Abstract
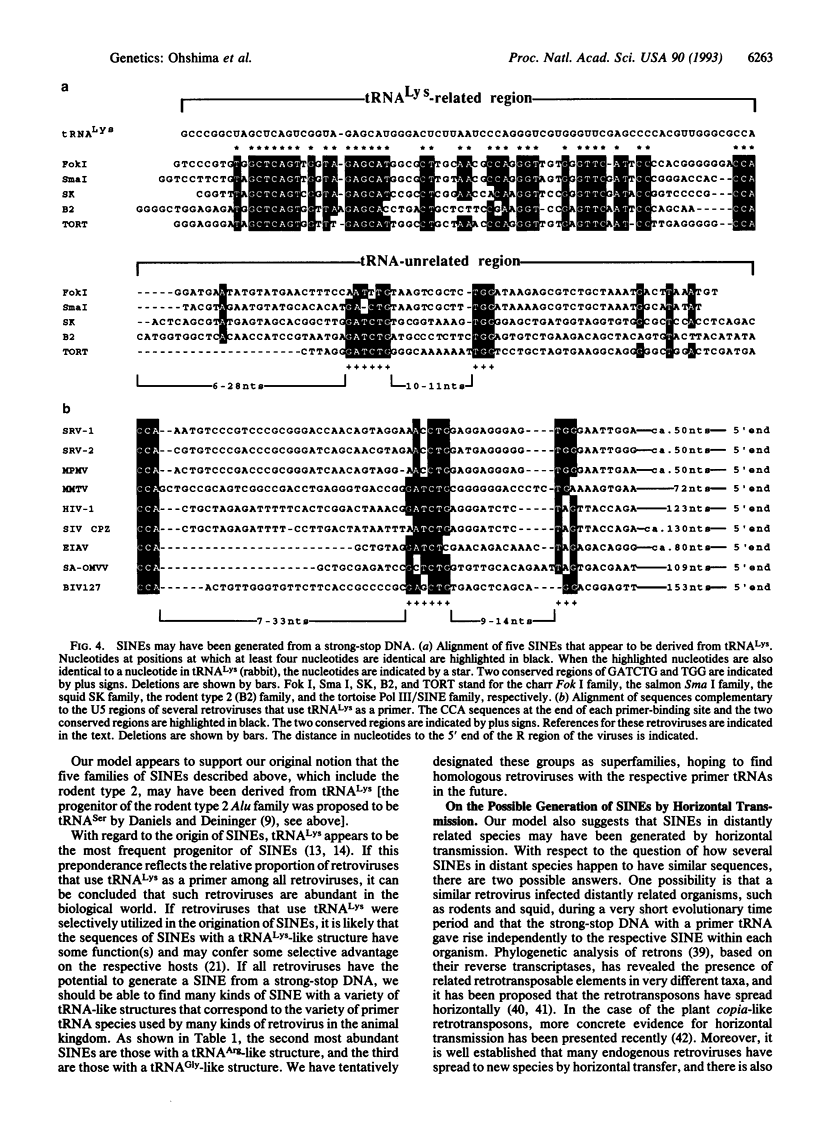
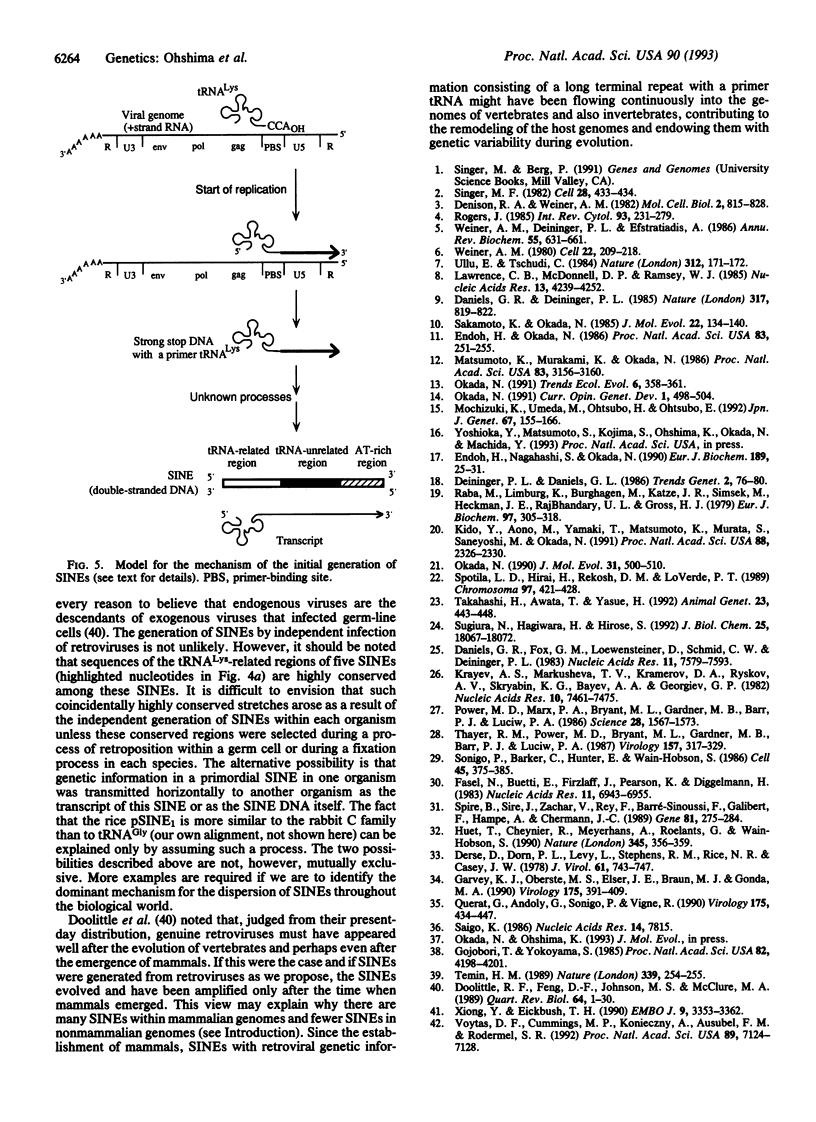
Using labeled transcripts generated in vitro from squid total genomic DNA as a probe, we isolated and characterized a SINE that is present in the squid genome. The squid SINE appears to be derived from a tRNA(Lys). When the consensus sequences of five different SINEs with a tRNA(Lys)-like structure from distantly related species, including squid, were aligned, we found in the tRNA-unrelated region two sequence motifs that were almost identical among these five SINEs. This observation suggests a common evolutionary origin for these SINEs and/or some function(s) for these motifs. Similar sequences were unexpectedly found to be present in sequences complementary to the U5 regions of several mammalian retroviruses whose primer is a tRNA(Lys). On the basis of these findings, we present a model for the generation of SINEs. We propose that they are derived from a "strong-stop DNA" with a primer tRNA(Lys) that is an intermediate in the reverse transcription of certain retroviruses. Our model suggests that a certain group of SINEs may have been generated by horizontal transmission, although it is not clear whether information was transmitted via a similar retrovirus or via an RNA or DNA of a SINE.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Fox G. M., Loewensteiner D., Schmid C. W., Deininger P. L. Species-specific homogeneity of the primate Alu family of repeated DNA sequences. Nucleic Acids Res. 1983 Nov 11;11(21):7579–7593. doi: 10.1093/nar/11.21.7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison R. A., Weiner A. M. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol. 1982 Jul;2(7):815–828. doi: 10.1128/mcb.2.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Dorn P. L., Levy L., Stephens R. M., Rice N. R., Casey J. W. Characterization of equine infectious anemia virus long terminal repeat. J Virol. 1987 Mar;61(3):743–747. doi: 10.1128/jvi.61.3.743-747.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Endoh H., Nagahashi S., Okada N. A highly repetitive and transcribable sequence in the tortoise genome is probably a retroposon. Eur J Biochem. 1990 Apr 20;189(1):25–31. doi: 10.1111/j.1432-1033.1990.tb15455.x. [DOI] [PubMed] [Google Scholar]
