Abstract
Amyloid-forming proteins undergo a structural transition from α-helical to disordered conformations and, ultimately, cross-β fibrils. The unfolding and aggregation of the amyloid β-peptide (Aβ) have been implicated in the development and progression of Alzheimer’s disease (AD) and cerebral amyloid angiopathy (CAA). However, the events underlying the initial structural transition leading to the disease state remain unclear. Although most cases are sporadic, several genetic variants exist that alter the electrostatic properties of Aβ and lead to more rapid unfolding and more severe phenotypes. In the present study, the enhanced unfolding is shown to be due to the mutated side chains altering the local peptide-bond dipole moments leading to local destabilization of the α-helix, as determined from polarizable molecular dynamics (MD) simulations of wild-type (WT) Aβ fragments and several common mutations. The local perturbation of the helix then leads to progressive unwinding of the α-helix in a cooperative fashion due to decreases in adjacent (i ± 1) and hydrogen-bonded (i + 4) peptide-bond dipole moments. Side-chain dynamics, subsequent variations in dipole moments, and ultimately the response in the peptide-bond dipole moments are all modulated by solvent dielectric properties based on simulations in water versus ethanol. The polarizable simulation results, along with simulations using the additive CHARMM36 force field, further indicate that cooperativity due to the alignment of peptide bonds leading to enhanced dipole moments is a fundamental force in stabilizing α-helices.
Introduction
Pathological protein aggregation is associated with numerous diseases,1,2 many of which feature the formation of amyloid aggregates, which are characterized by a parallel, cross-β structure.3 An example is amyloid β-peptide (Aβ), which is the principal protein component of intra- and extracellular oligomers, fibrils, and plaques in Alzheimer’s disease (AD) and cerebral amyloid angiopathy (CAA). Notably, specific mutations of Aβ have been shown to lead to severe Alzheimer’s phenotypes.4−6 Aβ and many other amyloidogenic peptide sequences are natively α-helical but unfold to give rise to β-strand structures. Although the Aβ monomer is nontoxic,7 understanding the factors contributing to helical instability on the monomer level is of fundamental interest in elucidating the initial events in these disease pathways, as well as providing crucial insight into the stabilizing forces in protein folding.
Protein folding is a cooperative process.8 This cooperativity drives unfolded proteins toward the native, folded state to avoid aggregation and amyloid formation.9,10 The folding of α-helices is driven in part by cooperativity arising from the alignment of peptide bonds through hydrogen bonding,11 as the peptide-bond dipole moments enhance each other during helix formation.12 The alignment of peptide-bond dipoles also gives rise to a helix macrodipole, with excess positive charge at the N-terminal end of the helix and excess negative charge toward the C-terminus. The macrodipole has been implicated in helical stability and ligand binding,13 and induced polarization arising from hydrogen bonding is an important contributing factor to this phenomenon,12 as recently shown for the folding of an α-helical peptide using molecular dynamics simulations in conjunction with a polarizable force field.14
Folding studies of the Aβ monomer have shown that its transition from a predominantly α-helical form to a disordered ensemble is modulated by pH and solution environment.15−17 At physiological pH, acidic groups are predominantly deprotonated, contributing to charge repulsion that can drive the peptide toward a more disordered ensemble. Coles et al. hypothesized that disorder might originate at the Glu22–Asp23 acidic dyad toward the C-terminal end of the first helix in the Aβ structure,15 given that the Aβ structure at a membrane–water interface was not completely helical. Positions 22 and 23 in the Aβ sequence are particularly interesting because they are the sites of several mutations that give rise to altered aggregation properties and greater cytotoxicity.4−6 The most widely studied mutant Aβ forms are the D23N “Iowa”,18 E22Q “Dutch”,19,20 E22G “Arctic”,21 and E22K “Italian”22 species. Each of these mutations alters the electrostatic nature of the peptide; thus, an investigation of the microscopic details underlying their unfolding pathways is of particular interest.
In the present work, wild-type (WT) and mutant Aβ15–27 sequences were used in a systematic study of α-helix unfolding at the level of the Aβ monomer based on MD simulations performed using a polarizable force field based on the classical Drude oscillator model.23 The Aβ15–27 region of the peptide was chosen as it contains three full helical turns while remaining a computationally tractable system for MD simulations in explicit solvent, similar to previous studies on fragments such as Aβ12–24,24 Aβ12–28,25,26 and Aβ13–26.27 Although N-terminal residues have been implicated in modulating the conformational ensemble of Aβ,28,29 such fragments serve as useful models for critical regions in Aβ; the unfolding of the α-helix in this central region of Aβ is known to be important in aggregation and toxicity.30,31 Given that Aβ is proteolytically cleaved in the membrane from the larger amyloid precursor protein (APP)32 and unfolding takes place in solution and at membrane–water interfaces, the effects of solvent polarity were examined using ethanol as a membrane surrogate. The findings have implications not only for the unfolding mechanism in AD and CAA directly but also for factors governing α-helical stability in general.
Results
Presented are simulations of the unfolding of WT Aβ15–27 along with several mutants [Figure S1, Supporting Information (SI)] performed using a polarizable force field based on the classical Drude oscillator model, allowing for an understanding of the role of the induction of dipole moments on helix unwinding. In addition, to verify that the observed unfolding mechanism was associated with explicit polarization in the force field, several of the simulations were repeated using the polarizable model and performed with the additive CHARMM36 force field.33 To facilitate analysis of the induction of the peptide backbone dipoles in the polarizable simulations, we define the dipole moment along the helical axis, μpara,, as described in Figure S2 (SI).
