in mild to moderate COPD. Hum Genet. 2013;132(1):79–90. doi: 10.1007/s00439-012-1219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44(8):886–9. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheadle C, Berger AE, Mathai SC, Grigoryev DN, Watkins TN, Sugawara Y, et al. Erythroid-specific transcriptional changes in PBMCs from pulmonary hypertension patients. PLoS One. 2012;7(4):e34951. doi: 10.1371/journal.pone.0034951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSS-B. 1995;57(1):289–300. [Google Scholar]
- 22.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AHA medical/scientific statement. 1994 revisions to classification of functional capacity and objective assessment of patients with diseases of the heart. Circulation. 1994;90(1):644–5. [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieude P, Guedj M, Wipff J, Ruiz B, Riemekasten G, Matucci-Cerinic M, et al. Association of the TNFAIP3 rs5029939 variant with systemic sclerosis in the European Caucasian population. Ann Rheum Dis. 2010;69(11):1958–64. doi: 10.1136/ard.2009.127928. [DOI] [PubMed] [Google Scholar]
- 26.Austin ED, Ma L, LeDuc C, Berman Rosenzweig E, Borczuk A, Phillips JA, 3rd, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ Cardiovasc Genet. 2012;5(3):336–43. doi: 10.1161/CIRCGENETICS.111.961888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Roman-Campos D, Austin ED, Eyries M, Sampson KS, Soubrier F, et al. A novel channelopathy in pulmonary arterial hypertension. N Engl J Med. 2013;369(4):351–61. doi: 10.1056/NEJMoa1211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt KA, Mistry V, Bockett NA, Ahmad T, Ban M, Barker JN, et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498(7453):232–5. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 30.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim Biophys Acta. 2012;1821(8):1068–77. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–66. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 33.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5(4):282–95. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 34.Gadsby DC, Bezanilla F, Rakowski RF, De Weer P, Holmgren M. The dynamic relationships between the three events that release individual Na(+) ions from the Na(+)/K(+)-ATPase. Nat Commun. 2012;3:669. doi: 10.1038/ncomms1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Mil SW, Klomp LW, Bull LN, Houwen RH. FIC1 disease: a spectrum of intrahepatic cholestatic disorders. Semin Liver Dis. 2001;21(4):535–44. doi: 10.1055/s-2001-19034. [DOI] [PubMed] [Google Scholar]
- 36.Wortmann SB, Vaz FM, Gardeitchik T, Vissers LE, Renkema GH, Schuurs-Hoeijmakers JH, et al. Mutations in the phospholipid remodeling gene SERAC1 impair mitochondrial function and intracellular cholesterol trafficking and cause dystonia and deafness. Nat Genet. 2012;44(7):797–802. doi: 10.1038/ng.2325. [DOI] [PubMed] [Google Scholar]
- 37.Ray NB, Durairaj L, Chen BB, McVerry BJ, Ryan AJ, Donahoe M, et al. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat Med. 2010;16(10):1120–7. doi: 10.1038/nm.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saini-Chohan HK, Dakshinamurti S, Taylor WA, Shen GX, Murphy R, Sparagna GC, et al. Persistent pulmonary hypertension results in reduced tetralinoleoyl-cardiolipin and mitochondrial complex II + III during the development of right ventricular hypertrophy in the neonatal pig heart. Am J Physiol Heart Circ Physiol. 2010;301(4):H1415–24. doi: 10.1152/ajpheart.00247.2011. [DOI] [PubMed] [Google Scholar]
- 39.Avouac J, Cagnard N, Distler JH, Schoindre Y, Ruiz B, Couraud PO, et al. Insights into the pathogenesis of systemic sclerosis based on the gene expression profile of progenitor-derived endothelial cells. Arthritis Rheum. 2011;63(11):3552–62. doi: 10.1002/art.30536. [DOI] [PubMed] [Google Scholar]
- 40.Nishimura Y, Tadokoro S, Tanaka M, Hirashima N. Detection of asymmetric distribution of phospholipids by fluorescence resonance energy transfer. Biochemical and biophysical research communications. 2012;420(4):926–30. doi: 10.1016/j.bbrc.2012.03.106. [DOI] [PubMed] [Google Scholar]
- 41.Fadeel B, Xue D. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Critical reviews in biochemistry and molecular biology. 2009;44(5):264–77. doi: 10.1080/10409230903193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Method of the year 2013. Nat Methods. 2014;11(1):1. doi: 10.1038/nmeth.2801. [DOI] [PubMed] [Google Scholar]
- 43.Eberwine J, Sul JY, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11(1):25–7. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]
- 44.Keller KE, Yang YF, Sun YY, Sykes R, Acott TS, Wirtz MK. Ankyrin repeat and suppressor of cytokine signaling box containing protein-10 is associated with ubiquitin-mediated degradation pathways in trabecular meshwork cells. Mol Vis. 2013;19:1639–55. [PMC free article] [PubMed] [Google Scholar]
- 45.Beavo JA, Brunton LL. Cyclic nucleotide research -- still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3(9):710–8. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 46.Ghofrani HA, Osterloh IH, Grimminger F. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat Rev Drug Discov. 2006;5(8):689–702. doi: 10.1038/nrd2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.