- Endoh H., Okada N. Total DNA transcription in vitro: a procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci U S A. 1986 Jan;83(2):251–255. doi: 10.1073/pnas.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Buetti E., Firzlaff J., Pearson K., Diggelmann H. Nucleotide sequence of the 5' noncoding region and part of the gag gene of mouse mammary tumor virus; identification of the 5' splicing site for subgenomic mRNAs. Nucleic Acids Res. 1983 Oct 25;11(20):6943–6955. doi: 10.1093/nar/11.20.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey K. J., Oberste M. S., Elser J. E., Braun M. J., Gonda M. A. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology. 1990 Apr;175(2):391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Yokoyama S. Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4198–4201. doi: 10.1073/pnas.82.12.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Cheynier R., Meyerhans A., Roelants G., Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990 May 24;345(6273):356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B., McDonnell D. P., Ramsey W. J. Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res. 1985 Jun 25;13(12):4239–4252. doi: 10.1093/nar/13.12.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Murakami K., Okada N. Gene for lysine tRNA1 may be a progenitor of the highly repetitive and transcribable sequences present in the salmon genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3156–3160. doi: 10.1073/pnas.83.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Umeda M., Ohtsubo H., Ohtsubo E. Characterization of a plant SINE, p-SINE1, in rice genomes. Jpn J Genet. 1992 Apr;67(2):155–166. doi: 10.1266/jjg.67.155. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Okada N. Transfer RNA-like structure of the human Alu family: implications of its generation mechanism and possible functions. J Mol Evol. 1990 Dec;31(6):500–510. doi: 10.1007/BF02102077. [DOI] [PubMed] [Google Scholar]
- Power M. D., Marx P. A., Bryant M. L., Gardner M. B., Barr P. J., Luciw P. A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986 Mar 28;231(4745):1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- Querat G., Audoly G., Sonigo P., Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1990 Apr;175(2):434–447. doi: 10.1016/0042-6822(90)90428-t. [DOI] [PubMed] [Google Scholar]
