Abstract
Objective
Engagement of signaling lymphocytic activation molecule family member 4 (SLAMF4, CD244, 2B4) by its ligand SLAMF2 (CD48) modulates function and expansion of both NK cells and a subset of cytotoxic CD8+ T cells. As the cytotoxicity of CD8+ T lymphocytes isolated from systemic lupus erythematosus (SLE) patients is known to be impaired, we assess here whether expression and function of the checkpoint regulator SLAMF4 is altered on SLE CD8+ T cells.
Methods
Expression of SLAMF4 by healthy and SLE T cells was determined by Q-PCR and flow cytometry. T cells were activated with anti-CD3 antibody and degranulation activity was monitored by the surface expression of LAMP-1 (CD107a). The SLAMF4+ and SLAMF4− CD8 T cell subpopulations were characterized by LAMP-1, perforin and granzyme B expression and viral peptide-induced proliferation.
Results
SLAMF4 gene and surface protein expression is downregulated in CD8+ T cells from SLE patients as compared to cells obtained from healthy donors. Importantly, SLE patients have significantly fewer SLAMF4+ CD8+ T cells compared to healthy subjects. SLAMF4− CD8+ T cells from SLE patients have a decreased cytotoxic capacity and proliferative responses to viral peptides. The loss of memory SLAMF4+ CD8+ T cells in SLE patients is linked to the fact that they lose CD8 expression and become double negative T cells.
Conclusion
A selective loss of SLAMF4+ CD8+ T cells contributes to the compromised ability of SLE T cells to fight against infections.
Genetic, environmental as well as hormonal and immunoregulatory factors contribute to the pathogenesis and clinical manifestations of systemic lupus erythematosus (SLE) (1). CD4+ T cells are the main drivers of the B cell-dependent autoantibody response in lupus (2) and display molecular and biochemical abnormalities, which account for their aberrant function (3). However, the role of CD8+ T cells in autoimmunity have been less well understood despite the fact that their cytotoxic function is known to be compromised for a long time and considered to contribute to the increased infection rates among patients with SLE (4–6).
The signaling lymphocytic activation molecule family member 4 (SLAMF4, CD244, 2B4) is expressed on the surface of human natural killer (NK) cells, γδ T cells, basophils, monocytes and a subset of effector memory CD8+ αβ T cells (7, 8). SLAMF4 is a type I trans-membrane glycoprotein. The extracellular region of SLAMF4 is comprised of an N-terminal V-Ig and a C-terminal C2-Ig domain, whereas the cytoplasmic tail of SLAMF4 contains 4 intracellular tyrosine switch motifs (ITSM). Although most SLAM family receptors engage in homotypic interactions, SLAMF4 interacts with high affinity with SLAMF2 (CD48). Upon SLAMF4-SLAMF2 interaction, the SLAM-associated protein (SAP, SH2D1A), a small Src homology 2-domain containing adaptor molecule, is recruited to the ITSMs in the cytoplasmic region of SLAMF4 and mediates downstream signaling (9, 10). Engagement of SLAMF4 can either promote or restrain NK and CD8+ T cell function (reviewed in (11)).
The expression of SLAMF4 on CD8+ T cells correlates with T cell activation, cytotoxic T lymphocyte differentiation and exhaustion (7, 12, 13). SLAMF4+ CD8+ T cells do not express CD62L, CD28 and CCR7 but they produce perforin, granzyme B and IFN-γ (7, 12, 14). At the clinical level, SLAMF4 and the adapter protein SAP have been described increased in CD8+ T cells from HTLV-I-infected patients with neurologic manifestations (15). Expansion of cytotoxic CD8+ T cells has been documented in SLE patients in correlation with disease activity (16, 17). A splice variant of SLAMF4 has been reported to be preferentially expressed in peripheral blood mononuclear cells from patients with SLE (18). Moreover, a single nucleotide polymorphism of the SLAMF4 has been associated with the presence of renal and neuropsychiatric lupus manifestations (19). The percentage of SLAMF4-expressing NK cells and monocytes are reduced in patients with SLE compared to healthy controls (18, 20). Also, IL-7Rαlow memory CD8+ T cells have been reported to be increased in patients with SLE and to express higher levels of SLAMF4 compared with IL-7Rαhigh memory CD8+ T cells; engagement of SLAMF4 enhanced cytotoxic function of IL-7Rαlow EM CD8+ T cells against target cells (21).
We report here that SLE patients have significantly fewer SLAMF4+ CD8+ T cells compared to healthy donor T cells with decreased SAP expression and impaired cytotoxic activity. The selective loss of SLAMF4+ CD8+ T cells may explain the decreased cytotoxic cell responses in patients with SLE and the increased rate of infections.
Materials and methods
Human subjects, T cell isolation and treatment
Healthy donors and patients fulfilling the American College of Rheumatology-established criteria for the diagnosis of SLE were included. The disease activity was measured using the SLE Disease activity index (SLEDAI). The study was approved by the Institutional Review Board of Beth Israel Deaconess Medical Center. Peripheral blood mononuclear cells (PBMC) were isolated using gradient centrifugation. Total T cells were isolated by negative selection using the RosetteSep Kit (Stem Cell Technologies, Vancouver, Canada). T cell purity was always ≥95%. CD8+ T cells were purified using the Human CD8 T Cell Isolation Kit from Miltenyi Biotec (Auburn, CA) according to the manufacturer’s instructions (purity ≥93%). SLAMF4− and SLAMF4+ CD8+ T cells were sorted by Aria (BD Biosciences, San Jose, CA). Cells were cultured in RPMI medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum and antibiotics. Activation was achieved using immobilized anti-CD3 (0.5 µg/ml) and anti-CD28 (5 µg/ml) antibodies (both purchased from BioXcell). Viral peptide activation of CD8+ T cells used PBMCs incubated with was achieved by treating healthy and SLE PBMCs with a mix of CMV, EBV and influenza peptides (CEF) at 2 µg/ml final concentration (optimal dose was determined in preliminary experiments – data not shown) for 6 days in the presence of 50 U/ml IL-2 (PeproTech, Rocky Hill, NJ) and cell proliferation was determined measuring carboxyfluorescein succinimidyl ester (CFSE) dilution by flow cytometry.
Flow cytometry
The phenotype of T cell subpopulations was monitored by flow cytometric analysis using fluorochrome-conjugated antibodies against CCR7, CD45RA, SLAMF4, perforin, granzyme B, interferon γ as well as isotype-matched control antibodies (Biolegend, San Diego, CA). To check cell viability Zombie UV Fixable Viability Kit was used and the positive cells were excluded as dead (Biolegend). To quantify lytic granule exocytosis we used CD107a/LAMP-1 staining (Biolegend). Surface expression of CD107a reflects the exocytosis of lytic granules. The proliferative capacity of CEF peptide mix-treated CD8+ T cells was monitored by CFSE dilution (Sigma Aldrich, St. Louis, MO). Fluorescence intensities were measured and analyzed by LSR II SORP flow cytometer (BD Biosciences Immunocytometry Systems, Franklin Lakes, NJ). Data analysis was performed using FlowJo version X.0.7 (Ashland, OR). Analysis and presentation of distributions was performed using SPICE version 5.1, downloaded from http://exon.niaid.nih.gov (22).
