Abstract
MicroRNAs (miRNAs) are short non coding RNAs aberrantly expressed in solid and hematopoietic malignancies where they play a pivotal function as post-transcriptional regulators of gene expression. Recent reports have unveiled a central role of miRNAs in multiple myeloma onset and progression and preclinical findings are progressively disclosing their potential therapeutic value as drugs or targets. In this review, we provide the basic insights of miRNA biology and function, showing how these molecules are extensively dysregulated in malignant plasma cells (PC) and related microenvironment, thus favoring clone survival and proliferation. We here describe how these critical activities have recently been evaluated to design miRNA-based therapies against multiple myeloma cells and its surrounding microenvironment.
Keywords: MicroRNAs (miRNAs), non coding RNAs, miRNA therapy, multiple myeloma (MM) bone disease
Introduction
Multiple myeloma (MM) is characterized by proliferation of malignant plasma cells (PCs) within the bone marrow (BM) and it is commonly preceded by the premalignant monoclonal gammopathy of undetermined significance (MGUS) (1). It represents about 10% of all hematologic malignancies being characterized by relevant clinical end organ damage signs that can be life threatening and/or severely affect quality of life (2). MM onset and development may be due to a B cell clone (3) that differentiates into malignant PCs in a permissive BM microenvironment (BMM). Although the actual occurrence of a pre-PC B cell clone remains elusive (see below), MM cells rely for survival and proliferation on a complex network of interactions with surrounding BMM cell components, including BM stromal cells (BMSCs), immune cells, osteoclasts (OCLs) and osteoblasts (OBLs). Genomic abnormalities of malignant PCs are classified into complex non-hyperdiploid (nHD) or hyperdiploid karyotypes (HD). The former include typical translocations such as t(4;14), t(14;16), chromosomal deletions (1q, 13q and 17p) and gains (1q), which carry a poor prognosis (4). Both nHD and HD karyotypes are observed already in MGUS, whereas additional chromosomal aberrations eventually arise during transition to asymptomatic MM and smouldering MM, involving c-MYC, p53, RAS and NF-κB pathway (2).
Treatment of MM has radically changed since the introduction of novel agents (proteasome inhibitors, PIs and immunomodulatory drugs, ImiDs) and their combination with conventional chemotherapeutics (CCs). The current treatment is based on a combination of CCs with PIs and/or ImiDs. According to patient age and performance status, this therapy may be followed by stem cell autologous transplantation. However, even with these new drugs, the disease invariably relapses and requires additional treatments. Although the median PFS and OS for MM patients have dramatically improved in recent years (5) and new generation PIs and ImiDs have been made recently available for clinicians, to date MM is still incurable. This scenario prompted investigators to explore innovative therapeutic strategies. While molecularly targeted therapies have not yet satisfied this need (6) and immunotherapy, although promising, is still in its infancy (7), and a plethora of new agents/strategies are approaching to advanced phases (7-16), the recent achievements on the role of microRNAs (miRNAs) in cancer development and progression have opened a new avenue of investigation in cancer research and therapy. Here, we provide an overview on miRNA involvement in the MM pathobiology and we review the most recent preclinical findings based on miRNA as drug or targets as potential new therapeutics for this important disease.
Overview of miRNA biology
miRNAs belong to the family of short non coding RNAs of about 18-24 nt length, that target the 3'-untranslated region (3'-UTR) of mRNAs, inhibiting protein translation, and account for approximately 1% of the non coding genome sequences in different species. Expression and targeting activity of miRNAs sharing the same seed sequence can be different and tissue specific (17). Mature miRNAs derive from a complex process, where a pri-miRNA (70–100 nt length) is transcribed by RNA pol II and cleaved in the nucleus by the ribonuclease DROSHA to originate a 60–70 nt length pre-miRNA, which is exported to the cytoplasm and undergoes a second cleavage by RNA POL III family member, DICER, to become a mature 18–24 nt length miRNA duplex. One of the 2 strands can be driven within the miRNA-containing RNA-induced silencing complex (miRISC) to target 3’-UTR of mRNAs (17). This specific activity confers to miRNAs the capability to control gene expression at a post-transcriptional level, thus regulating intracellular pathways at different levels with a spectrum of validated and putative activities that range from cell cycle to apoptosis, cell metabolism and ageing (18). The complexity of miRNA functions are far from being definitely clarified and additional findings are emerging, contributing to define the whole scenario of miRNA biology. However, such a complexity has opened the question on the role of miRNAs in the development of diseases including cancer. Indeed, it is now clear that miRNA homeostasis is deeply dysregulated in human cancers, either at the onset or along the progression of the disease (19). In MM, several studies have analyzed miRNA expression and activity in comparison with MGUS patients or healthy individuals. In the next section, we will describe the most relevant findings on this topic.
miRNA deregulation in multiple myeloma (MM)
Early studies evaluated the miRNA expression in healthy individuals, MGUS and MM patients. miRNA dysregulation has been shown to progressively increase from healthy individuals to MGUS and MM patients (20,21). Specific miRNA signatures for MGUS and MM were also determined. Among others, miR-21 and miR-106a-92 were found up-regulated both in MGUS and MM, while miR-32 and miR-17-92 were mostly up-regulated in MM. Some authors performed a similar analysis in relapsed/refractory MM patients, identifying miR-15 and -16 as down-regulated (see below), while miRs-221/222, -181a, -181b were found up-regulated. These data indicated that miRNA modulation can be involved in the transition from MGUS to MM (22).
