Abstract
INTRODUCTION
We performed linkage analyses in Caribbean Hispanic families with multiple late-onset Alzheimer’s disease (LOAD) cases to identify regions that may contain disease causative variants.
METHODS
We selected 67 LOAD families to perform genome-wide linkage scan. Analysis of the linked regions was repeated using the entire sample of 282 families. Validated chromosomal regions were analyzed using joint linkage and association.
RESULTS
We identified 26 regions linked to LOAD (HLOD ≥ 3.6). We validated thirteen of the regions (HLOD ≥ 2.5) using the entire family sample. The strongest signal was at 11q12.3 (rs2232932: HLODmax= 4.7, Pjoint= 6.6 × 10−6), a locus located ~2Mb upstream of the MS4A gene cluster. We additional identified a locus at 7p14.3 (rs10255835: HLODmax= 4.9, Pjoint= 1.2 × 10−5), a region harboring genes associated with the nervous system (GARS, GHRHR and NEUROD6).
DISCUSSION
Future sequencing efforts should focus on these regions since they may harbor familial LOAD causative mutations.
Keywords: Caribbean Hispanic families, Late Onset Alzheimer’s Disease, linkage analysis, joint linkage and association
INTRODUCTION
Late onset Alzheimer disease (LOAD) is a highly prevalent, progressive, neurodegenerative disorder characterized by a slow transition from mild memory impairment to severe cognitive loss. Genetic variants in the amyloid precursor protein (APP) gene [1] and presenilins 1 (PSEN1) [2] and 2 (PSEN2) [3, 4] genes cause early onset AD and enhance generation or aggregation of the amyloid β peptide [5, 6]. Simultaneously, the apolipoprotein E (APOE) ε4 allele decreases age of onset and increases disease susceptibility[7]. Several large-scale genome-wide association studies (GWAS) using dense SNP arrays identified additional susceptibility loci for LOAD: CLU, PICALM, CR1, BIN1, MS4A4A, ABCA7, CD2AP, CD33, EPHA1, HLA-DRB5/HLA-DRB1, PTK2B, SORL1, SLC24A4, DSG2, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2 and CASS4. As is the case for other complex genetic disorders, the effect sizes of these loci are small (odds ratios: 1.0–1.2). In large multiplex pedigrees with LOAD, targeted exome sequencing identified missense, nonsense and splice-site variants in APP, PSEN1, PSEN2, APOE, GRN and MAPT9. Other genes with rare variants (RVs) for LOAD include SORL1 [8], ADAM10 [9], PICALM [10], PLD3 [11] and TREM2 [12]. Overall, the majority of the genes identified to date cluster in four pathways, namely inflammation and immune response, lipid metabolism, APP generation and metabolism, and endocytosis and intracellular vesicle trafficking.
Much work lies ahead to determine the specific roles these loci play in LOAD and if so, which mutations or variants are truly causal. In addition, the identified loci only explain a fraction of the population attributable risk leaving additional genetic variants to be identified. Because large families enriched with AD cases are more likely to harbor causal variants, we performed parametric and non-parametric linkage analyses in 67 Caribbean Hispanic LOAD families with five or more affecteds genotyped individuals per family, with low frequency of APOE-ε4 (defined as families where less than half of its members were APOE-ε4 carriers), without mutations in known genes and with available genomewide genotype data. We subsequently evaluated the identified loci in the entire sample of 282 LOAD families.
MATERIALS AND METHODS
Ethics statement
Study participants were recruited as part of the EFIGA Study (Estudio Familiar de Influencia Genetica de Alzheimer).Written informed consent for the study was obtained from all subjects and/or authorized representatives and study partners. EFIGA Study was approved by the institutional review board of the New York State Psychiatric Institute.
Study samples
A set of 67 families with 469 genotyped members was selected from a total sample of 282 families with 1,060 members, to perform the initial linkage analysis. The selection criteria was based on there being a large number of affecteds in the family (an average of five or more affected individuals per family), a low APOE-ε4 allele frequency (17.6%), absence of known mutations in PSEN1, PSEN2 or APP and the availability of GWAS data. The 282 additional families (591 subjects) had lower density of affecteds per family but similar phenotyping, low APOE-ε4 allele frequency (26%), and GWAS data were available. These 282 Caribbean Hispanic families were from a family-based and case-control study of LOAD, the Estudio Familiar de Influencia Genetica de Alzheimer (EFIGA) [13].
EFIGA participants have been recruited since January 1998 from clinics in the Dominican Republic and Puerto Rico, as well as the Alzheimer Disease Research Center Memory Disorders Clinic at Columbia University in New York City. Participants are followed-up every 18 months. Each participant completed a standardized assessment at approximately 18 month intervals, including medical history, physical and neurological examination and an extensive neuropsychological battery for evaluation of cognitive impairment [14]. This battery measured cognitive function in key domains affected by aging and dementia, including memory, visuospatial ability, psychomotor speed, and executive function. The measures include the Selective Reminding Test [15], the Benton Visual Retention Test recognition and matching trials [16], the Rosen Drawing Test [17], the Boston Naming Test [18], the Controlled Oral Word Association Test [19], the Category Fluency Test [20], the Color Trails Test [21], the Similarities subtest from the Wechsler Adult Intelligence Scale [22] and the orientation items from the Mini-Mental State Examination [23]. Brief tests of writing and reading comprehension, and formal measures of reading recognition were administered including the Reading subtest of the Wide Range Achievement Test 3 [24], the Letter-Word Identification Subtest of the Woodcock-Johnson Spanish Psycho-Educational Battery [25] and the Word Accentuation Test [26]. Functional status was assessed using the Disability and Functional Limitation Instrument [27] which contains self- and observer ratings in the following areas: Instrumental activities, such as using the telephone, handling money, and completing chores; personal self-maintenance activities, such as bathing, dressing, using the toilet; perceived difficulty with memory, language, and visuospatial function, mobility, activities and social participation. The Clinical Dementia Rating Scale (CDR) [28] was completed. The diagnosis of LOAD was made at a consensus conference of physicians and neuropsychologists based on guidelines from the National Institute of Neurological and Communicative Disorders and Stroke–the Alzheimer Disease and Related Disorders Association (NINDS-ADRDA) [29].
Genotyping and quality control procedures
Genome-wide genotype data were obtained using the Illumina Human Hap 650k or Illumina 1M arrays, therefore analysis was performed using the subset of SNPs that were common to the different genetic platforms (583,002 SNPs). Single-nucleotide polymorphisms with minor allele frequencies (MAFs) less than 0.01, call rates less than 98%, or in which were not in Hardy-Weinberg equilibrium (P < 10-6 in controls) were excluded. Participants whose reported sex differed from the sex assignment determined by analysis of the X-chromosome SNPs using PLINK version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml) were excluded. Reported relationships among individuals within a family were confirmed by pairwise genome-wide estimates of IBD allele sharing. All discrepancies were reviewed with the available clinical and pedigree data to determine the most likely relationship consistent with IBD estimates.