Real-time quantitative reverse transcriptase-polymerase chain reaction (Q-RT-PCR)
Total RNA was isolated from T cells by RNeasy Mini Kit (Qiagen, Valencia, CA). Reverse transcription was performed at 37 °C for 120 min from 100 ng total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCRs for SLAMF4, SAP, perforin and granzyme B genes were performed (Light Cycler 480, Roche, Indianapolis, IN) with 40 cycles at 94 °C for 12 sec and 60 °C for 60 sec using Taqman assays (Applied Biosystems). All PCR reactions were run in triplicates with a control reaction containing no RT enzyme. The comparative Ct method was used to quantify transcripts relative to the endogenous control gene large ribosomal protein.
Western blotting
Total protein was extracted from sorted SLAMF4− and SLAMF4+ T cells using RIPA buffer (Boston Bioproducts, Ashland, MA) supplemented with protease inhibitors (5 mM sodium fluoride, 4 mM sodium vanadate, aprotinin, leupeptin, 1 mM dithiothreitol and 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride). Equal amounts of proteins (15 µg) were resolved on a 4–12% Bis-Tris NuPAGE precast gel at 200 V for 1h and transferred to PVDF membrane at 30 V for 2h, blocked in 5% non-fat milk in Tris-buffered saline with Tween 20 (TBS-T) for 3h, incubated with primary anti-SAP or anti-β-Actin antibody (Santa Cruz, Dallas, TX) overnight, washed three times with TBS-T, incubated with HRP-conjugated goat anti-mouse or goat anti-rat secondary antibody for 1h, washed three times with TBS-T buffer, incubated for 5 min with ECL reagent (GE Healthcare, Piscataway, NJ) and visualized by Fuji LAS-3000 scanner.
Statistical analysis
Paired Student one- and two-tailed t tests were used (* p≤0.05, ** p≤0.01, *** p≤0.005). For all experiments, the mean and the standard deviation (SD) are reported for at least n=3 independent experiments. Statistical comparison among T cell subset distributions was performed using χ2 tests in SPICE 5.1.
Results
Patients with SLE display decreased levels of SLAMF4 and reduced percentage of SLAMF4+ CD8+ T cells compared to healthy donors
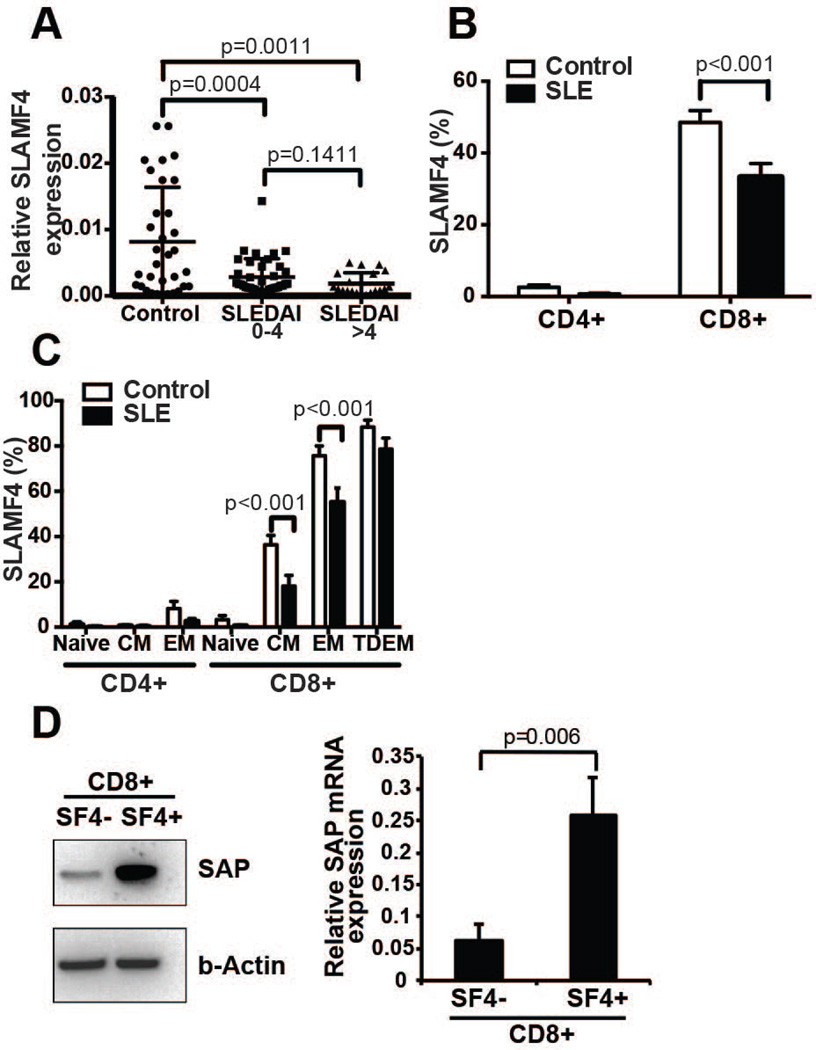
Total T cells were isolated from the peripheral blood of healthy subjects and patients with SLE and the SLAMF4 mRNA expression was measured by real-time quantitative PCR. SLE T cells expressed decreased levels of SLAMF4 mRNA compared to healthy T cells. Although there was a tendency towards lower SLAMF4 mRNA expression in T cells from patients with more severe (SLEDAI >4) as compared to patients with inactive (SLEDAI 0–4) disease, the difference did not reach statistical significance (Figure 1A). SLAMF4 is mainly expressed on CD8+ T cells and the percentage of SLE CD8+ T cells expressing SLAMF4 molecules on their cell surface was significantly lower compared to healthy individuals, measured by flow cytometry (Figure 1B). The expression of SLAMF4 was assessed within each differentiated CD8+ T cells subsets i.e. naïve (CCR7+ CD45RA+), central memory (CCR7+ CD45RA−), effector memory (CCR7− CD45RA−) and terminally differentiated effector memory (CCR7− CD45RA+) populations. Central memory (CCR7+ CD45RA−) and effector memory (CCR7− CD45RA−) CD8+ T cell populations from SLE patients displayed significantly less SLAMF4 expression compared to controls (Figure 1C). Western blot analysis of SLAMF4− CD8+ cell lysates from normal donors revealed reduced SAP expression compared to lysates of SLAMF4+ CD8+ T cells (Figure 1D). Moreover, SLAMF4− CD8+ displayed less SAP mRNA expression compared to SLAMF4+ CD8+ T cells (Figure 1D).