Unfolding of WT Aβ15–27
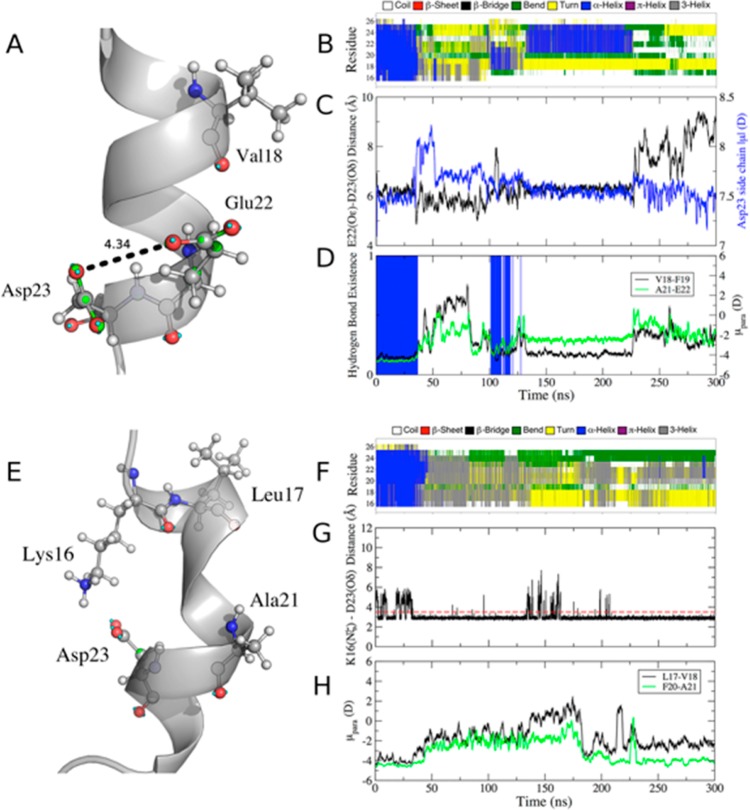
The unfolding of the WT helix in the polarizable simulation proceeded rapidly in both water and ethanol (Figure 1B,F), yielding an average α-helical content of less than 20% (Table S1, SI). In ethanol, 310-helices were observed, whereas in water, one-turn α-helices re-formed transiently, with α-helical conformations lost by the end of the 300-ns simulation in both cases. Helix unwinding in water began when the Glu22 and Asp23 side chains came in close proximity (<4.5 Å, Figure 1A,C) and, as a consequence of the repulsive interaction, the dipole moment of the Asp23 side chain increased by ∼1.5 D (Figure 1C). In response, the component of the dipole moments of the Glu22–Asp23 and Val18–Phe19 peptide bonds parallel to the helix axis (μpara,) decreased in magnitude before reversing in direction (Figure 1D), leading to the unwinding of this turn of the helix. The reduction in μpara coincided with the loss of the (i, i + 4) hydrogen bond between Val18 and Glu22 (Figure 1D), as well as the Phe19–Asp23 hydrogen bond (Figure S3, SI). The simultaneous change in direction of μpara and breaking of backbone hydrogen bonds in both cases indicates that the local peptide-bond dipole moments respond quickly to variations in the side-chain dipole moments, leading to alterations in helix hydrogen bonding. Whereas the electrostatic repulsion between neighboring anionic side chains is intuitively unfavorable, the subsequent destabilization of backbone hydrogen bonding and dipole-moment cooperativity up to four residues away, through the hydrogen-bonding network and encompassing an entire α-helical turn, emphasizes the specific role of side-chain dynamics on the stability of the α-helix through alteration of the peptide-bond dipole moments. Interestingly, water molecules that mediated the ion–ion interaction between the Glu22 and Asp23 side chains were also perturbed. Water molecules that bridged the Glu22 and Asp23 carboxylate groups (defined as any water with an oxygen atom simultaneously within 3.5 Å of a carboxylate oxygen of each residue) had an average μ value of 2.53 ± 0.17 D, slightly above the bulk value (2.46 ± 0.17 D). This outcome indicates that the repulsion between the two negatively charge side chains is somewhat dissipated by hydrating water.
Figure 1.
Unfolding of WT Aβ15–27 in water and ethanol and response of dipole moments to side-chain dynamics. Atoms are colored by element (C in gray, O in red, N in blue, H in white), lone pairs in cyan, and Drude oscillators in green. (A) Close contact of Glu22 and Asp23 side chains in water that leads to destabilization of the helical structure from a snapshot at 36 ns. (B) Secondary structure over time in water according to Dictionary of Secondary Structure of Proteins (DSSP). (C) Dipole moment of the Asp23 side chain over time (blue) and minimum distance between Glu22 and Asp23 carboxylate O atoms (black). (D) Val18 to Glu22 i to i + 4 backbone hydrogen-bond existence (blue spikes indicate the presence of a hydrogen bond, i.e., existence = 1) and time series for the component of the dipole moment parallel to the helix axis (μpara; Figure S2, SI) for the indicated peptide bonds. (E) Salt bridge formed by Lys16 and Asp23 with the resulting distortion of the α-helix in ethanol from a snapshot at 40 ns. (F) Secondary structure over time in ethanol. (G) Time series of the minimum distance between Lys16(Nζ) and Asp23(Oδ1/Oδ2) atoms. Any value less than or equal to 3.5 Å (red dashed line) indicates an intact salt bridge. (H) Time series of μpara for the indicated peptide bonds, showing the loss of dipole-moment alignment upon stable formation of the Lys16–Asp23 salt bridge. The dipole moment and distance time series in panels C, D, G, and H are shown as 1-ns running averages for clarity. A negative value of μpara indicates alignment of C=O with the helix axis.
In ethanol, the WT Aβ15–27 peptide also unfolded rapidly (Figure 1F), although the mechanism was different. Helical unwinding began around Leu17 and proceeded toward the C-terminal end of the α-helix, culminating in a total loss of α-helicity by ∼45 ns. Unwinding was initiated when the side chain of Lys16 formed a stable salt bridge with Asp23 (Figure 1E,G), straining the backbone and perturbing the Leu17–Ala21 backbone hydrogen bond. This strain led to a reversal of μpara in the Leu17–Val18 and Phe20–Ala21 peptide bonds (Figure 1H), indicating a loss of dipole alignment and hydrogen bonding. The association of opposing charges in ethanol is more favorable because of the lower solvent polarity (ε = 22) and resulted in the salt bridge remaining intact for ∼90% of the simulation (Figure 1G). In water, Lys16–Asp23 interactions were screened by the large dielectric constant of the medium and did not contribute to the destabilization of the helix, as the salt bridge was intact for less than 0.2% of the trajectory. Even though Lys16 is able to sequester the side chain of Asp23 away from that of Glu22 to prevent unfavorable side-chain interactions as in the case of the simulation in water, instability still resulted. Taken together, the results of the WT Aβ15–27 simulations suggest that its α-helical structure is prone to disorder in environments with either high or low polarity.