- Raba M., Limburg K., Burghagen M., Katze J. R., Simsek M., Heckman J. E., Rajbhandary U. L., Gross H. J. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem. 1979 Jun;97(1):305–318. doi: 10.1111/j.1432-1033.1979.tb13115.x. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Saigo K. A copia primer pseudogene possibly generated by an aberrant reverse transcription of a copia-related element in Drosophila. Nucleic Acids Res. 1986 Oct 10;14(19):7815–7815. doi: 10.1093/nar/14.19.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Okada N. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol. 1985;22(2):134–140. doi: 10.1007/BF02101691. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Barker C., Hunter E., Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986 May 9;45(3):375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Spire B., Sire J., Zachar V., Rey F., Barré-Sinoussi F., Galibert F., Hampe A., Chermann J. C. Nucleotide sequence of HIV1-NDK: a highly cytopathic strain of the human immunodeficiency virus. Gene. 1989 Sep 30;81(2):275–284. doi: 10.1016/0378-1119(89)90188-1. [DOI] [PubMed] [Google Scholar]
- Spotila L. D., Hirai H., Rekosh D. M., Lo Verde P. T. A retroposon-like short repetitive DNA element in the genome of the human blood fluke, Schistosoma mansoni. Chromosoma. 1989 May;97(6):421–428. doi: 10.1007/BF00295025. [DOI] [PubMed] [Google Scholar]
- Sugiura N., Hagiwara H., Hirose S. Molecular cloning of porcine soluble angiotensin-binding protein. J Biol Chem. 1992 Sep 5;267(25):18067–18072. [PubMed] [Google Scholar]
- Takahashi H., Awata T., Yasue H. Characterization of swine short interspersed repetitive sequences. Anim Genet. 1992;23(5):443–448. doi: 10.1111/j.1365-2052.1992.tb02164.x. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Thayer R. M., Power M. D., Bryant M. L., Gardner M. B., Barr P. J., Luciw P. A. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987 Apr;157(2):317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Voytas D. F., Cummings M. P., Koniczny A., Ausubel F. M., Rodermel S. R. copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980 Nov;22(1 Pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]