Figure 1. SLE T cells have decreased expression of SLAMF4 and the SLAM adaptor molecule SAP.
mRNA levels of SLAMF4 in total T cells of controls and SLE patients measured by Q-PCR (A). Cell surface protein expression of SLAMF4 on different subpopulations of CD4+ and CD8+ T cells in controls vs. SLE patients measured by flow cytometry showing a significant decrease in SLAMF4+ CD8+ T cells in SLE (B, C) (n=10 controls and 10 SLE samples; gated on total T cells; CM: central memory, EM: effector memory, TDEM: terminally differentiated effector memory T cells). Western blot (one representative example) and gene expression (n=3) of sorted SLAMF4− and SLAMF4+ CD8+ T cells show significant downregulation of SAP levels in SLAMF4− T cells (D).
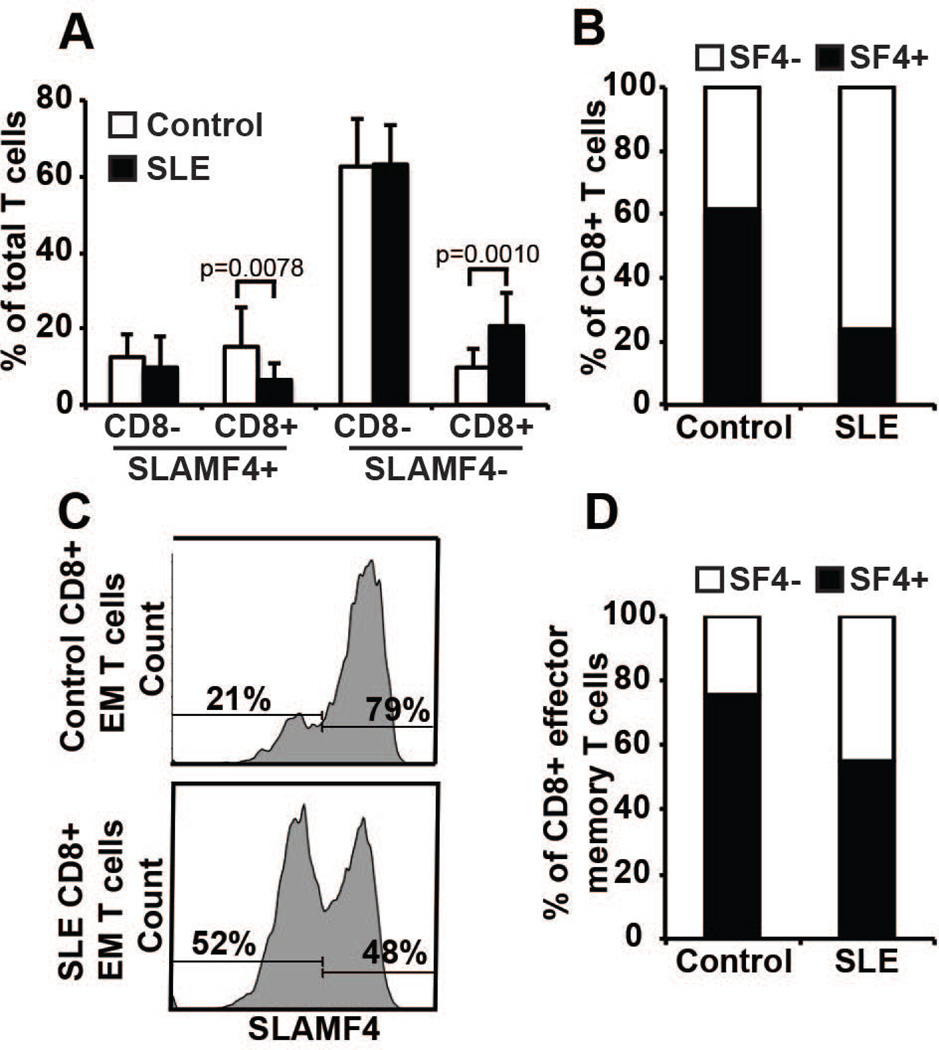
With the exception of CD4− CD8− T cells, which are expanded in SLE patients, all CD4+ and CD8+ subset distribution was comparable between SLE patients and healthy subjects (Supplementary Figure 1). Interestingly, the SLAMF4− CD8+ T cell population was expanded in SLE patients compared to that of healthy controls (Figure 2A) resulting in decreased ratio of SLAMF4+:SLAMF4− CD8+ T cells in SLE favoring the SLAMF4− CD8+ subset (Figure 2B). The expression of SLAMF4 was enriched within the effector memory CD8+ T cells subset compared to the naïve population. Moreover, in SLE patients the SLAMF4− effector memory CD8+ T cell population was expanded compared to healthy controls (Figure 2C and D). Collectively, these results show that the effector memory CD8+ T cell population is phenotypically different in SLE patients than in healthy subjects, especially in terms of the absence or reduced expression of SLAMF4.
Figure 2. SLE CD8+ T cells have an altered SLAMF4+:SLAMF4− ratio favoring SLAMF4− CD8+ T cells.
Flow cytometry analysis (A) and ratio calculations (B) of SLAMF4 expression showing a significant decrease of SLAMF4+ CD8+ and a parallel upregulation of SLAMF4− CD8+ T cells in SLE T cells compared to normal controls (n=10 controls and 10 SLE samples; gated on total T cells). SLAMF4 expression of control and SLE effector/memory CD8+ T cells in one characteristic experiment (C) and cumulative ratio calculation from 10 sample pairs (D) (gated on CD8+ effector memory T cells).