Charge-Neutralizing D23N and E22Q Aβ15–27 Mutants
Experiments have shown that these mutants unfold and aggregate more rapidly than the WT peptide,4−6 suggesting that factors beyond the side-chain charge–charge interactions seen in the WT peptide contribute to the instability of the Aβ helical structure. In water, the helical turn of residues 22–26 in the D23N peptide was lost almost immediately (Figure 2B), initiated by the rotation of the χ1 dihedral angle of Asn23 (N–Cα-Cβ-Cγ) from trans (t) to gauche– (g–, Figure 2A,C). This rotation led to a hydrogen bond between its side-chain amide and the backbone carbonyl group of Phe19 (Figure 2A), causing a significant decrease in the magnitude of μpara of the Glu22–Asn23 peptide-bond dipole (Figure 2D) and leading to disorder in this region of the peptide at ∼10 ns. The remainder of the peptide remained helical (Figure 2B), and μpara of the Phe19–Phe20 peptide bond remained aligned with the helix axis out to ∼160 ns (Figure 2D). Subsequently, the remainder of the helical content was lost (Figure 2B), coinciding with the loss in magnitude and then a switch in the orientation of μpara of the Phe19–Phe20 peptide bond. In ethanol, this outcome was not observed, and the peptide remained ∼70% α-helical for the duration of the simulation (Table S1, SI) despite the fact that the Asn23 side-chain χ1 distribution was nearly identical to that in water (Figure S4, SI), indicating that interactions with the Phe19 backbone carbonyl were still occurring in ethanol but did not lead to destabilization. In water, the average Asn23 side-chain dipole moment was 6.27 ± 0.47 D (average ± root-mean-square fluctuation), whereas in ethanol, it was only 5.17 ± 0.45 D. The larger dipole in water allowed the side-chain amide of Asn23 to more effectively compete for the backbone hydrogen bond between Phe19 and Asn23, leading to unwinding, whereas in ethanol, the smaller side-chain dipole moment did not have this effect. Moreover, the transition from t → g– in water led to an increase in the Asn23 side-chain dipole moment (inset, Figure S4, SI), suggesting environment- and conformation-dependent behavior. Considering only α-helical conformations, in water, the t conformation of Asn23 had an average dipole moment of 5.68 ± 0.48 D, whereas rotation to g– led to an increase to 5.96 ± 0.46 D. In ethanol, the side-chain dipole moment was insensitive to conformation, being 5.17 ± 0.44 D in t and 5.17 ± 0.42 D in g–.
Figure 2.
Dynamics of charge-neutralizing mutants D23N and E22Q in water. (A) Snapshot of D23N in water at 11.81 ns showing hydrogen-bond formation. (B) Secondary structure evolution of D23N in water. (C) Side-chain χ1 time series for Asn23. (D) Time series of μpara for the peptide bonds involved in hydrogen-bond exchange. (E) Snapshot of E22Q in water at 124.56 ns showing hydrogen-bond formation leading to dipole enhancement. (F) Secondary structure evolution of E22Q in water. (G) Time series of μpara for the peptide bonds affected by the Gln22–Asp23 side-chain hydrogen bond, the formation of which is indicated by black spikes. (H) Side-chain dipole moment of Gln22, with values for each frame shown as circles and a 1-ns running average shown as a red line. The μpara time series in panels D and G are also shown as 1-ns running averages for clarity.
Side-chain–backbone interactions also figured prominently in unfolding of the E22Q peptide in water. Helical instability from Leu17 to Phe20 appeared within ∼40 ns of simulation time (Figure 2F). Transient hydrogen-bond formation between the side-chain Gln22 amide and Asp23 carboxylate early in the trajectory led to short-lived decreases in the magnitude of μpara of several nearby residues (Figure 2G), with the loss of helix from Leu17 to Phe20 corresponding to a decrease in μpara for the Val18–Phe19 and Ala21–Gln22 peptide bonds. The remaining helical structure was lost at ∼124 ns when the Gln22–Asp23 side-chain hydrogen bond (Figure 2G) persisted. This interaction arose when the χ1 and χ2 (Cα–Cβ–Cγ–Cδ) dihedral angles of Gln22 were both gauche+ (g+). In this hydrogen-bonded conformation (Figure 2E), which persisted for ∼18 ns (124–142 ns in the trajectory, Figure 2G), the side-chain dipole moment of Gln22 increased by ∼2 D (Figure 2H), an effect subsequently transmitted to the backbone peptide bonds, leading to a reversal in the direction of μpara of the Ala21–Gln22 and Gln22–Asp23 peptide bonds (Figure 2G). This reversal of μpara was transmitted to the Val18–Phe19 (i – 4) peptide bond, leading to unwinding of this turn of the α-helix (Figure 2F,G).
The reversal in μpara of the Gln22–Asn22 peptide bond in water was a direct result of interactions with the Gln22 side chain. Figure S5C,D (SI) shows the relationship between μpara of the Gln22 side chain and μpara of the Gln22–Asp23 peptide bond. In water, the backbone μpara responded to the fluctuations in the side-chain μpara (Figure S5C, SI), which are due to changes in side-chain orientation and concomitant Gln22 and Asp23 side-chain–side-chain hydrogen-bond formation (Figure S5E, SI). In ethanol, the backbone μpara was not sensitive to this effect (Figure S5D, SI), despite conformational variations in χ1 that lead to hydrogen bonding (Figure S5F, SI). It is also important to highlight the fact that, whereas Gln22–Asp23 hydrogen-bond formation in water resulted in a large dipole response [increase in the total side-chain μ value of ∼2 D, Figure 2H, and ∼4 D in μpara, Figure S5C (SI)], there was no systematic response in μpara in ethanol (Figure S5D, SI). Thus, the backbone μpara remained aligned with the helix axis, and the majority of the helix persisted throughout the simulation (Figure S5B, SI).
Salt-Bridge Formation in E22K Aβ15–27
NMR experiments by Masuda et al. showed that, in the E22K Aβ peptide, a salt bridge forms between Lys22 and Asp23, promoting the formation of a local bend instead of a helix.34 We observed this phenomenon in the simulations of E22K Aβ15–27 in water. Upon formation of the salt bridge at ∼25 ns (Figure 3B), the magnitude of Lys22–Asp23 μpara decreased, which was transmitted to the neighboring Ala21–Lys22 peptide bond (Figure 3D), contributing to the loss of the α-helical turn that includes Lys22 and Asp23 (Figure 3A). Throughout the remainder of the simulation, the Lys22–Asp23 salt bridge broke and formed multiple times (Figure 3B), leading to sampling of both g+ and t conformations of χ1 of Lys22 (Figure 3C). In contrast, in ethanol, the salt bridge persisted (Figure S6D, SI), with Lys22 χ1 heavily favoring the t conformation (Figure S6C, SI); in this case, the helix was stable throughout the simulation (Figure S6A, SI) with only transient deformation due to the Lys22 g+ state (Figure S6B, SI).