Power analysis
To estimate the power of the set of 67 Hispanics families in detecting linkage, we performed simulations of genotypes for 100 replicates using SLINK [30, 31]. We assumed age dependent penetrance models (dominant and recessive) as described in the manuscript and a recombination fraction ranging 0.0–0.45. We considered the minor allele frequencies (MAFs) of the SNPs identified as genome-wide linked to LOAD in the 67 Hispanics families as described in Table 3. The MAFs ranged from 10% to 48% with an average MAF of 27%.
Table 3.
Chromosomal regions with HLODs>=3.6 in the 67 families’ dataset and HLODs>=2.5 in the entire sample of 282 LOAD families.
| Chr | SNP | bp | band | 67 LOAD families | 215 LOAD families | 282 LOAD families | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M0 | α | M2 | α | M0 | α | M2 | α | M0 | α | M2 | α | ||||
| 2 | rs4665809 | 26,251,361 | 2p23.3 | 2.5 | 0.50 | 3.6 | 0.88 | 0.0 | 0.11 | 0.2 | 1.00 | 2.3 | 0.40 | 3.9 | 0.88 |
| 2 | rs10173959 | 31,028,945 | 2p23.1 | 3.3 | 0.47 | 4.5 | 0.85 | 0.0 | 0.10 | 0.0 | 0.10 | 3.0 | 0.37 | 4.2 | 0.75 |
| 2 | rs2071048 | 138,721,597 | 2q22.1 | 3.4 | 0.55 | 3.7 | 0.95 | 0.1 | 0.09 | 0.3 | 0.67 | 2.8 | 0.35 | 3.9 | 0.89 |
| 3 | rs1709965 | 106,659,501 | 3q13.12 | 3.1 | 0.63 | 4.0 | 1.00 | 0.1 | 0.10 | 0.1 | 0.33 | 2.2 | 0.37 | 3.7 | 0.84 |
| 3 | rs6438213 | 114,329,961 | 3q13.31 | 4.0 | 0.73 | 4.9 | 1.00 | 0.0 | 0.05 | 0.1 | 0.41 | 2.6 | 0.43 | 4.9 | 1.00 |
| 5 | rs7700370 | 33,702,830 | 5p13.3 | 3.7 | 0.64 | 2.4 | 0.71 | 0.3 | 0.21 | 0.7 | 1.00 | 3.4 | 0.46 | 3.0 | 0.75 |
| 6 | rs9390610 | 149,113,186 | 6q25.1 | 2.9 | 0.76 | 3.7 | 1.00 | 0.0 | 0.00 | 0.0 | 0.00 | 1.1 | 0.31 | 2.9 | 1.00 |
| 6 | rs9390611 | 149,114,811 | 6q25.1 | 3.3 | 0.64 | 4.3 | 1.00 | 0.0 | 0.00 | 0.0 | 0.00 | 2.2 | 0.37 | 3.7 | 1.00 |
| 6 | rs11966489 | 157,414,029 | 6q25.3 | 3.4 | 0.70 | 4.1 | 1.00 | 0.0 | 0.00 | 0.0 | 0.00 | 1.7 | 0.34 | 2.5 | 0.79 |
| 7 | rs2240401 | 30,673,345 | 7p14.3 | 3.1 | 0.48 | 3.7 | 0.83 | 0.3 | 0.26 | 0.3 | 0.68 | 3.3 | 0.40 | 3.9 | 0.81 |
| 7 | rs10255835 | 31,044,646 | 7p14.3 | 3.5 | 0.74 | 4.7 | 1.00 | 0.2 | 0.22 | 0.2 | 0.81 | 3.1 | 0.50 | 4.9 | 1.00 |
| 7 | rs218097 | 31,497,237 | 7p14.3 | 3.2 | 0.50 | 3.8 | 0.90 | 0.0 | 0.00 | 0.0 | 0.00 | 2.3 | 0.31 | 3.1 | 0.74 |
| 7 | rs6651030 | 156,876,217 | 7q36.3 | 4.3 | 0.91 | 3.5 | 1.00 | 0.0 | 0.03 | 0.6 | 1.00 | 2.4 | 0.51 | 4.1 | 1.00 |
| 9 | rs3739553 | 129,984,107 | 9q33.3 | 3.7 | 0.89 | 3.5 | 1.00 | 0.1 | 0.14 | 0.0 | 0.24 | 2.3 | 0.53 | 3.2 | 1.00 |
| 10 | rs12360009 | 14,333,240 | 10p13 | 2.5 | 0.51 | 4.3 | 1.00 | 0.1 | 0.14 | 0.0 | 0.01 | 2.3 | 0.41 | 4.0 | 1.00 |
| 11 | rs2232932 | 61,957,137 | 11q12.3 | 2.6 | 0.89 | 3.6 | 1.00 | 1.0 | 0.51 | 1.1 | 1.00 | 3.3 | 0.69 | 4.7 | 1.00 |
M0 corresponds to affecteds only model; M2 corresponds to age-dependent penetrance model; α corresponds to the estimated proportion of families linked to the locus.
Statistical Analyses
Prior to the genome scan, we used independent SNPs markers (correlation coefficient R2=1) and with minimum allele frequencies equal or greater than 1% to examine for relationships among family members in the 67 families dataset (6,043 SNPs) and the entire set of families (5,865 SNPs) using the program PREST [32]. Based on the results, we corrected the relationships (e.g., full sibling to half-sibling), selected one from each monozygotic twin, and excluded individuals who were determined to be biologically unrelated. We then performed parametric two-point affected-only and two-point age-penetrance models for LOAD in the 67 families dataset using MERLIN (http://www.sph.umich.edu/csg/abecasis/Merlin/) setting the disease allele frequency at 0.001 and with a penetrance function of (0.01, 0.01, 0.90), and (0.01, 0.90, 0.90) for recessive and dominant models. A heterogeneity model for linkage analysis was applied to account for possible genetic heterogeneity across families since causal genes may differ among families. Subsequently, we repeated these analyses in the entire dataset of 282 families for regions with genome-wide significant evidence of linkage (HLOD =3.6) [33]. To narrow down the candidate regions, we selected SNPs showing a suggestive evidence of linkage (HLODs ≥ 2.5) [33] and performed joint linkage and association analyses using PSEUDOMARKER [34]. PSEUDOMARKER uses parametric inheritance models and exact likelihood computations to evaluate the evidence for linkage, LD, neither, or both between a putative trait locus and a set of genotyped markers providing a framework for testing hypotheses about linkage and LD. It yields statistics that are stochastically equivalent to several popular model-free methods if applied to simple family structures, for instance mother-father-child triads, case-control samples, or affected sib-pairs. PSEUDOMARKER, however, has substantial advantages over the simpler nonparametric methods when analyzing more complex family structures such as the present sample of multigenerational Caribbean Hispanic families. Adjustment for multiple testing in the joint linkage and association analysis was carried out using Bonferroni correction. The threshold for significance was established as adjusted P =0.003.