SLAMF4 is a marker for CD8+ T cells with strong cytotoxic capacity
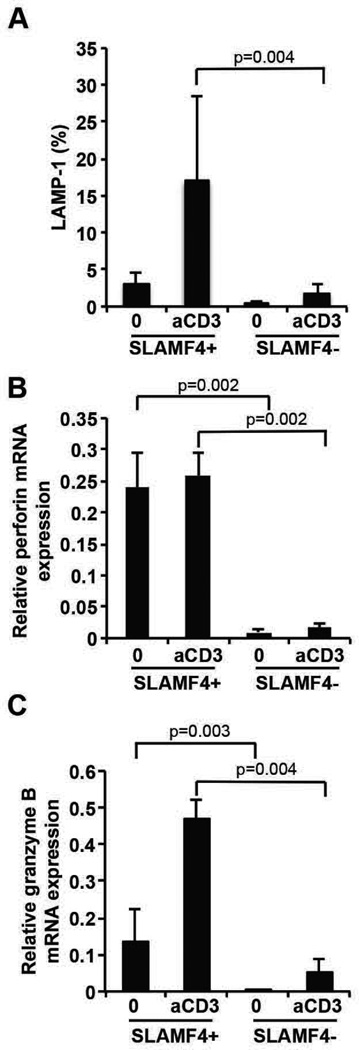
To prove that SLAMF4+ CD8+ T cells are more potent cytotoxic cells than their SLAMF4− counterparts we sorted these two populations from healthy donors and compared their degranulation capacity following activation with an anti-CD3 antibody. SLAMF4+ CD8+ T cells were able to significantly upregulate the LAMP-1 molecule on their cell surface after activation showing effective degranulation activity compared to SLAMF4− CD8+ T cells (Figure 3A). Gene expression of perforin and granzyme B was also evaluated in sorted SLAMF4+ and SLAMF4− CD8+ T cells in normal donors before and following stimulation with plate-bound anti-CD3 antibody. We conclude that isolated SLAMF4+ CD8+ T cells express significantly higher amounts of perforin and granzyme B compared to their SLAMF4− counterparts (Figure 3B and C), as suggested previously (12, 23).
Figure 3. SLAMF4 marks CD8+ T cells with effective cytotoxic capacity.
Sorted SLAMF4+ and SLAMF4− CD8+ T cells were activated by anti-CD3 antibody for 2h and degranulation was measured by the expression of LAMP-1. SLAMF4+ CD8+ T cells degranulate more effectively compared to the SLAMF4− counterparts (A) (gated on CD8+ T cells). SLAMF4+ CD8+ T cells express more perforin and granzyme B genes compared to the SLAMF4− population after 6h of anti-CD3 antibody activation, providing effective cytotoxic machinery (B, C). n=3.
Upon activation SLE CD8+ T cells degranulate at a decreased rate and their effector memory sub-population contains fewer perforin-granzyme B double positive cells
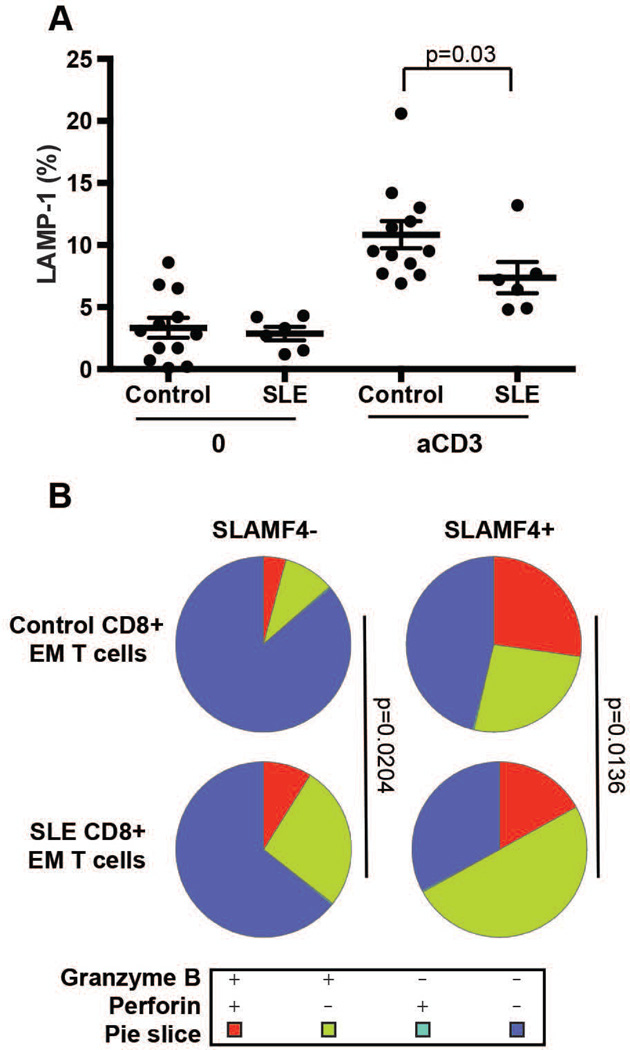
Based on the observed differences in the ratios of SLAMF4+ and SLAMF4− CD8+ T cell populations of healthy and SLE patients in the next set of experiments we characterized these cells functionally. Degranulation of purified CD8+ T cells from patients with SLE and normal donors was investigated following stimulation with anti-CD3 antibody and the cell surface expression of LAMP-1 molecules was measured by flow cytometry. Following anti-CD3-mediated stimulation, the percentage of SLE CD8+ T cells that upregulated cell-surface LAMP-1 was significantly lower compared to normal CD8+ T cells, indicating that degranulation activity is defective in patients with SLE (Figure 4A).
Figure 4. SLE T cells show less degranulation upon activation and SLE SLAMF4+ CD8+ effector memory T cell pool has decreased amounts of perforin/granzyme B double positive population.
Control and SLE CD8+ T cells were activated with anti-CD3 antibody for 2h and degranulation was measured by LAMP-1 expression (A) (n=12 control and 6 SLE; gated on CD8+ T cells, mean ± SEM). Resting control and SLE effector/memory (EM) CD8+ T cells were labeled with anti-perforin and anti-granzyme B antibodies. Data were collected by flow cytometry and analyzed by using the SPICE program (B) (n=9 control and 10 SLE).
Next we compared the SLAMF4+ and SLAMF4− CD8+ effector memory populations isolated from control and SLE individuals and found that SLAMF4+ effector memory cells produced more granzyme B and perforin compared to SLAMF4− cells. Among the SLAMF4+ subsets more cells were able to produce both perforin and granzyme B from healthy donors while SLE T cells showed higher percentage of granzyme B single positive staining (Figure 4B and Supplementary Figure 3). The data suggests that defects in cytotoxicity observed in SLE may be at least partly explained by the altered ratio of SLAMF4+/SLAMF4− CD8+ effector/memory T cells, favoring the presence of SLAMF4− cells with decreased killing capacity.