Figure 3.

Salt-bridge dynamics in E22K Aβ15–27 perturb nearby dipole moments in water. (A) Secondary structure evolution according to DSSP. (B) Time series of the minimum distance between Lys22 Nζ and Asp23 Oδ1/Oδ2 atoms, shown as a 1-ns running average for clarity. A value of ≤3.5 Å (dashed blue line) was used as an indicator of an intact salt bridge. (C) Time series of the Lys22 χ1 dihedral angle. (D) Time series of μpara for the indicated peptide bonds, illustrating their response to the side-chain dynamics. (E) Snapshot from 22.30 ns of the simulation in water, with an intact Lys22–Asp23 salt bridge, with the Lys22 χ1 in the t conformation, such that Asp23 is sequestered away from the peptide bond. (F) Lys22 in the g+ conformation from a snapshot at 25.01 ns of the simulation in water, interacting directly with Asp23, which is brought in close contact with the peptide bond.
Table 1 reports the average μpara values for the Ala21–Lys22 and Lys22–Asp23 peptide bonds as a function of Lys22 χ1 in α-helical states of the Ala21–Lys22–Asp23 sequence. In both water and ethanol, rotation of Lys22 χ1 to the g+ conformation led to a decrease in the magnitude of μpara in both of these peptide bonds (Table 1), which destabilized the helix. The reason for this destabilization is that the g+ state of Lys22 χ1 brought the Asp23 carboxylate in close proximity (3.98 ± 0.48 Å on average in helical states) to the carbonyl oxygen of the Lys22–Asp23 peptide bond (Figure 3F), whereas the t conformation of Lys22 χ1 sequestered the acid moiety away (4.47 ± 0.20 Å, Figure 3E), although the Lys22–Asp23 side-chain interaction is present in both the g+ and t χ1 conformations. Thus, although the Lys22–Asp23 salt bridge can form in both water and ethanol, the differences in the dielectric constants of the solvents lead to increased opening and closing in the former that allows for interactions of the Asp23 side chain with the backbone while the salt bridge persists in ethanol with χ1 in t dominating, thereby not destabilizing the helix. Unlike in the WT peptide, in which water molecules between Glu22 and Asp23 dissipated some of the ion–ion repulsion, in the case of water-mediated salt bridges, there was no shift in the water dipole moments. They behaved exactly like bulk water, with an average value of μ = 2.46 ± 0.17 D. In the case of water mediating interactions between unlike charges, there is no dissipation of the dipole response.
Table 1. μpara Values for the Ala21–Lys22 and Lys22–Asp23 Peptide Bonds As a Function of Lys22 χ1 Conformationa.
| peptide bond | Lys22 χ1 | water | ethanol |
|---|---|---|---|
| Ala21–Lys22 | t | –4.22 ± 0.70 | –4.69 ± 0.23 |
| g+ | –3.92 ± 0.55 | –4.54 ± 0.27 | |
| g– | –4.29 ± 0.35 | – | |
| Lys22–Asp23 | t | –4.20 ± 0.45 | –4.67 ± 0.28 |
| g+ | –3.85 ± 0.53 | –4.38 ± 0.37 | |
| g– | –4.00 ± 0.46 | – |
Values (average ± root-mean-square fluctuation) calculated only for frames in which Ala21–Lys22–Asp23 were in an α-helical conformation.
Impact of Side-Chain Deletion in E22G Aβ15–27
The Aβ15–27 E22G mutant rapidly unfolded in water (Figure S7B, SI), whereas unfolding in ethanol proceeded more slowly (Figure S8D, SI), such that 43% total α-helical content was retained during the simulation (Table S1, SI). In the E22G peptide, close packing between Gly25 and Gly22 in water arose because of the absence of side-chain atoms at position 22 (Figure S7A, SI). This packing persisted for ∼50 ns, during which time the α-helix was in a distorted but metastable state in which backbone hydrogen bonds deviated from linearity (Figure S7A, SI). Conversion of these two Gly residues between right- and left-handed helical conformations, with peptide-bond dipole moments responding accordingly (Figure S7C,D, SI), initiated unfolding by perturbing the Ala21–Gly25 backbone hydrogen bond (Figure S7A, SI). Unfolding then propagated through the backbone hydrogen-bonding network as μpara reversed along the helix axis (Figure S7C,D, SI). In ethanol, Gly–Gly packing was not observed, although several residues in the E22G peptide were disordered for the majority of the simulation (Figure S8, SI). The intrinsic flexibility of Gly25 allowed Asp23 to reorient such that the Asp23–Val24 peptide bond was oriented in the opposite direction of the helix, with μpara reflecting this change in conformation (Figure S8A,C, SI) and subsequently perturbing nearby peptide-bond dipoles to transiently destabilize the helix at ∼100 ns (Figure S8C,D, SI). The disordered state of Asp23 was stabilized by a tight turn around Gly25, allowing nearby amide groups to form favorable interactions with the carboxylate moiety of Asp23 (Figure S8B, SI).
Reproducibility of the Drude Results and Comparison with Additive CHARMM36 Simulations
The WT and D23N peptides were each simulated two additional times with the Drude model for 100 ns, initiated from the same starting configurations but different velocities, to test the reproducibility of the observed unfolding mechanism. The results are summarized in Figures S9 and S10 (SI) for the WT and D23N systems, respectively. All runs produced compatible results; that is, for the WT peptide, proximity of Glu22 and Asp23 acid groups resulted in spikes of the Asp23 side-chain μ value, concomitant with the reversal of μpara along the helix axis and destabilization of backbone hydrogen bonding (Figure S9, SI). Similarly, the rotation of Asn23 into a g– χ1 state destabilized Phe19–Asn23 backbone hydrogen bonding and resulted in unfolding of the D23N peptide (Figure S10, SI). All of these events occurred within the first 100 ns of each replicate simulation, thus the results described above from the initial simulation are consistently observed and the role of the polarization response is emphasized.