Brain expression eQTL analyses
To determine brain expression profiles of the identified candidate LOAD genes within the linkage regions, we used the publically available data from the United Kingdom Brain Expression Consortium (UKBEC) repository which is based on tissue from 12 brain regions from 134 individuals free of neurodegenerative disorders analyzed using the Affymetrix Exon 1.0 ST Array (http://www.braineac.org/). The datasets used contain the information for SNPs and indels found within 1Mb of the transcription start site of the transcript (total span of 2Mb). Markers have been restricted to those with good imputation quality (Rsq > 0.5) and minor allele frequency (MAF) ≥ 5%.
In silico gene functional interaction network
We also constructed a composite gene-gene functional interaction network to further investigate the relationships between reported LOAD susceptibility genes in GWAS meta-analysis [35] and the candidate genes we have identified. We used the publically available bioinformatic web-interface GeneMANIA. This interface uses a linear regression-based algorithm to calculate a composite functional association network from multiple networks derived from different proteomic or genomic data sources.
While several hundred genes under each of the linkage peaks were possible candidates for eQTL and functional networks analyses, we selected those only genes that were expressed in brain, highly conserved and that represented biologically plausible LOAD candidate genes.
RESULTS
Supplemental Figure 1 summarizes the analysis pipeline.
Results from quality control metrics excluded one subject on the basis of pedigree relationships discrepancies and 5,568 SNPs on the basis of call rate and deviations from Hardy-Weinberg equilibrium.
Characteristics of the family cohort are shown in Table 1. In the 67 families’ dataset, the mean age at onset in LOAD cases was 74 ± 10, the age of unaffecteds at last evaluation was 68 ± 10, and the APOE-ε4 allele frequency was 17.6%. Note that the APOE-ε4 allele frequency for cases in the original 67 families’ dataset was low because we selected these families where less than half of the members were APOE-ε4 carriers. In the 215 remaining families, the mean age at LOAD onset was 74 ± 9, the age of unaffecteds at last evaluation was 70 ± 8 and the APOE-ε4 allele frequency was 26.1%.
Table 1.
Late Onset Caribbean Hispanics family sample characteristics
| 67 Families | Additional 215 Families | |
|---|---|---|
| Families, n | 67 | 215 |
| Subjects, n | 469 | 591 |
| Affection status, n (%) | ||
| Affected | 354 (75.5) | 434(73.4) |
| Average (SD) of affected individuals per family | 5.1 (1.8) | 2.1 (0.8) |
| Unaffected | 114 (24.3) | 156(26.4) |
| Average (SD) of unaffected individuals per family | 2.2 (1.5) | 1.5 (0.9) |
| Unknown | 1 (0.2) | 1(0.2) |
| Females, n(%) | 276 (5.9) | 385(65.1) |
| Age at onset of affecteds, mean (SD) | 74 (9.6) | 74.1(9.5) |
| Age at last examination of unaffecteds, mean (SD) | 68 (9.7) | 69.6(8.3) |
| APOE allele frequency, n (%) | ||
| ε4 | 165 (17.6) | 309(26.1) |
| ε3 | 735 (78.4) | 821(69.5) |
| ε2 | 38 (4.0) | 52(4.4) |
For both dominant and recessive models, simulation results indicated that, with the linked SNP minor allele frequency of 10%, there was >80% power to detect LOD scores exceeding the Lander and Kruglyak threshold for genome-wide linkage (HLOD ≥ 3.6) [33]. If the minor allele frequency is ≥27%, the power to detect linkage increased to 100% (Supplemental Table 1).
In the initial phase, we used two-point affected-only or two-point age-dependent penetrance models. We identified 26 linkage regions with HLOD scores exceeding 3.6 (Table 2). Because the 67 families’ dataset was selected for clustering of affected subjects who were predominantly APOE-ε4 allele negative, we did not observe linkage at the APOE region of 19q13.2. The highest linkage peak (HLOD= 5.0) occurs at SNP marker rs12589681 on chromosome 14q12 (27cM, 33Mb) under the age dependent penetrance model. The 14q12 locus was located ~40Mb upstream of PSEN1 (These families screened negative for PSEN1 mutations). The linked SNP rs12589681 is an intronic variant of AKAP6 gene, an ion channel that binds to protein kinase A and anchors it to the nuclear membrane or sarcoplasmic reticulum. Interestingly, a recent report has identified two rare variants in the AKAP9 gene, in the same gene family and similar function to AKAP6 that significantly increase the risk of Alzheimer ’s disease in African-Americans [36].
Table 2.
Chromosomal regions with HLODs>=3.6 in the 67 families’ dataset.
| Chr | SNP | bp | band | HLOD_M0 | HLOD_M2 |
|---|---|---|---|---|---|
| 1 | rs17505638 | 90,388,187 | 1p22.2 | 3.7 | 2.7 |
| 2 | rs4665809 | 26,251,361 | 2p23.3 | 2.5 | 3.6 |
| 2 | rs10173959 | 31,028,945 | 2p23.1 | 3.3 | 4.5 |
| 2 | rs2071048 | 138,721,597 | 2q22.1 | 3.4 | 3.7 |
| 2 | rs12694971 | 159,700,075 | 2q24.1 | 3.6 | 2.3 |
| 2 | rs7589614 | 166,189,640 | 2q24.3 | 3.6 | 3.0 |
| 3 | rs1709965 | 106,659,501 | 3q13.12 | 3.1 | 4.0 |
| 3 | rs6438213 | 114,329,961 | 3q13.31 | 4.0 | 4.9 |
| 3 | rs1272257 | 120,216,892 | 3q13.33 | 3.3 | 3.6 |
| 3 | rs1902576 | 137,573,307 | 3q22.3 | 3.7 | 1.5 |
| 3 | rs4687347 | 192,326,678 | 3q29 | 2.5 | 3.6 |
| 5 | rs7700370 | 33,702,830 | 5p13.3 | 3.7 | 2.4 |
| 6 | rs10945192 | 70,746,249 | 6q13 | 4.0 | 2.8 |
| 6 | rs13191198 | 122,927,060 | 6q22.31 | 3.5 | 3.9 |
| 6 | rs9390610 | 149,113,186 | 6q25.1 | 2.9 | 3.7 |
| 6 | rs9390611 | 149,114,811 | 6q25.1 | 3.3 | 4.3 |
| 6 | rs11966489 | 157,414,029 | 6q25.3 | 3.4 | 4.1 |
| 7 | rs2240401 | 30,673,345 | 7p14.3 | 3.1 | 3.7 |
| 7 | rs10255835 | 31,044,646 | 7p14.3 | 3.5 | 4.7 |
| 7 | rs218097 | 31,497,237 | 7p14.3 | 3.2 | 3.8 |
| 7 | rs6464774 | 146,528,042 | 7q35 | 3.7 | 3.3 |
| 7 | rs6651030 | 156,876,217 | 7q36.3 | 4.3 | 3.5 |
| 9 | rs10982035 | 116,782,921 | 9q32 | 1.6 | 4.8 |
| 9 | rs3739553 | 129,984,107 | 9q33.3 | 3.7 | 3.5 |
| 10 | rs11258995 | 14,332,431 | 10p13 | 2.1 | 3.7 |
| 10 | rs12360009 | 14,333,240 | 10p13 | 2.5 | 4.3 |
| 11 | rs2232932 | 61,957,137 | 11q12.3 | 2.6 | 3.6 |
| 11 | rs11607134 | 131,661,985 | 11q25 | 3.7 | 2.6 |
| 14 | rs12589681 | 33,263,851 | 14q12 | 4.2 | 5.0 |
| 15 | rs3784518 | 101,854,764 | 15q26.3 | 3.7 | 2.7 |
M0 corresponds to affecteds only model; M2 corresponds to age-dependent penetrance model.