SLE CD8+ T cells respond poorly to viral peptides
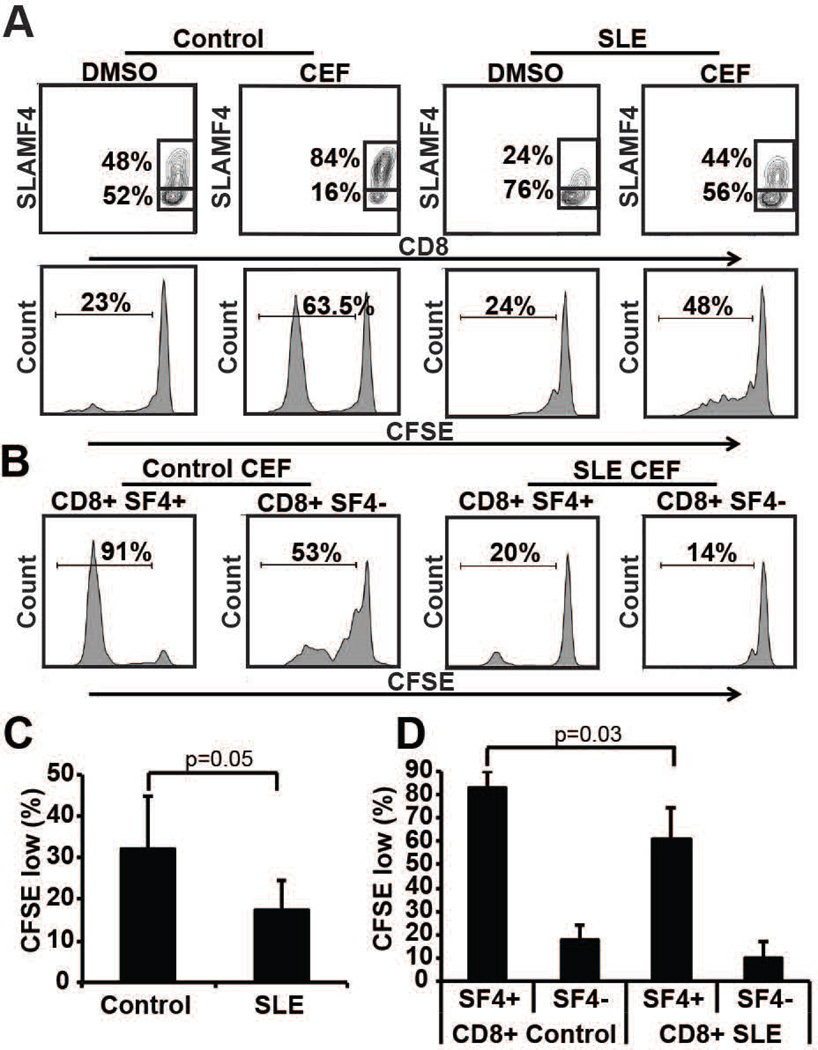
We next determined the ability of SLE CD8+ cells to respond to viral peptides. To this end we used a viral peptide mix (CMV, EBV and influenza – CEF) to activate healthy and SLE PBMCs. After 6 days of activation the proliferation of CFSE-labeled total PBMCs and CD8+ T cells was tested by flow cytometry. CEF-stimulated PBMCs displayed an increased SLAMF4−/SLAMF4+ CD8+ T cell ratio compared to the dissolvent control-treated PBMCs (Figure 5A). Healthy CEF-treated cells proliferated at higher rates compared to SLE cells as judged by the CFSE dilution of total PBMCs (Figure 5A and C) and of SLAMF4− and SLAMF4+ CD8+ T cells (Figure 5B and D). Only SLAMF4+ CD8+ T cells, but not the SLAMF4− counterparts, were able to proliferate effectively upon CEF treatment (Figure 5B and D). In contrast, when anti-CD3 treatment was used to activate healthy and SLE T cells, both the total and the SLAMF4−/SLAMF4+ CD8+ T cell populations proliferated effectively showing that SLE patients fail to activate their virus-specific CD8+ T cell populations but they can respond well to non-specific T cell activation (Supplementary Figure 2). These results further support the concept that the selective loss of SLAMF4+ CD8+ T cells in SLE might be the one of the reasons for the elevated infection rates in this condition.
Figure 5. SLE CD8+ T cells are less responsive to viral peptide stimulation.
Control and SLE PBMCs were labeled with CFSE and treated with CMV, EBV and influenza viral peptide mix (CEF). After 6 days in culture the SLAMF4+/SLAMF4− CD8+ T cell population ratio was investigated (gated on CD8+ T cells) as well as total (A) and CD8+ T cell (B) proliferation was measured by CFSE dilution (one typical experiment is shown). Cumulative proliferation of normal and SLE PBMCs (C) and SLAMF4+/SLAMF4− CD8+ T cells (D) show significantly less proliferation of SLE cells compared to normal controls (n=5).
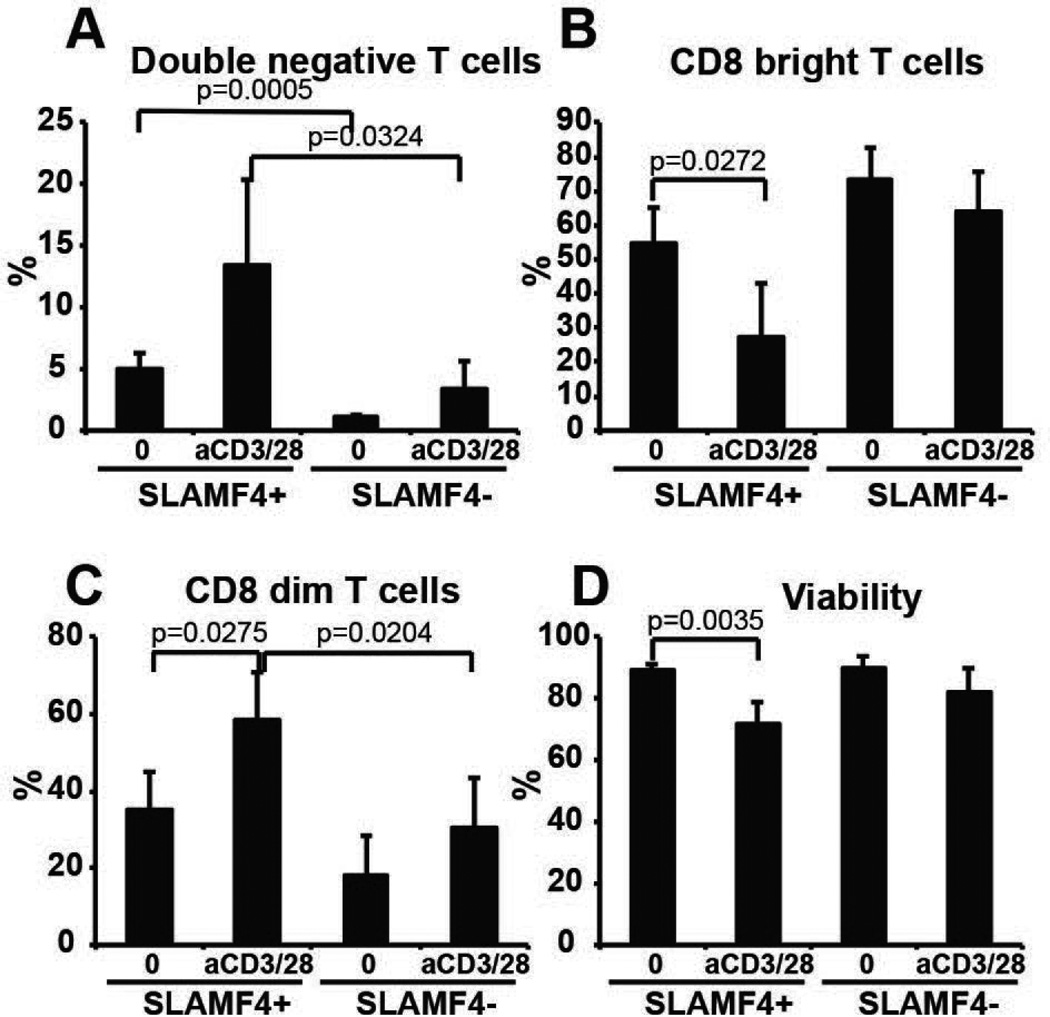
SLAMF4+ CD8+ T cells are more likely become double negative T cells
Human CD4 CD8 double negative T cells can derive from activated CD8+ T cells (24). In the effort to find an explanation for the selective loss of SLAMF4+ CD8+ T cells in SLE we activated sorted cells obtained from normal donors with anti-CD3 and anti-CD28 antibodies for 5 days and measured the percentage of double negative cells (24). SLAMF4+ CD8+ T cells were more likely become double negative cells spontaneously as well as upon activation compared to SLAMF4− T cells (Figure 6A). Also, the percentage of CD8bright cells in the SLAMF4+ population decreased and the percentage of CD8dim cells increased compared to SLAMF4− cells, showing the ongoing process of losing CD8 molecule expression (Figure 6B and C). Moreover, the cell viability was significantly decreased in the SLAMF4+ population upon activation (Figure 6D). Collectively these results show that SLAMF4+ CD8+ T cells are more exposed to change their phenotype and/or less viable after activation.