To provide additional context for the findings using the Drude polarizable force field, we performed the same simulations with the additive CHARMM36 force field.33 By definition, dipole response will not be observed in this fixed-charge model (beyond geometry changes such as bond stretching or dihedral-angle rotation), so the additive simulations serve as an important reference for establishing the role of induced polarization in the simulations described above. In water, all of the Aβ15–27 peptides disordered to some extent (Table S1 and Figure S11, SI), although the WT, D23N, and E22K peptides were more helical with CHARMM36 than they were in the Drude simulations. In the additive systems, unfolding of all of the peptides proceeded in a similar manner. The N- and/or C-terminal residues initially became disordered, allowing the side-chain amide groups of Gln15 and Asn27 to interact with various side chains in the Aβ15–27 sequence. In none of these cases did these side chains compete for backbone hydrogen bonding. Instead, hydrogen bonds transiently formed between groups already destabilized and exposed to water, thus stabilizing the unfolded state but not contributing to the unfolding pathway. This is a nonspecific effect that occurred in all of the simulations, which, in the majority of cases, did not propagate to the rest of the structure as observed in the polarizable simulations.
The WT simulations in water with CHARMM36, the Glu22 and Asp23 side chains to come in close contact of <3.5 Å at 3.23 ns. However, no perturbation of the helical structure occurred (Figure S11, SI), which is assumed to be due to the lack of a change in the Asp23 side-chain μ value. The absence of a destabilizing effect by Glu22–Asp23 interactions is inconsistent with experimental evidence that they are responsible for destabilizing helical structure as a function of pH.15 In the D23N mutant, the Asn23 side chain heavily sampled the g– conformation and formed hydrogen bonds with the Phe19 carbonyl oxygen. However, as changes in the dipole of the side chain were limited to the impact of dihedral-angle rotation (Figure S12, SI), helicity was preserved for most of the trajectory (Figure S11, SI). As with the Drude systems, the WT and D23N simulations in water were repeated twice more for 100 ns, with nonspecific unfolding occurring in all cases (Figure S11, SI). We note that the additive TIP3P water diffuses approximately 3 times too fast, which might enhance the rate at which conformational changes occur in a nonspecific manner. In the E22K simulation, the Lys22–Asp23 salt bridge formed only infrequently (6.9% of the time) and was associated with transient destabilization of the helical turn involving only these two residues (Figure S11, SI), as a result of a small change in peptide-bond alignment rather than an electrostatic response. These results suggest that there is an intrinsic strain on the helical structure associated with the formation of the salt bridge between these neighboring residues, although the CHARMM36 simulation suggests that it is not a strong destabilizing force, whereas the polarizable simulations described above clearly showed propagating effects of dipole reorientation that destabilized the α-helical structure. Both the E22G and E22Q peptides disordered rapidly in water with the additive force field (Figure S11, SI); the disordering of the termini led to fast unfolding that was not associated with any specific interactions arising from the mutations.
In ethanol, all peptides with the additive force field remained predominantly helical, with generally a larger α-helical content than the Drude force field (Table S1 and Figure S11, SI). N-terminal residues were the only ones to disorder in these simulations, and the helical turns encompassing the sites of mutations remained helical throughout all of the simulations (Figure S11, SI). Thus, the additive simulations would lead to the conclusion that, in ethanol, there is little to no disordering associated with the mutagenic region of Aβ. Taken together, the simulations using CHARMM36 suggest only a nonspecific fraying at the ends of the Aβ15–27 peptide occurs in ethanol and water, with this fraying leading to unfolding in water whereas specific behaviors related to Glu22, Asp23, or any of the mutations, were not observed.
Discussion
The present study involved a systematic investigation of the underlying contributions to helical unfolding in the amyloidogenic Aβ15–27 fragment and examined the effects of both mutations and solvent on the unfolding of this peptide. Similar Aβ fragments have been used to efficiently examine unfolding phenomena.25−27,35−38 Moreover, by studying systems in water (ε = 79) and ethanol (ε = 22), solvent effects can be evaluated and related back to unfolding in aqueous solution or at membrane–water interfaces, as ethanol is a reasonable model of the glycerol region of the membrane–water interface given its dielectric constant.39
Previous studies have concluded that mutant Aβ peptides have enhanced cytotoxicity, aggregate more rapidly,4−6,18 and have altered fibril structures.40 The present work did not seek to resolve the details of these pathways, instead focusing on the early events in the unfolding of the helix that is generally believed to be intact upon liberation of Aβ from the helical transmembrane domain of APP. The NMR structure from Coles et al.15 is a logical initial model, as it was determined in the presence of detergent micelles, thus approximating a membrane–water interface.
In agreement with the mechanism hypothesized by Coles et al.,15 our results indicate that side-chain electrostatic interactions are principally responsible for unfolding of the WT peptide, with Glu22–Asp23 repulsion driving unfolding in water. This repulsion perturbs the Asp23 side-chain dipole that is communicated to the dipole moment of the adjacent peptide bond and propagated through the backbone hydrogen-bonding network to perturb additional peptide-bond dipoles. Salt-bridge formation also plays a role in helical destabilization of the WT peptide in ethanol and in the E22K peptide in water, but in contrast, the salt bridge stabilized the α-helicity of E22K in ethanol. Thus, the present results show how side-chain dynamics affect peptide-bond dipole moments, in turn contributing to the instability of α-helical structure of the Aβ15–27 peptide. Electrostatic interactions between charged side chains are the most important intrapeptide interactions that affect helix stability, suggesting that our findings might also extend to intrinsically disordered proteins, which are often highly charged.41
Similar to the Lys22–Asp23 dynamics, in the E22Q peptide, hydrogen-bond dynamics between Gln22 and Asp23 led to helical instability in water. Although the formation of this side-chain hydrogen led to a large (∼2 D) increase in the Gln22 side-chain dipole moment, only in water was the Gln22 side-chain amide group aligned such that its μpara opposed the helix axis, causing a similar response in the backbone and subsequent helical unwinding. The effect on peptide-bond dipoles was not observed in ethanol, suggesting that the polarity of the solvent modulates the susceptibility of the backbone to side-chain dipole dynamics. Hydrogen bonding and competition for native interactions contribute to the helical instability of the D23N peptide in water. The side-chain amide of Asn23 destabilizes its own backbone hydrogen bonding, leading to rapid unwinding in water. The solvent polarity and subsequent response of the side chain are important factors, as in water, Asn23 is more strongly polarized than in ethanol, allowing it to more effectively compete for backbone hydrogen bonding. The side-chain response to solvent polarity and nearby dipoles is a feature unique to the Drude polarizable model, highlighting the importance of explicit polarization in MD simulations, especially mechanistic studies such as this one.