To narrow down the genomic regions putatively containing causative variants, we evaluated the SNPs found in the 67 families’ dataset by using the entire sample of 282 LOAD families. Thirteen of the identified chromosomal regions had 16 SNPs with evidence of significant linkage to LOAD in the 67 families’ dataset, and were also suggestive for linkage (HLOD= 2.5) in the entire sample of families (Table 3). To refine the signals at these thirteen loci, we also conducted additional joint linkage and association analyses (Table 4). We identified five linkage regions where SNPs showed significant association with LOAD: 11q12.3, 7p14.3, 10p13, 3q13.31 and 2p23.3. The strongest signal corresponds to chromosome 11q12.3. In the 67 families’ dataset, rs2232932 yielded a maximum HLOD of 3.6 under the age dependent penetrance model. When assessed in the entire sample of families, the maximum HLOD was 4.7 under the age dependent penetrance model. Results from the joint linkage and association analysis demonstrated that under the recessive model, rs2232932 is significantly associated with LOAD (Pjoint=6.6 × 10−6) after correction for multiple testing. The ~2Mb region encompassing the linkage peak harbors among other genes, the membrane-spanning 4A (MS4A) cluster gene, previously reported as associated with susceptibility to AD [37].
Table 4.
Results for the thirteen chromosomal candidate regions using joint linkage and association analysis in the entire sample of 282 LOAD families
| Chr | SNP | bp | band | 67 LOAD fams | 215 LOAD fams | 282 LOAD fams | |||
|---|---|---|---|---|---|---|---|---|---|
| Pjoint | Pjoint | Pjoint | |||||||
| Dom | Rec | Dom | Rec | Dom | Rec | ||||
| 2 | rs4665809 | 26,251,361 | 2p23.3 | 1.2 × 10−4 | 0.001 | 0.474 | 0.420 | 2.8 x10−4 | 0.001 |
| 2 | rs10173959 | 31,028,945 | 2p23.1 | 5.9 × 10−5 | 0.005 | 0.998 | 0.881 | 0.001 | 0.013 |
| 2 | rs2071048 | 138,721,597 | 2q22.1 | 1.7 × 10−4 | 0.151 | 0.126 | 0.111 | 0.003 | 0.039 |
| 3 | rs1709965 | 106,659,501 | 3q13.12 | 2.9 × 10−5 | 0.003 | 0.924 | 0.326 | 0.005 | 0.027 |
| 3 | rs6438213 | 114,329,961 | 3q13.31 | 7.0 × 10−6 | 3.7 × 10−5 | 0.759 | 0.779 | 2.7 x10−4 | 0.002 |
| 5 | rs7700370 | 33,702,830 | 5p13.3 | 0.001 | 0.033 | 0.695 | 0.596 | 0.014 | 0.042 |
| 6 | rs9390610 | 149,113,186 | 6q25.1 | 4.4 × 10−4 | 0.032 | 0.818 | 0.190 | 0.149 | 0.055 |
| 6 | rs9390611 | 149,114,811 | 6q25.1 | 0.001 | 0.001 | 0.650 | 0.719 | 0.019 | 0.022 |
| 6 | rs11966489 | 157,414,029 | 6q25.3 | 1.5 × 10−4 | 0.001 | 0.358 | 0.364 | 0.009 | 0.043 |
| 7 | rs2240401 | 30,673,345 | 7p14.3 | 7.0 × 10−5 | 4.3 × 10−4 | 0.096 | 0.049 | 2.8 ×10−5 | 7.4 ×10−5 |
| 7 | rs10255835 | 31,044,646 | 7p14.3 | 3.0 × 10−5 | 5.0 × 10−6 | 0.189 | 0.239 | 3 ×10−5 | 1.2 ×10−5 |
| 7 | rs218097 | 31,497,237 | 7p14.3 | 0.001 | 0.016 | 0.885 | 0.804 | 0.016 | 0.095 |
| 7 | rs6651030 | 156,876,217 | 7q36.3 | 2.0 x10−4 | 0.013 | 0.559 | 0.680 | 0.013 | 0.037 |
| 9 | rs3739553 | 129,984,107 | 9q33.3 | 4.9 × 10−4 | 0.005 | 0.359 | 0.584 | 0.005 | 0.034 |
| 10 | rs12360009 | 14,333,240 | 10p13 | 3.1 × 10−4 | 0.001 | 0.151 | 0.222 | 7.9 x10−5 | 3.4 ×10−4 |
| 11 | rs2232932 | 61,957,137 | 11q12.3 | 3.1 × 10−5 | 1.9 × 10−4 | 0.046 | 0.060 | 6.6 x10−6 | 3.9 ×10−5 |
Pjoint correspond to the P-value of the joint linkage and association analysis. Regions with significant SNPs after Bonferroni’s adjustment (P ≤ 0.003) under both models are shown in bold and italics.
Two-point linkage analysis results for the 215 and 282 families are provided in Supplemental Figure 2. Multipoint linkage results for the three sets of families (67, 215 and 282 families) are provided in Suppl. Figures 3, 4 and 5.
In the published IGAP meta-analysis of GWAS LOAD susceptibility genes [35], SNP rs983392 at MS4A6A was reported as significantly associated with LOAD. Because this SNP is not part of our genotyping platform, we selected a subset of 5 SNPs in strong linkage disequilibrium (R2 ≥ 0.8) with rs983392 and carried out joint linkage and association analysis in the entire set of 282 families. Our results showed that none of the SNP markers appear to be associated with LOAD, suggesting that there may be allelic heterogeneity at this locus between Caucasian subjects-source of IGAP and Hispanics.
We additionally investigated the SNPs identified in the joint linkage and association analysis in the IGAP meta-analysis GWAS [35] (Supplemental Table 2) and found that the strongest IGAP association corresponds to SNP rs218097 in 7p14.3 (P-value =0.002).
The 7p14.3 region also yielded strong linkage and association. In the 67 families’ dataset, rs10255835 yielded a maximum HLOD of 4.7 under the age dependent penetrance model. When assessed in the entire sample of families, the maximum HLOD was 4.9 under the age dependent penetrance model. Results from the joint linkage and association analysis demonstrated that under the recessive model, rs10255835 is significantly associated with LOAD (Pjoint=1.2 × 10−5) after correction for multiple testing. The SNP is located within a region that harbors several biological candidate genes. The SNP is located ~12Kb upstream of GHRHR the growth hormone releasing receptor, ~332Kb downstream of the transcription factor NEUROD6 and ~400Kb downstream of GARS, a gene which encodes a glycyl-tRNA synthetase.