Figure 6. SLAMF4+ CD8+ T cells more likely become double negative T cells compared to SLAMF4− CD8+ T cells.
CD8+ T cells were sorted based on SLAMF4 expression and were activated with anti-CD3/anti-CD28 antibodies. Higher percentages of SLAMF4+ CD8+ T cells become double negative (A–C) and die (D) after 5 days of culture compared to SLAMF4− counterparts. n=3.
Discussion
CD8+ T cells are able to promote or prevent autoimmune diseases (reviewed in (25)). The role of CD8+ T cells in SLE has not been well characterized despite the fact that cytotoxic cell responses have been invariably reported to be defective (4, 26, 27).
We have presented evidence that central memory and effector memory CD8+ T cells from SLE patients have reduced expression of the receptor SLAMF4 and defective cytotoxic granule exocytosis. Importantly, the response of SLAMF4+ CD8+ T cells was decreased compared to that of control cells and because this subset is compromised and the non-responding SLAMF4− CD8+ subset is expanded, the overall cytotoxic response of CD8+ T cells in SLE is bound to be limited.
Kim et al. assessed the expression of SLAMF4 in a CD8+ T cell subpopulation defined as CCR7− IL-7Rαlow and found it increased in SLE patients (21). They did not distinguish the CD45RA+ effector memory and CD45RA− terminally differentiated effector memory populations among the CD8+ CCR7− T cells. In comparison, in the present study we assessed the expression of SLAMF4 in all differentiated subsets of CD8+ T cells (shown in Figure 1C): naïve (CD45RA+, CCR7+), central memory (CD45RA−, CCR7+), effector memory (CD45RA−, CCR7−) and terminally differentiated effector memory (CD45RA+, CCR7−), without considering the expression of IL-7Rα. We found decreased expression of SLAMF4 in the CD45RA− CCR7− population in SLE patients compared to healthy controls. The discrepancy in SLAMF4 expression between SLE patients and controls may be explained by the consideration of two different CD8+ T cell subpopulations (Kim et al: CD8+, CCR7−, IL7Rαlow; our study: CD8+, CD45RA−, CCR7−).
This study brings forward another interesting point, which is the decreased response to a mixture of three foreign viral antigen-defined peptides (CMV, EBV and influenza). SLE patients are more susceptible to viral infections (14, 28) and EBV viral load is reportedly elevated in patients with SLE (27, 29). Viral antigen-specific responses are also decreased in lupus-prone mice (30). These observations support the possibility of decreased cytotoxic T cell functions in SLE. The capacity of EBV-specific CD8+ T cells to secrete IFNγ is reduced in SLE patients compared to healthy individuals (27, 31, 32) and CD8+ T cells have impaired capacity to degranulate cytotoxic granules (32).
SLAMF4 - SLAMF2 interaction can mediate both activating and inhibitory signals depending on the cell type, i.e. human vs. mouse NK cells, and setting, e.g. during viral infection. Upon ligand binding SLAMF4 may serve as a co-stimulatory molecule promoting NK and CD8+ T cell functions such as proliferation, cytokine production and degranulation (12, 15, 33–37). However, other studies have also reported an inhibitory role of SLAMF4, suggesting that this molecule may have a dual checkpoint function (38, 39). More recently, SLAMF4 is characterized as an exhaustion/inhibitory molecule on virus-specific CD8+ T cells (40, 41). Chronic antigen stimulation can quickly and selectively eliminate memory but not naïve CD8+ T cells by blocking cell proliferation as well as modulating inhibitory receptor expression and reliance on help from CD4+ T cells (42). It has been reported that signaling via SLAMF4 controls the expansion of cytotoxic CD8αβ+ intraepithelial T lymphocytes in mice (43). Data from SLAMF4-deficient mice have also shown that SLAMF4 is an important inhibitor of NK cell cytotoxic functions (44) and it has an NK cell-independent negative regulatory role in lupus pathogenesis (45). Based on these information and our findings (Figure 3) SLAMF4 expression marks cells with effective cytotoxic capabilities and may serve as a built-in negative feedback mechanism on these potentially harmful cells.
It is not clear why SLE patients have less SLAMF4-expressing CD8+ T cells and/or loose SLAMF4 from their surface. The expression of surface molecules on SLE T cells is controlled at many levels (46) and it is quite possible that SLAMF4 is decreased because of limited transcription or because it is degraded faster. Moreover, a rapid downregulation of SLAMF4 cell surface expression on CD8+ T cells has been reported upon simultaneous signaling via both T cell receptor and SLAMF4 (47). The presence of persistent (auto)antigen challenge of CD8+ memory T cells in SLE also can contribute to the selective loss of this SLAMF4+ population (42). Although these possibilities were not addressed in this study, we found that upon stimulation, SLAMF4+ CD8+ T cells have an increased propensity to loose CD8 from their surface and become CD3+ CD4− CD8− double negative cells. We have shown before that this process accounts for the expansion of DN in SLE patients (24, 48). Finally, it is possible that SLE patients have anti-SLAMF4 antibodies that engage and down-regulate its surface expression. Previously, it was shown that stimulation of human NK cells with an anti-SLAMF4 antibody led to a downregulation of surface SLAMF4 by reducing the activity of its promoter and by promoting its internalization (49, 50).