Conclusions
The present work has provided new insights into the driving forces for Aβ unfolding by modeling a peptide fragment that is known to harbor mutations that lead to severe disease phenotypes. Explicit inclusion of induced polarization, an effect that is not frequently included in MD simulations, is a critical component of the physical behavior, as evidenced by the different mechanisms of unfolding seen with the additive CHARMM36 force field. Whereas the additive force field undergoes generic fraying of the N- and C-termini, leading to unfolding in water, polarization effects manifest themselves on the local backbone dipoles, leading to sequence-dependent effects, a behavior that has been suggested previously for other proteins.42 The polarizable model also shows clear differences in the mechanism of unfolding as a function of solvent, and the results have implications for understanding Aβ unfolding in different environments, such as the membrane or in aqueous solution. Further studies of full-length Aβ and other amyloidogenic peptides using models that include explicit treatment of electronic polarization in water and membranes would be necessary to more fully describe the unfolding and aggregation pathways, and perhaps mechanisms of membrane perturbation.
Methods
The initial coordinates for the Aβ15–27 fragment were taken from the first model of the NMR structure of Aβ40 of Coles et al.,15 in which the fragment forms a continuous α-helix, flanked by disordered residues at the N-terminal end and a bend at its C-terminal end. To negate end effects from charged termini, the N- and C-termini of the Aβ15–27 fragment were capped with acetyl and amide groups, respectively. The sequence of the WT fragment is Ac-NKVLFFAEDVGSN-NH2 [see Figure S1 (SI) for additional sequence information]. Mutations (D23N, E22Q, E22G, and E22K) were introduced by deleting conflicting side-chain atoms and rebuilding the missing atoms of the mutated residue using the internal coordinate builder in CHARMM.43 Each peptide was solvated in a 43-Å cubic box of water (SWM4-NDP44 for Drude systems or TIP3P45−47 for CHARMM36 systems) or ethanol.48 To neutralize the net charge of the WT peptide, one Na+ ion was added to the system. Similarly, one Cl– ion was added to the E22K system. Coordinates and topologies were prepared by CHARMM-GUI49 using the Drude-201323 or CHARMM3633 force fields for proteins.
Each system was energy-minimized and equilibrated under an NPT ensemble for 1 ns with restraints on all peptide non-hydrogen atoms (5.0 kcal mol–1 Å–2). Given the considerable change in size and electronic nature at position 22 of the E22K mutation, this peptide was equilibrated for an additional 1 ns with restraints on backbone non-hydrogen atoms only, allowing the side chains to reorient to avoid any initial bias from the in silico mutation. Following equilibration, unrestrained MD simulations were carried out for 300 ns. All polarizable simulations were performed using NAMD with the Langevin integration scheme for polarizable simulations50,51 or the standard leapfrog integrator for additive simulations. Neighbor lists were updated within 16.0 Å, and short-range van der Waals interactions were switched smoothly to zero over a range of 10.0–12.0 Å. Electrostatic interactions were calculated using the particle mesh Ewald (PME) method,52,53 with a 1-Å grid spacing throughout the unit cell. For polarizable systems, a dual Langevin thermostat50 was used to regulate the temperature of the real atoms at 298 K and the Drude oscillators at 1 K, using friction coefficients of 5.0 and 20.0 ps–1, respectively. The Langevin piston method54,55 was used to regulate the pressure at 1.0 atm with a decay period of 0.1 ps and an oscillation period of 0.2 ps. Bonds involving hydrogen atoms were constrained with SHAKE,56 and the time step was set to 1 fs. A “hard-wall” constraint57 was used to prevent displacements larger than 0.2 Å between the Drude oscillators and their parent atoms. For additive systems, the Langevin piston method was used to regulate temperature and pressure, with the same relaxation times as in the Drude simulations. The integration time step for additive simulations was 2 fs.
Analysis was performed using facilities in CHARMM43 or the GROMACS 5.058 interface to the Dictionary of Secondary Structure of Proteins (DSSP).59 For dipole analysis, the peptide bond of residue i is defined as the carbonyl functional group (including real atoms, Drude oscillators, and lone pairs) of residue i, and the N, H, Cα, and Hα atoms and connected Drude oscillators in residue i+1 (Figure S2, SI). The total dipole moments were decomposed in three dimensions to provide insight into their orientations. The peptides were aligned along the x axis based on a least-squares fit to Cα atoms, after which dipole moments were calculated. The x component of the dipole moment is thus parallel to the helix axis and is referred to in the present work as μpara. Values of μpara < 0 indicate alignment with the helix axis (Figure S2, SI).
Acknowledgments
Financial support for the present work was provided by National Institutes of Health Grants F32GM109632 (to J.A.L.) and GM072558 and GM051501 (to A.D.M.). Computing resources were provided by the University of Maryland Computer-Aided Drug Design Center.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.5b09978.