On chromosome 10p13, an intronic variant of FMRD4A gene, SNP rs12360009, yielded significant linkage and association (HLODmax=4.3 in the 67 families’ dataset, HLODmax=4.0 in the entire family sample, both scores under the age dependent penetrance model, and Pjoint=7.9 × 10−5 under the dominant model). A genome-wide haplotype association study has reported FMRD4A gene as a risk locus for Alzheimer's disease [38].
SNP rs6438213 on 3q13.31 appeared significantly linked and associated with AD, with a HLODmax of 4.9 in the entire sample of 282 families under the age-dependent penetrance model and Pjoint=2.7 × 10−4 under the dominant model. The SNP is located in ZBTB20 gene, zinc finger gene that in mice is expressed in newborn pyramidal neurons of the hippocampus [39].
Within the 2p23.3 region, rs4665809 yielded a HLODmax=3.6 in the 67 families’ dataset, HLODmax=3.9 in the entire family sample (both scores under the age dependent penetrance model) and Pjoint=2.8 × 10−4 under the recessive model. The locus is not near a known gene, however, it is located ~5Kb upstream of RAB10 gene, a member of the RAS superfamily of small GTPases, involved in axon development in hippocampal neurons Interestingly, another cargo protein, RAB7A, showed significant evidence of association with AD [40], suggesting a direct link between the activity of the retromer complex and the pathogenesis of AD.
Results from eQTL analysis using the Brain Almanac repository (Supplemental Table 3) indicated that our LOAD candidate genes displayed brain region-specific expression patterns. As shown in Supplemental Figure 6, NEUROD6 gene appears to be highly expressed in hippocampus while the expression level of SLC30A3 gene is signifincalty higher in temporal cortex. For each of the genes, the most significant fold change in brain expression between brain regions is also shown, i.e. hippocampal expression of NEUROD6 is 11.2 fold higher than its expression in the thalamus (p=1 x10−60).
The functional network analysis using GeneMANIA confirmed the functional association between our candidate genes and several susceptibility LOAD genes (Supplemental Table 4, Supplemental Figure 7).
DISCUSSION
In a subset of multiplex Caribbean Hispanic families multiply affected by LOAD, we identified four regions with HLOD scores=3.6. In testing regions in a larger dataset of Caribbean Hispanics families we were able to generalize (HLOD =2.5) the finding and identify significant evidence for association (Pjoint =0.003).
The selection of the enriched families as the initial cohort for linkage analysis confirmed their utility to identify genetic variants. As the alpha values demonstrated, more than 80% of the families in the 67 families’ dataset were linked to the identified loci, while the remaining set of 215 families appeared to be more heterogeneous with the proportion of families linked to the locus ranging from 14% to 51 % (Supplemental Table 5), except for SNP at 2p23.3 that in both families’ datasets reaches alpha values of 100%.
The strongest signal corresponds to SNP rs2232932 at chromosome 11q12.3 with a HLODmax=4.7 in the entire sample of 282 families and strong evidence for joint linkage and association (Pjoint= 6.6 x10−6). The SNP is located 1.5Mb upstream of MS4A4E gene. An increased risk of developing Alzheimer’s Disease has previously been found to be associated with several variants within membrane-spanning 4-domains subfamily A (MS4A) gene cluster[37]. As cell membrane proteins, MS4A family members are found to participate in the regulation of calcium signaling which have been widely discussed in neurodegeneration and AD[41]. An important role in immunity has already been identified for several members of this cluster indicating the possible involvement of MS4A gene cluster in AD pathogenesis [42].
SNP rs10255835 at 7p14.3 (HLODmax=4.9 in the entire sample of 282 LOAD families and Pjoint=1.2 × 10−5) lies in a region harboring several genes of biological relevance. Approximately 12Kb upstream is the gene encoding the growth hormone releasing receptor, GHRHR. It has been reported that neuroendocrine mechanisms play a role in LOAD development [43]. The region also harbors helix-loop transcription factor NEUROD6 that is involved in the development and differentiation of the nervous system. It has been previously reported that in human AD brains, NEUROD6 expression levels were significantly altered, i.e., significantly downregulated in hippocampus [44]. Finally, the 7p14 region encompasses GARS gene, a glycyl-tRNA synthetase that catalyzes the attachment of glycine to tRNA. Several GARS mutations have been identified in individuals with distal hereditary motor neuropathy type V and in individuals with a form of Charcot-Marie-Tooth disease [45]. A reduction in glycyl-tRNA synthetase activity may impair the ability of axons to transmit nerve impulses. Whether these genes are involved in LOAD or simply lie in a region that contains another, as yet unidentified gene remains to be determined. Remarkably, 7p14.1 region has also been reported in a meta-analysis of a sample of 2,206 LOAD affected individuals from Caucasian and Caribbean Hispanic ancestry families [46].
We replicated the association of FMRD4A gene on 10p13 and AD, intronic SNP rs12360009 yielded strong evidence of both linkage and association (Pjoint=7.9 x10−5). Genome-wide haplotype analysis identified FMRD4A as consistently associated with AD risk in seven independent European populations. Interestingly, this locus is included in the large AD linkage region regularly identified on chromosome 10 [38]. FRMD4A belongs to the FERM super family, which includes ubiquitous components of the cytocortex involved in cell structure, transport and signaling functions. FRMD4A gene product interacts with Arf6 which was recently reported to control APP processing[47] suggesting that FRMD4A could also be implied in this metabolism. This hypothesis is sustained by the observation of an association of the FRMD4A locus with plasma Aβ1–42/Aβ1–40, at once reinforcing the plausibility of the association of this gene with AD risk and its potential implication in a subtle control of the APP metabolism.
The SNP rs6438213 at 3q13.31 is an intronic variant of ZBTB20 gene, a zinc finger transcription factor that has been associated with hippocampal function. In mice, ZBTB20 is expressed in newborn pyramidal neurons of the hippocampus and it is implicated in the regulation of cortical neurogenesis [39]. Gene expression studies in human brain samples showed that is highly expressed in the hippocampal, cerebellum and white matter regions of the brain and suggest that ZBTB20 plays an essential role in the development and regionalization of the human hippocampus [48].
SNP rs4665809 within the 2p23.3 region maps ~ 5Kb upstream of RAB10 gene, a member of to the RAS superfamily of small GTPases. They GTPases are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. In neurons, it is involved in axonogenesis through regulation of vesicular membrane trafficking toward the axonal plasma membrane. The 2p23.3 region also harbors SLC30A3 gene, a synaptic vesicle transporter responsible for neuronal export of zinc into the synaptic space. In a transgenic mouse model of AD, a dramatic reduction of cerebral amyloid angiopathy occurs after targeted disruption of SLC30A3 [49]. It has also been recently reported that its activity is critical for synaptic targeting of Aβ oligomers [50]. Several previous genome screens identified 2p23–24 as candidate region contributing to LOAD risk [51, 52].
Researchers in accompanying study in non-Hispanic Caucasian LOAD families found strong evidence for linkage and association in 11q12.3 locus. To further investigate the region in the 67 families dataset, we run parametric multipoint analysis in the 4Mb/2cM region encompassing the linkage peak for a 1-cM grid using MERLIN. Multipoint HLOD scores were lower than the two-point scores and the low alpha values suggested high genetic heterogeneity. When family specific LOD scores were computed within each of the regions, the largest scores were achieved by a small number of families segregating with these loci.