In conclusion, here we present cellular abnormalities in CD8+ SLE T cells that involve the loss of SLAMF4+ population and/or downregulation of SLAMF4 from the surface and suppression of their ability to respond to antigenic stimulation. Our findings advance the understanding of the cell pathways, which account for decreased cytotoxic cell activity in SLE patients and the increased susceptibility to infections.
Supplementary Material
Acknowledgments
This work was supported by the NIH grant 2P01AI065687-06A1.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Vratsanos GS, Jung S, Park YM, Craft J. CD4(+) T cells from lupus-prone mice are hyperresponsive to T cell receptor engagement with low and high affinity peptide antigens: a model to explain spontaneous T cell activation in lupus. J Exp Med. 2001;193(3):329–337. doi: 10.1084/jem.193.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moulton VR, Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res Ther. 2011;13(2):207. doi: 10.1186/ar3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsokos GC, Magrath IT, Balow JE. Epstein-Barr virus induces normal B cell responses but defective suppressor T cell responses in patients with systemic lupus erythematosus. J Immunol. 1983;131(4):1797–1801. [PubMed] [Google Scholar]
- 5.Mok CC, Lau CS. Pathogenesis of systemic lupus erythematosus. J Clin Pathol. 2003;56(7):481–490. doi: 10.1136/jcp.56.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, et al. Morbidity and mortality in systemic lupus erythematosus during a 10-year period: a comparison of early and late manifestations in a cohort of 1,000 patients. Medicine (Baltimore) 2003;82(5):299–308. doi: 10.1097/01.md.0000091181.93122.55. [DOI] [PubMed] [Google Scholar]
- 7.Valiante NM, Trinchieri G. Identification of a novel signal transduction surface molecule on human cytotoxic lymphocytes. J Exp Med. 1993;178(4):1397–1406. doi: 10.1084/jem.178.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non-MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol. 2005;42(4):489–494. doi: 10.1016/j.molimm.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Altvater B, Landmeier S, Pscherer S, Temme J, Juergens H, Pule M, et al. 2B4 (CD244) signaling via chimeric receptors costimulates tumor-antigen specific proliferation and in vitro expansion of human T cells. Cancer Immunol Immunother. 2009;58(12):1991–2001. doi: 10.1007/s00262-009-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat Immunol. 2009;10(9):973–980. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 11.Waggoner SN, Kumar V. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front Immunol. 2012;3:377. doi: 10.3389/fimmu.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speiser DE, Colonna M, Ayyoub M, Cella M, Pittet MJ, Batard P, et al. The activatory receptor 2B4 is expressed in vivo by human CD8+ effector alpha beta T cells. J Immunol. 2001;167(11):6165–6170. doi: 10.4049/jimmunol.167.11.6165. [DOI] [PubMed] [Google Scholar]
- 13.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petri M. Infection in systemic lupus erythematosus. Rheum Dis Clin North Am. 1998;24(2):423–456. doi: 10.1016/s0889-857x(05)70016-8. [DOI] [PubMed] [Google Scholar]
- 15.Enose-Akahata Y, Matsuura E, Oh U, Jacobson S. High expression of CD244 and SAP regulated CD8 T cell responses of patients with HTLV-I associated neurologic disease. PLoS Pathog. 2009;5(12):e1000682. doi: 10.1371/journal.ppat.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco P, Pitard V, Viallard JF, Taupin JL, Pellegrin JL, Moreau JF. Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):201–211. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 17.Fritsch RD, Shen X, Illei GG, Yarboro CH, Prussin C, Hathcock KS, et al. Abnormal differentiation of memory T cells in systemic lupus erythematosus. Arthritis Rheum. 2006;54(7):2184–2197. doi: 10.1002/art.21943. [DOI] [PubMed] [Google Scholar]
- 18.Kim JR, Mathew SO, Patel RK, Pertusi RM, Mathew PA. Altered expression of signalling lymphocyte activation molecule (SLAM) family receptors CS1 (CD319) and 2B4 (CD244) in patients with systemic lupus erythematosus. Clin Exp Immunol. 2010;160(3):348–358. doi: 10.1111/j.1365-2249.2010.04116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ota Y, Kawaguchi Y, Takagi K, Tochimoto A, Kawamoto M, Katsumata Y, et al. Single nucleotide polymorphisms of CD244 gene predispose to renal and neuropsychiatric manifestations with systemic lupus erythematosus. Mod Rheumatol. 2010;20(5):427–431. doi: 10.1007/s10165-010-0302-x. [DOI] [PubMed] [Google Scholar]
- 20.Puxeddu I, Bongiorni F, Chimenti D, Bombardieri S, Moretta A, Bottino C, et al. Cell surface expression of activating receptors and co-receptors on peripheral blood NK cells in systemic autoimmune diseases. Scand J Rheumatol. 2012;41(4):298–304. doi: 10.3109/03009742.2011.648657. [DOI] [PubMed] [Google Scholar]
- 21.Kim JS, Cho BA, Sim JH, Shah K, Woo CM, Lee EB, et al. IL-7Ralpha low memory CD8+ T cells are significantly elevated in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2012;51(9):1587–1594. doi: 10.1093/rheumatology/kes100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79(2):167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garni-Wagner BA, Purohit A, Mathew PA, Bennett M, Kumar V. A novel function-associated molecule related to non-MHC-restricted cytotoxicity mediated by activated natural killer cells and T cells. J Immunol. 1993;151(1):60–70. [PubMed] [Google Scholar]
- 24.Crispin JC, Tsokos GC. Human TCR-alpha beta+ CD4− CD8− T cells can derive from CD8+ T cells and display an inflammatory effector phenotype. J Immunol. 2009;183(7):4675–4681. doi: 10.4049/jimmunol.0901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravano DM, Hoyer KK. Promotion and prevention of autoimmune disease by CD8+ T cells. J Autoimmun. 2013;45:68–79. doi: 10.1016/j.jaut.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Stohl W, Elliott JE, Li L, Podack ER, Lynch DH, Jacob CO. Impaired nonrestricted cytolytic activity in systemic lupus erythematosus: involvement of a pathway independent of Fas, tumor necrosis factor, and extracellular ATP that is associated with little detectable perforin. Arthritis Rheum. 1997;40(6):1130–1137. doi: 10.1002/1529-0131(199706)40:6<1130::AID-ART17>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Kang I, Quan T, Nolasco H, Park SH, Hong MS, Crouch J, et al. Defective control of latent Epstein-Barr virus infection in systemic lupus erythematosus. J Immunol. 2004;172(2):1287–1294. doi: 10.4049/jimmunol.172.2.1287. [DOI] [PubMed] [Google Scholar]