Twelve additional figures and one table (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Soto C. Unfolding the Role of Protein Misfolding in Neurodegenerative Diseases. Nat. Rev. Neurosci. 2003, 4, 49–60 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Sipe J. D.; Cohen A. S. Review: History of the Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–98 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- Tycko R. Solid-State NMR Studies of Amyloid Fibril Structure. Annu. Rev. Phys. Chem. 2011, 62, 279–299 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor J. P.; McVoy L.; Van Nostrand W. E. Charge Alterations of E22 Enhance the Pathogenic Properties of the Amyloid β-Protein. J. Neurochem. 2000, 74, 2209–2212 10.1046/j.1471-4159.2000.0742209.x. [DOI] [PubMed] [Google Scholar]
- Miravalle L.; Tokuda T.; Chiarle R.; Giaccone G.; Bugiani O.; Tagliavini F.; Frangione B.; Ghiso J. Substitutions at Codon 22 of Alzheimer’s Aβ Peptide Induce Diverse Conformational Changes and Apoptotic Effects in Human Cerebral Endothelial Cells. J. Biol. Chem. 2000, 275, 27110–27116 10.1074/jbc.M003154200. [DOI] [PubMed] [Google Scholar]
- Van Nostrand W. E.; Melchor J. P.; Cho H. S.; Greenberg S. M.; Rebeck G. W. Pathogenic Effects of the D23N Iowa Mutant Amyloid β-Protein. J. Biol. Chem. 2001, 276, 32860–32866 10.1074/jbc.M104135200. [DOI] [PubMed] [Google Scholar]
- Cizas P.; Budvytyte R.; Morkuniene R.; Moldovan R.; Broccio M.; Lösche M.; Niaura G.; Valincius G.; Borutaite V. Size-dependent Neurotoxicity of β-Amyloid Oligomers. Arch. Biochem. Biophys. 2010, 496, 84–92 10.1016/j.abb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranker A. D.; Dobson C. M. Collapse and Cooperativity in Protein Folding. Curr. Opin. Struct. Biol. 1996, 6, 31–42 10.1016/S0959-440X(96)80092-3. [DOI] [PubMed] [Google Scholar]
- Booth D. R.; Sunde M.; Bellotti V.; Robinson C. V.; Hutchinson W. L.; Fraser P. E.; Hawkins P. N.; Dobson C. M.; Radford S. E.; Blake C. C. F.; et al. Instability, Unfolding and Aggregation of Human Lysozyme Variants Underlying Amyloid Fibrillogenesis. Nature 1997, 385, 787–793 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- Dumoulin M.; Last A. M.; Desmyter A.; Decanniere K.; Canet D.; Larsson G.; Spencer A.; Archer D. B.; Sasse J.; Muyldermans S.; et al. A Camelid Antibody Fragment Inhibits the Formation of Amyloid Fibrils by Human Lysozyme. Nature 2003, 424, 783–788 10.1038/nature01870. [DOI] [PubMed] [Google Scholar]
- Guo H.; Karplus M. Solvent Influence on the Stability of the Peptide Hydrogen Bond: A Supramolecular Cooperative Effect. J. Phys. Chem. 1994, 98, 7104–7105 10.1021/j100080a002. [DOI] [Google Scholar]
- Wada A. The Alpha-helix as an Electric Macro-dipole. Adv. Biophys. 1976, 9, 1–63. [PubMed] [Google Scholar]
- Hol W. G. J.; van Duijnen P. T.; Berendsen H. J. C. The α-Helix Dipole and the Properties of Proteins. Nature 1978, 273, 443–446 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Huang J.; MacKerell A. D. Jr. Induction of Peptide Bond Dipoles Drives Cooperative Helix Formation in the (AAQAA)3 Peptide. Biophys. J. 2014, 107, 991–997 10.1016/j.bpj.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles M.; Bicknell W.; Watson A. A.; Fairlie D. P.; Craik D. J. Solution Structure of Amyloid β-Peptide(1–40) in a Water-Micelle Environment: Is the Membrane-Spanning Domain Where We Think It Is?. Biochemistry 1998, 37, 11064–11077 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- Zagorski M. G.; Barrow C. J. NMR Studies of Amyloid β-Peptides: Protein Assignments, Secondary Structure, and Mechanism of an α-Helix → β-Sheet Conversion for a Homologous, 28-Residue, N-terminal Fragment. Biochemistry 1992, 31, 5621–5631 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- Crescenzi O.; Tomaselli S.; Guerrini R.; Salvadori S.; D'Ursi A. M.; Temussi P. A.; Picone D. Solution Structure of the Alzheimer Amyloid β-Peptide (1–42) in an Apolar Microenvironment. Eur. J. Biochem. 2002, 269, 5642–5648 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- Grabowski T. J.; Cho H. S.; Vonsattel J. P. G.; Rebeck G. W.; Greenberg S. M. Novel Amyloid Precursor Protein Mutation in an Iowa Family with Dementia and Severe Cerebral Amyloid Angiopathy. Ann. Neurol. 2001, 49, 697–705 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- van Broeckhoven C.; Haan J.; Bakker E.; Hardy J. A.; Van Hul W.; Wehnert A.; Vegter-Van der Vlis M.; Roos R. A. Amyloid beta Protein Precursor Gene and Hereditary Cerebral Hemorrhage with Amyloidosis (Dutch). Science 1990, 248, 1120–1122 10.1126/science.1971458. [DOI] [PubMed] [Google Scholar]
- Levy E.; Carman M. D.; Fernandez-Madrid I. J.; Power M. D.; Lieberburg I.; van Duinen S. G.; Bots G. T.; Luyendijk W.; Frangione B. Mutation of the Alzheimer’s Disease Amyloid Gene in Hereditary Cerebral Hemorrhage, Dutch Type. Science 1990, 248, 1124–1126 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- Kamino K.; Orr H. T.; Payami H.; Wijsman E. M.; Alonso M. E.; Pulst S. M.; Anderson L.; O’Dahl S.; Nemens E.; White J. A.; et al. Linkage and Mutational Analysis of Familial Alzheimer Disease Kindreds for the APP Gene Region. Am. J. Hum. Genet. 1992, 51, 998–1014. [PMC free article] [PubMed] [Google Scholar]
- Tagliavini F.; Rossi G.; Padovani A.; Magoni M.; Andora G.; Sgarzi M.; Bizzi A.; Savoiardo M.; Carella F.; Morbin M.; et al. A new βPP mutation related to hereditary cerebral haemorrhage. Alzheimer's Rep. 1999, 2, S28. [Google Scholar]
- Lopes P. E. M.; Huang J.; Shim J.; Luo Y.; Li H.; Roux B.; MacKerell A. D. Jr. Polarizable Force Field for Peptides and Proteins Based on the Classical Drude Oscillator. J. Chem. Theory Comput. 2013, 9, 5430–5449 10.1021/ct400781b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Zhang C.; Derreumaux P.; Gräslund A.; Morozova-Roche L.; Mu Y. Intrinsic Determinants of Aβ12–24 pH-Dependent Self-Assembly Revealed by Combined Computational and Experimental Studies. PLoS One 2011, 6, e24329. 10.1371/journal.pone.0024329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertino M.; Vitalis A.; Caflisch A. Disordered Binding of Small Molecules to Aβ(12–28). J. Biol. Chem. 2011, 286, 41578–41588 10.1074/jbc.M111.285957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumketner A.; Shea J.-E. Folding Landscapes of the Alzheimer Amyloid-β(12–28) Peptide. J. Mol. Biol. 2006, 362, 567–579 10.1016/j.jmb.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Ito M.; Johansson J.; Strömberg R.; Nilsson L. Unfolding of the Amyloid β-Peptide Central Helix: Mechanistic Insights from Molecular Dynamics Simulations. PLoS One 2011, 6, e17587. 10.1371/journal.pone.0017587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P. H.; Tarus B.; Derreumaux P. Familial Alzheimer A2V Mutation Reduces the Intrinsic Disorder and Completely Changes the Free Energy Landscape of the Aβ1–28 Monomer. J. Phys. Chem. B 2014, 118, 501–510 10.1021/jp4115404. [DOI] [PubMed] [Google Scholar]