Linkage analysis of the 67 families under the age-dependent penetrance model (M2), yielded seven loci out of the total thirteen that showed higher HLOD scores after the addition of the 215 family set, i.e., the HLOD for SNP rs2232932 at 11q12.3 increased from 3.6 to 4.7. If we consider the affected only analysis model (M0), two of the thirteen loci increased their evidence for linkage after the addition of the 215 families. Our findings suggest that the linkage evidence at these loci is mostly driven by the initial set of 67 families and the addition of 215 families supports the evidence of linkage in some of these loci. As previously reported, methods were LOD score is maximized over disease model are more powerful compared with affecteds-only methods [53], therefore is expected that confirmation of the linkage signals is stronger under M2 model.
The decrease in HLOD scores observed for some of the linkage signals after the addition of the 215 families suggest allelic/locus heterogeneity in these families. The main source of heterogeneity is our selection criteria for the families. The initial 67 Hispanics families were selected because they had multiple LOAD cases, did not carry known mutations in established LOAD genes and had low allele frequency of APOE-ε4. As previously reported, ascertainment of highly selected extended families potentially increases homogeneity and therefore improve the power to detect linkage [54]. Moreover, these families contain significant more linkage information. Previous studies have demonstrated that small sample sizes can identify genomic regions of interest [55]. As an example, the genetic analysis that demonstrated LOAD linkage to chromosome 19, and subsequently permitted the identification of APOE locus as a major susceptibility locus, was conducted using only 32 pedigrees [56]. These criteria were not applied to the additional set of 215 families, these families were selected on the basis of at least two affected individuals with genetic data available.
In brief, sources of heterogeneity are: i) the size of the families (the average pedigree size in the 67 families was 11 individuals versus six in the 215 families), ii) the number of affected individuals per family (the average number of affected members in the 67 families was five with AD versus two with AD in the 215 families) (Table 1) and iii) the selection was also not based on APOE-ε4 (the frequency of APOE-ε4 allele was 18% in the 67 families versus a frequency of 26% in the 215 families) (Table 1).
Analysis of SNPs in strong linkage disequilibrium with IGAP LOAD susceptibility loci previously reported at MS4A4E suggested allelic heterogeneity for this locus when Caucasian and Caribbean Hispanic populations were compared. However, several of the identified SNPs in the other chromosomal regions show nominally significant results, suggesting that some of the identified loci may be generalized to Caucasian populations.
We observed suggestive, but not definitive region-specific patterns of brain expression of the LOAD candidate genes when analyzing publically available eQTL datasets in human brain. Moreover, in silico analysis of potential functional networks further support their interaction with several reported susceptibility LOAD genes.
Taken together, these linkage analyses in multiplex Caribbean Hispanic families identified several reported and novel regions with suggestive linkage to LOAD that contain candidate genes, some of which are biologically highly plausible. However, SNPs that influence cis-acting regulatory elements often do not influence the expression of the closest gene, and the true LOAD-related gene may be several genes up or downstream. Follow-up sequence analyses in these families should help to identify rare causative sequence variants in these regions shared within or across sequenced families.
The 67 families we described and analyzed in this manuscript are part of the national Alzheimer’s Disease Sequencing Project (ADSP). Along with the non-Hispanic White families reported in the companion paper (Kunkle et al), they are undergoing whole-genome sequencing (WGS). The linkage regions identified in this manuscript will be prioritized in the ADSP analysis, that is, the WGS data will be used to identify exonic and regulatory variants under these linkage peaks and subsequently investigate the role of these rare variants in the susceptibility to LOAD. To further identify causative/protective variants segregating with LOAD in these linkage regions, additional analysis of the WGS data will be carry out as part of the ADSP project, i.e., joint linkage and association analyses, gene based analyses.
Alzheimer’s Disease Sequencing Project (ADSP) data sharing
All data from the analyses in this manuscript, including quality control documentation, GWAS array data and phenotype data for each family, and linkage analyses results, is available for download at The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site’s ADSP website (https://www.niagads.org/adsp/content/home). Applicants must submit a data access request to dbGaP. Applications are reviewed by the ADSP Data Access Committee (DAC) and the NIAGADS Data Use Committee (DUC). ADSP phenotype and sequence data are made available to the research community at large in keeping with the NIH Genomics Data Sharing Policy http://gds.nih.gov/. NIA has established the National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS) as a national genetics data repository in order to facilitate access by qualified investigators to genotypic and phenotypic data for the study of the genetics of late-onset Alzheimer's disease. NIAGADS is working in partnership with dbGaP (ADSP at dbGaP) to provide ADSP data to the research community. Data can be requested either from dbGaP or NIAGADS. Instructions for application for ADSP data and an explanation of the review process can be found at: ADSP at dbGaP and NIAGADS ADSP Application Instructions.
The ADSP has in place a memorandum of understanding https://www.niagads.org/sites/all/public_files/ADSPdocs/ADSP-MOU.pdf. In the spirit of the clear benefit that ensues from converting such data sets into community resources as rapidly as possible, and in keeping with community expectations for the use of unpublished genome sequence data, it is expected for the first phase of the study called the Discovery Phase, that users of the data will withhold publication until the producers of the data have published their findings. ADSP participants will publish their data in an expeditious fashion in at least one major paper reporting the results of the ADSP to be jointly submitted by all of the members.
Supplementary Material
Suppl. Table 1 . Power calculations for detecting linkage in the 67 Caribbean Hispanic families
Suppl. Table 2. IGAP results for the identified LOAD SNPs
Suppl. Table 3. Characteristics of the BRAINEAC dataset used for the LOAD candidate genes.
Suppl. Table 4. GeneMANIA functional interactions of the LOAD candidate genes and IGAP GWAS susceptibility LOAD loci
Suppl. Table 5. HLODs and alpha values for the chromosomal regions identified in the 67 families’ dataset and in the remaining 215 families
Supplemental Figure 1. Flow analysis chart
Supplemental Figure 2. Two-point genome-wide linkage plots for the sets of 215 and 282 Hispanics families. HLODs were computed under the affected only model (M0) and age dependent penetrance model (M2). A) two-point HLODs for the 215 fams under the affected only model; B) two-point HLODs for the 215 fams under the age dependent penetrance model; C) two-point HLODs for the 282 fams under the affected only model; D) two-point HLODs for the 282 fams under the age dependent penetrance model.
Supplemental Figure 3. Multi-point genome-wide linkage plots for the set of 67 Hispanics families
Supplemental Figure 4. Multi-point genome-wide linkage plots for the set of 215 Hispanics families
Supplemental Figure 5. Multi-point genome-wide linkage plots for the set of 282 Hispanics families
Supplemental Figure 6. BRAINEAC brain expression profiles for the LOAD candidate genes
Supplemental Figure 7. GeneMANIA functional network for LOAD candidate genes and IGAP GWAS LOAD susceptibility genes
Supplemental Figure 8. Principal component analysis results for the set of 215 Hispanic families. The figure shows the plots for three first principal components. Each solid circle represents an individual. a) samples are represented on principal components 1 and 2 estimated on the genotype data; b) samples are represented on principal components 1 and 3; c) samples are represented on principal components 2 and 3. In all three comparisons between the estimated principal components, the majority of the family members belong to a single cluster, demonstrating their homogeneity in terms of ethnic background.