- 28.Kang I, Park SH. Infectious complications in SLE after immunosuppressive therapies. Curr Opin Rheumatol. 2003;15(5):528–534. doi: 10.1097/00002281-200309000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Moon UY, Park SJ, Oh ST, Kim WU, Park SH, Lee SH, et al. Patients with systemic lupus erythematosus have abnormally elevated Epstein-Barr virus load in blood. Arthritis Res Ther. 2004;6(4):R295–R302. doi: 10.1186/ar1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman LA, Tsokos GC. Lupus-prone mice fail to raise antigen-specific T cell responses to intracellular infection. PLoS One. 2014;9(10):e111382. doi: 10.1371/journal.pone.0111382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berner BR, Tary-Lehmann M, Yonkers NL, Askari AD, Lehmann PV, Anthony DD. Phenotypic and functional analysis of EBV-specific memory CD8 cells in SLE. Cell Immunol. 2005;235(1):29–38. doi: 10.1016/j.cellimm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Larsen M, Sauce D, Deback C, Arnaud L, Mathian A, Miyara M, et al. Exhausted cytotoxic control of Epstein-Barr virus in human lupus. PLoS Pathog. 2011;7(10):e1002328. doi: 10.1371/journal.ppat.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messmer B, Eissmann P, Stark S, Watzl C. CD48 stimulation by 2B4 (CD244)-expressing targets activates human NK cells. J Immunol. 2006;176(8):4646–4650. doi: 10.4049/jimmunol.176.8.4646. [DOI] [PubMed] [Google Scholar]
- 34.Lee KM, Bhawan S, Majima T, Wei H, Nishimura MI, Yagita H, et al. Cutting edge: the NK cell receptor 2B4 augments antigen-specific T cell cytotoxicity through CD48 ligation on neighboring T cells. J Immunol. 2003;170(10):4881–4885. doi: 10.4049/jimmunol.170.10.4881. [DOI] [PubMed] [Google Scholar]
- 35.Sivori S, Parolini S, Falco M, Marcenaro E, Biassoni R, Bottino C, et al. 2B4 functions as a co-receptor in human NK cell activation. Eur J Immunol. 2000;30(3):787–793. doi: 10.1002/1521-4141(200003)30:3<787::AID-IMMU787>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 36.Johnson LA, Goldfarb RH, Mathew PA. Regulation of IFN-gamma production following 2B4 activation in human NK cells. In Vivo. 2000;14(5):625–629. [PubMed] [Google Scholar]
- 37.Dupre L, Andolfi G, Tangye SG, Clementi R, Locatelli F, Arico M, et al. SAP controls the cytolytic activity of CD8+ T cells against EBV-infected cells. Blood. 2005;105(11):4383–4389. doi: 10.1182/blood-2004-08-3269. [DOI] [PubMed] [Google Scholar]
- 38.Lee KM, McNerney ME, Stepp SE, Mathew PA, Schatzle JD, Bennett M, et al. 2B4 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J Exp Med. 2004;199(9):1245–1254. doi: 10.1084/jem.20031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacca P, Pietra G, Falco M, Romeo E, Bottino C, Bellora F, et al. Analysis of natural killer cells isolated from human decidua: Evidence that 2B4 (CD244) functions as an inhibitory receptor and blocks NK-cell function. Blood. 2006;108(13):4078–4085. doi: 10.1182/blood-2006-04-017343. [DOI] [PubMed] [Google Scholar]
- 40.Youngblood B, Wherry EJ, Ahmed R. Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Curr Opin HIV AIDS. 2012;7(1):50–57. doi: 10.1097/COH.0b013e32834ddcf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezinne CC, Yoshimitsu M, White Y, Arima N. HTLV-1 specific CD8+ T cell function augmented by blockade of 2B4/CD48 interaction in HTLV-1 infection. PLoS One. 2014;9(2):e87631. doi: 10.1371/journal.pone.0087631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West EE, Youngblood B, Tan WG, Jin HT, Araki K, Alexe G, et al. Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35(2):285–298. doi: 10.1016/j.immuni.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Keeffe MS, Song JH, Liao G, De Calisto J, Halibozek PJ, Mora JR, et al. SLAMF4 Is a Negative Regulator of Expansion of Cytotoxic Intraepithelial CD8(+) T Cells That Maintains Homeostasis in the Small Intestine. Gastroenterology. 2015;148(5):991–1001. e4. doi: 10.1053/j.gastro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waggoner SN, Taniguchi RT, Mathew PA, Kumar V, Welsh RM. Absence of mouse 2B4 promotes NK cell-mediated killing of activated CD8+ T cells, leading to prolonged viral persistence and altered pathogenesis. J Clin Invest. 2010;120(6):1925–1938. doi: 10.1172/JCI41264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown DR, Calpe S, Keszei M, Wang N, McArdel S, Terhorst C, et al. Cutting edge: an NK cell-independent role for Slamf4 in controlling humoral autoimmunity. J Immunol. 2011;187(1):21–25. doi: 10.4049/jimmunol.1100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moulton VRT. G. C. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. The Journal of Clinical Investigation. 2015;125(6) doi: 10.1172/JCI78087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacheco Y, McLean AP, Rohrbach J, Porichis F, Kaufmann DE, Kavanagh DG. Simultaneous TCR and CD244 signals induce dynamic downmodulation of CD244 on human antiviral T cells. J Immunol. 2013;191(5):2072–2081. doi: 10.4049/jimmunol.1300435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hedrich CM, Rauen T, Crispin JC, Koga T, Ioannidis C, Zajdel M, et al. cAMP-responsive element modulator alpha (CREMalpha) trans-represses the transmembrane glycoprotein CD8 and contributes to the generation of CD3+ CD4− CD8− T cells in health and disease. J Biol Chem. 2013;288(44):31880–31887. doi: 10.1074/jbc.M113.508655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandusky MM, Messmer B, Watzl C. Regulation of 2B4 (CD244)-mediated NK cell activation by ligand-induced receptor modulation. Eur J Immunol. 2006;36(12):3268–3276. doi: 10.1002/eji.200636146. [DOI] [PubMed] [Google Scholar]
- 50.Mathew SO, Vaidya SV, Kim JR, Mathew PA. Human natural killer cell receptor 2B4 (CD244) down-regulates its own expression by reduced promoter activity at an Ets element. Biochem Biophys Res Commun. 2007;355(2):483–487. doi: 10.1016/j.bbrc.2007.01.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.