- Nasica-Labouze J.; Nguyen P. H.; Sterpone F.; Berthoumieu O.; Buchete N.-V.; Coté S.; De Simone A.; Doig A. J.; Faller P.; Garcia A.; et al. Amyloid β Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015, 115, 3518–3563 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Païviö A.; Nordling E.; Kallberg Y.; Thyberg J.; Johansson J. Stabilization of Discordant Helices in Amyloid Fibril-forming Proteins. Protein Sci. 2004, 13, 1251–1259 10.1110/ps.03442404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerelius C.; Sandegren A.; Sargsyan H.; Raunak R.; Leijonmarck H.; Chatterjee U.; Fisahn A.; Imarisio S.; Lomas D. A.; Crowther D. C.; et al. α-Helix Targeting Reduces Amyloid-β Peptide Toxicity. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 9191–9196 10.1073/pnas.0810364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G.; Koo E. H. Amyloid Precursor Protein Trafficking, Processing, and Function. J. Biol. Chem. 2008, 283, 29615–29619 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; MacKerell A. D. Jr. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y.; Irie K.; Murakami K.; Ohigashi H.; Ohashi R.; Takegoshi K.; Shimizu T.; Shirasawa T. Verification of the Turn at Positions 22 and 23 of the β-Amyloid Fibrils with Italian Mutation Using Solid-state NMR. Bioorg. Med. Chem. 2005, 13, 6803–6809 10.1016/j.bmc.2005.07.071. [DOI] [PubMed] [Google Scholar]
- Berhanu W. M.; Hansmann U. H. E. Side-chain Hydrophobicity and the Stability of Aβ16–22 Aggregates. Protein Sci. 2012, 21, 1837–1848 10.1002/pro.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja A.; Ito M.; Nilsson L. Implicit Solvent Models and Stabilizing Effects of Mutations and Ligands on the Unfolding of the Amyloid β-Peptide Central Helix. J. Chem. Theory Comput. 2013, 9, 834–846 10.1021/ct300941v. [DOI] [PubMed] [Google Scholar]
- Ma B.; Nussinov R. Stabilities and Conformations of Alzheimer’s β-Amyloid Peptide Oligomers (Aβ16–22, Aβ16–35, and Aβ10–35): Sequence Effects. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 14126–14131 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumketner A.; Krone M. G.; Shea J.-E. Role of the Familial Dutch Mutation E22Q in the Folding and Aggregation of the 15–28 Fragment of the Alzheimer Amyloid-β Protein. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6027–6032 10.1073/pnas.0708193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramse G.; Dols-Perez A.; Edwards M. A.; Fumagalli L.; Gomila G. Nanoscale Measurement of the Dielectric Constant of Supported Lipid Bilayers in Aqueous Solutions with Electrostatic Force Microscopy. Biophys. J. 2013, 104, 1257–1262 10.1016/j.bpj.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R.; Sciarretta K. L.; Orgel J. P. R. O.; Meredith S. C. Evidence for Novel β-Sheet Structures in Iowa Mutant β-Amyloid Fibrils. Biochemistry 2009, 48, 6072–6084 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N.; Gillespie J. R.; Fink A. L. Why Are ″Natively Unfolded″ Proteins Unstructured Under Physiologic Conditions?. Proteins: Struct., Funct., Genet. 2000, 41, 415–427. [DOI] [PubMed] [Google Scholar]
- Åqvist J.; Luecke H.; Quiocho F. A.; Warshel A. Dipoles Localized at Helix Termini of Proteins Stabilize Charges. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 2026–2030 10.1073/pnas.88.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. III; MacKerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux G.; Harder E.; Vorobyov I. V.; Roux B.; MacKerell A. D. Jr. A Polarizable Model of Water for Molecular Dynamics Simulations of Biomolecules. Chem. Phys. Lett. 2006, 418, 245–249 10.1016/j.cplett.2005.10.135. [DOI] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935 10.1063/1.445869. [DOI] [Google Scholar]
- Durell S. R.; Brooks B. R.; Ben-Naim A. Solvent-Induced Forces between Two Hydrophilic Groups. J. Phys. Chem. 1994, 98, 2198–2202 10.1021/j100059a038. [DOI] [Google Scholar]
- Neria E.; Fischer S.; Karplus M. Simulation of Activation Free Energies in Molecular Systems. J. Chem. Phys. 1996, 105, 1902. 10.1063/1.472061. [DOI] [Google Scholar]
- Anisimov V. M.; Vorobyov I. V.; Roux B.; MacKerell A. D. Jr. Polarizable Empirical Force Field for the Primary and Secondary Alcohol Series Based on the Classical Drude Model. J. Chem. Theory Comput. 2007, 3, 1927–1946 10.1021/ct700100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jiang W.; Hardy D. J.; Phillips J. C.; MacKerell A. D. Jr.; Schulten K.; Roux B. High-Performance Scalable Molecular Dynamics Simulations of a Polarizable Force Field Based on Classical Drude Oscillators in NAMD. J. Phys. Chem. Lett. 2011, 2, 87–92 10.1021/jz101461d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle Mesh Ewald: An N•log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092 10.1063/1.464397. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593 10.1063/1.470117. [DOI] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177. 10.1063/1.467468. [DOI] [Google Scholar]
- Feller S. E.; Zhang Y.; Pastor R. W.; Brooks B. R. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613. 10.1063/1.470648. [DOI] [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Chowdhary J.; Harder E.; Lopes P. E. M.; Huang L.; MacKerell A. D. Jr.; Roux B. A Polarizable Force Field of Dipalmitoylphosphatidylcholine Based on the Classical Drude Model for Molecular Dynamics Simulations of Lipids. J. Phys. Chem. B 2013, 117, 9142–9160 10.1021/jp402860e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; et al. GROMACS 4.5: a High-throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W.; Sander C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.