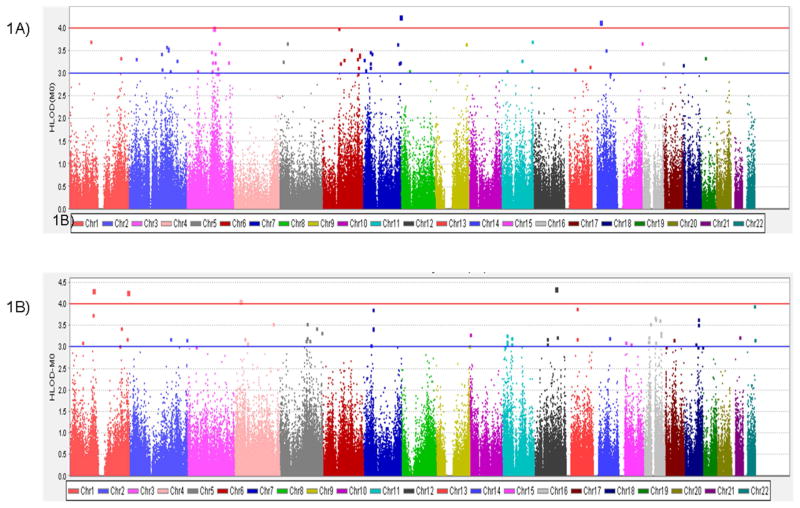
Figure 1.
Plot of the two-point linkage analysis in the 67 Caribbean Hispanics LOAD families. 1A) HLODs under the affected only model; 1B) HLODs under the age dependent penetrance model.
RESEARCH CONTEXT.
Genome-wide methods have led to the identification of risk loci for Late-Onset Alzheimer’s disease (LOAD). However, the loci explain a small fraction of LOAD heritability and the causal variants remain unidentified. Linkage analysis of large pedigrees with multiple affecteds is a powerful approach to identify genomic regions containing rare variants. Genome-wide scan of 67 Caribbean Hispanic families with multiple LOAD cases selected from an overall sample of 282 families identified 26 chromosomal regions with significant evidence of linkage. Thirteen of the regions were validated in the overall sample. Joint linkage and association identified locus on 11q12.3 located ~2Mb upstream of MS4A gene, a LOAD susceptibility gene (rs2232932, Pjoint= 6.6 × 10−6). Variants in 7p14.3 were associated with LOAD (rs10255835, Pjoint= 1.2 × 10−5), a region harboring genes implicated in development and differentiation of nervous system. Future sequencing efforts should focus on regions harboring rare variants contributing to familial LOAD risk.
Acknowledgments
This work was supported by National Institute of Health grants R37AG015473, UO1AG032984 and 5P50AG008702-25. Dr. Reitz was further supported by a Paul B. Beeson Career Development Award K23AG034550. Dr. Lindsay Farrer was supported by NIH grants R01-AG025259 and P30-AG13846. The funding sources had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The funding source had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
CONFLICT OF INTEREST
None of the authors has a conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 2.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–60. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 3.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–7. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 4.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–8. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 5.Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, Aulchenko Y, et al. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77:643–52. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St George-Hyslop PH, Myers RH, Haines JL, Farrer LA, Tanzi RE, Abe K, et al. Familial Alzheimer's disease: progress and problems. Neurobiol Aging. 1989;10:417–25. doi: 10.1016/0197-4580(89)90082-1. [DOI] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–9. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- 9.Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, et al. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet. 2009;18:3987–96. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnetz-Boutaud NC, Hoffman J, Coe JE, Murdock DG, Pericak-Vance MA, Haines JL. Identification and confirmation of an exonic splicing enhancer variation in exon 5 of the Alzheimer disease associated PICALM gene. Ann Hum Genet. 2012;76:448–53. doi: 10.1111/j.1469-1809.2012.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014;505:550–4. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romas SN, Santana V, Williamson J, Ciappa A, Lee JH, Rondon HZ, et al. Familial Alzheimer disease among Caribbean Hispanics: a reexamination of its association with APOE. Arch Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 14.Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, et al. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49:453–60. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- 15.Buschke H, Fuld PA. Evaluating storage, retention, and retrieval in disordered memory and learning. Neurology. 1974;24:1019–25. doi: 10.1212/wnl.24.11.1019. [DOI] [PubMed] [Google Scholar]
- 16.Benton AL. Development of finger-localization capacity in school children. Child Dev. 1955;26:225–30. [PubMed] [Google Scholar]
- 17.Rosen W. The Rosen Drawing Test. Veterans Administration Medical Center; Bronx, NY: 1981. [Google Scholar]
- 18.Kaplan E, Goodglass H, Weintraub S. Boston Naming Test. Lea & Febiger; Philadelphia, PA: 1983. [Google Scholar]
- 19.Benton A, Hamsher K. Multilingual Aphasia Examination. 1983 [Google Scholar]
- 20.Goodglass H, Kaplan E. The assesment of Aphasia and Related Disorders. Lea & Febiger; Philadelphia, PA: 1983. [Google Scholar]
- 21.D'Elia L, Satz P, Uchiyama C, White T. Color Trails Test Professional manual. Psychological Assesment Resources; Odessa, FL: 1994. [Google Scholar]
- 22.Wechsler D. Weschler Adult Intelligence Scale-Revised. The Psychological Corporation; New York, NY: 1981. [Google Scholar]
- 23.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson G. Wide Range Achievement Test 3- Administration Manual. Jastak Associates, Inc; Wilimington ,DE: 1993. [Google Scholar]
- 25.Woodcock R, Jonhson M. Woodcock & Jonhson Psycho-Educational Battery. DLM Teaching Resources; Allen, TX: 1977. [Google Scholar]
- 26.Del Ser T, Gonzalez-Montalvo JI, Martinez-Espinosa S, Delgado-Villapalos C, Bermejo F. Estimation of premorbid intelligence in Spanish people with the Word Accentuation Test and its application to the diagnosis of dementia. Brain Cogn. 1997;33:343–56. doi: 10.1006/brcg.1997.0877. [DOI] [PubMed] [Google Scholar]
- 27.Golden RR, Teresi JA, Gurland BJ. Development of indicator scales for the Comprehensive Assessment and Referral Evaluation (CARE) interview schedule. J Gerontol. 1984;39:138–46. doi: 10.1093/geronj/39.2.138. [DOI] [PubMed] [Google Scholar]
- 28.Morris JC, Edland S, Clark C, Galasko D, Koss E, Mohs R, et al. The consortium to establish a registry for Alzheimer's disease (CERAD). Part IV. Rates of cognitive change in the longitudinal assessment of probable Alzheimer's disease. Neurology. 1993;43:2457–65. doi: 10.1212/wnl.43.12.2457. [DOI] [PubMed] [Google Scholar]
- 29.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 30.Ott J. Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A. 1989;86:4175–8. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer AA, Lemire M, Ott J, Lathrop GM, Weeks DE. Coordinated conditional simulation with SLINK and SUP of many markers linked or associated to a trait in large pedigrees. Hum Hered. 2011;71:126–34. doi: 10.1159/000324177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–94. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 34.Gertz EM, Hiekkalinna T, Digabel SL, Audet C, Terwilliger JD, Schaffer AA. PSEUDOMARKER 2.0: efficient computation of likelihoods using NOMAD. BMC Bioinformatics. 2014;15:47. doi: 10.1186/1471-2105-15-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logue MW, Schu M, Vardarajan BN, Farrell J, Bennett DA, Buxbaum JD, et al. Two rare AKAP9 variants are associated with Alzheimer's disease in African Americans. Alzheimers Dement. 2014 doi: 10.1016/j.jalz.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert JC, Grenier-Boley B, Harold D, Zelenika D, Chouraki V, Kamatani Y, et al. Genome-wide haplotype association study identifies the FRMD4A gene as a risk locus for Alzheimer's disease. Mol Psychiatry. 2013;18:461–70. doi: 10.1038/mp.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development. 2007;134:1133–40. doi: 10.1242/dev.000265. [DOI] [PubMed] [Google Scholar]
- 40.Vardarajan BN, Bruesegem SY, Harbour ME, Inzelberg R, Friedland R, St George-Hyslop P, et al. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33:2231 e15–e30. doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermes M, Eichhoff G, Garaschuk O. Intracellular calcium signalling in Alzheimer's disease. J Cell Mol Med. 2010;14:30–41. doi: 10.1111/j.1582-4934.2009.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuccolo J, Deng L, Unruh TL, Sanyal R, Bau JA, Storek J, et al. Expression of MS4A and TMEM176 Genes in Human B Lymphocytes. Front Immunol. 2013;4:195. doi: 10.3389/fimmu.2013.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaszberenyi M, Rick FG, Szalontay L, Block NL, Zarandi M, Cai RZ, et al. Beneficial effects of novel antagonists of GHRH in different models of Alzheimer's disease. Aging (Albany NY) 2012;4:755–67. doi: 10.18632/aging.100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hokama M, Oka S, Leon J, Ninomiya T, Honda H, Sasaki K, et al. Altered expression of diabetes-related genes in Alzheimer's disease brains: the hisayama study. Cereb Cortex. 2014;24:2476–88. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pericak-Vance MA, Speer MC, Lennon F, West SG, Menold MM, Stajich JM, et al. Confirmation of a second locus for CMT2 and evidence for additional genetic heterogeneity. Neurogenetics. 1997;1:89–93. doi: 10.1007/s100480050013. [DOI] [PubMed] [Google Scholar]
- 46.Butler AW, Ng MY, Hamshere ML, Forabosco P, Wroe R, Al-Chalabi A, et al. Meta-analysis of linkage studies for Alzheimer's disease--a web resource. Neurobiol Aging. 2009;30:1037–47. doi: 10.1016/j.neurobiolaging.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Tepass U. FERM proteins in animal morphogenesis. Curr Opin Genet Dev. 2009;19:357–67. doi: 10.1016/j.gde.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Davies MN, Krause L, Bell JT, Gao F, Ward KJ, Wu H, et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 2014;15:R56. doi: 10.1186/gb-2014-15-4-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedlich AL, Lee JY, van Groen T, Cherny RA, Volitakis I, Cole TB, et al. Neuronal zinc exchange with the blood vessel wall promotes cerebral amyloid angiopathy in an animal model of Alzheimer's disease. J Neurosci. 2004;24:3453–9. doi: 10.1523/JNEUROSCI.0297-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshpande A, Kawai H, Metherate R, Glabe CG, Busciglio J. A role for synaptic zinc in activity-dependent Abeta oligomer formation and accumulation at excitatory synapses. J Neurosci. 2009;29:4004–15. doi: 10.1523/JNEUROSCI.5980-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–22. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 52.Hiltunen M, Mannermaa A, Thompson D, Easton D, Pirskanen M, Helisalmi S, et al. Genome-wide linkage disequilibrium mapping of late-onset Alzheimer's disease in Finland. Neurology. 2001;57:1663–8. doi: 10.1212/wnl.57.9.1663. [DOI] [PubMed] [Google Scholar]
- 53.Abreu PC, Greenberg DA, Hodge SE. Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet. 1999;65:847–57. doi: 10.1086/302536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger JD, Goring HH. Gene mapping in the 20th and 21st centuries: statistical methods, data analysis, and experimental design. Hum Biol. 2000;72:63–132. [PubMed] [Google Scholar]
- 55.Baron M. Optimal ascertainment strategies to detect linkage to common disease alleles. Am J Hum Genet. 1999;64:1243–8. doi: 10.1086/302336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pericak-Vance MA, Bebout JL, Gaskell PC, Jr, Yamaoka LH, Hung WY, Alberts MJ, et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48:1034–50. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table 1 . Power calculations for detecting linkage in the 67 Caribbean Hispanic families
Suppl. Table 2. IGAP results for the identified LOAD SNPs
Suppl. Table 3. Characteristics of the BRAINEAC dataset used for the LOAD candidate genes.
Suppl. Table 4. GeneMANIA functional interactions of the LOAD candidate genes and IGAP GWAS susceptibility LOAD loci
Suppl. Table 5. HLODs and alpha values for the chromosomal regions identified in the 67 families’ dataset and in the remaining 215 families
Supplemental Figure 1. Flow analysis chart
Supplemental Figure 2. Two-point genome-wide linkage plots for the sets of 215 and 282 Hispanics families. HLODs were computed under the affected only model (M0) and age dependent penetrance model (M2). A) two-point HLODs for the 215 fams under the affected only model; B) two-point HLODs for the 215 fams under the age dependent penetrance model; C) two-point HLODs for the 282 fams under the affected only model; D) two-point HLODs for the 282 fams under the age dependent penetrance model.
Supplemental Figure 3. Multi-point genome-wide linkage plots for the set of 67 Hispanics families
Supplemental Figure 4. Multi-point genome-wide linkage plots for the set of 215 Hispanics families
Supplemental Figure 5. Multi-point genome-wide linkage plots for the set of 282 Hispanics families
Supplemental Figure 6. BRAINEAC brain expression profiles for the LOAD candidate genes
Supplemental Figure 7. GeneMANIA functional network for LOAD candidate genes and IGAP GWAS LOAD susceptibility genes
Supplemental Figure 8. Principal component analysis results for the set of 215 Hispanic families. The figure shows the plots for three first principal components. Each solid circle represents an individual. a) samples are represented on principal components 1 and 2 estimated on the genotype data; b) samples are represented on principal components 1 and 3; c) samples are represented on principal components 2 and 3. In all three comparisons between the estimated principal components, the majority of the family members belong to a single cluster, demonstrating their homogeneity in terms of ethnic background.