The biological bases of differential miRNA expression can be partly related to the presence of complex chromosomal abnormalities as investigated by Lionetti et al. (23). In this study, 26 miRNAs were differentially expressed in the 5 translocation/cyclin (TC) groups, demonstrating the association between genomic aberrations with specific miRNA dysregulation. To further provide a potential clinical impact of this classification, an integrated score was built including gene expression profile (GEP) categories together with ISS and FISH classification. These data suggest a possible use of miRNA as prognostic tools to stratify MM patients in different risk classes.
Single dysregulated miRNAs in MGUS/MM may be directly involved in critical functions for malignant PCs such as cell cycle control, DNA repair and cross-talk with cell components of the surrounding microenvironment. In fact, cell cycle check-point proteins, such as p27Kip1 and p57Kip2, are suppressed by upregulation of miR-221/222 (24), while p53 is under the negative control of miR-181a/b, miR-106b-25 and miR-32, and miR-21 transcription depends on IL-6 signaling from BMSCs (25). IL-6 expression is negatively regulated by SOCS-1 that is targeted by miR-19a/b in MM (20).
As for other malignancies, the overall effect exerted by single miRNAs have led to consider up-regulated miRNAs as driver of PCs survival and expansion (oncomiRs), while down-regulated miRNAs as inhibitory signals for proliferation and progression (tumor suppressor miRNAs, TS-miRs).
Master regulators of miRNA function have been also shown as deeply dysregulated in MM. Indeed, Argonaut-2 protein (AGO-2), which is involved in most of the phases of miRNA processing until the release of mature miRNAs into the RISC complex, is up-regulated in high-risk MM samples (26). Another component of miRNA processing machinery, DICER, is progressively deregulated in MM as compared to MGUS and it is associated with reduced PFS (27). All together, these findings underpin the clinical relevance of miRNA modulation in the pathogenesis of MM.
The era of integromics: an innovative and comprehensive approach to evaluate miRNA regulation of cellular pathways
miRNA-dependent regulation of intracellular pathways is a dynamic process that implies a complex interplay among several factors. Indeed, novel technological platforms allow investigators to link data deriving from mRNA, miRNA and protein expression together with DNA copy number and SNPs data. The upcoming picture is a comprehensive view of miRNA mediated pathway modulation that goes beyond miRNA expression profiling and targeting information. In most cases the source of data to build miRNA network connections is founded on public available databases that can be interrogated by investigators. For instance, source data can be evaluated to identify critical components of the pathways, known as nodes (genes) and edges (interactions). Nodes are critical for pathway normal function as deactivation of a specific node can determine delays in pathway flow and/or its disconnection from other pathways. The importance of a node in a pathway is given by the number of connections and the type of partners connected.
Such a methodology has been applied by different authors also in MM setting, starting from pairing mRNA and miRNA expression data (i.e., transcriptional and post-transcriptional network regulators, respectively) to build up network connections and identify critical nodes for malignant PC pathways (28-31). For instance, the level of complexity can be further raised by introducing the evaluation of transcription factor expression data together with miRNA expression, which can compete for mRNA expression (32). Although these data are far from being conclusive, they disclose an innovative way to consider miRNA regulation of intracellular pathways in health and disease. The burden of information coming from the integrated analysis will require further evaluation to be successfully used as a prognostic tool and to design innovative therapeutic approaches.
Dissecting the therapeutical effects of miRNAs on multiple myeloma (MM) and related bone marrow microenvironment (BMM) components
If miRNAs retain a prominent role in MM development and progression, their enforced expression may be used as a therapeutic tool against the neoplastic PC clone and MM-dependent BMM (16,19,33-37). Indeed, the pleiotropic effects of miRNAs make them very suitable tools to design novel therapeutic strategies. The “paradigm” of miRNA based therapies was initially presented by Slack and Duchaine (38), who proposed to simply manipulate miRNA activity through replacement of suppressed miRNAs (miRNA mimic based therapy) or by inhibiting up-regulated miRNAs (antagomir approach). A consistent amount of preclinical data both in vitro and in vivo has been produced since then, showing that miRNA modulation is a feasible approach; the complexity of pathways under miRNA control has also suggested that the positive effect of miRNA-based therapies probably goes beyond the simple concept of replacement/antagonism as miRNA activity cannot be entirely predicted only on the base of known validated targets and related pathways.
Indeed, several intracellular pathways are crucial in regulating MM cell survival and proliferation. PI3K/AKT and MAPK pathways are often activated in human cancer (39-42) and play a central role in MM onset and development (6). On the other hand, miRNAs can be regulated by signaling molecules such as P53 and EGFR (43,44), thus showing the complexity of their intracellular modulating activities.
The general design of experimental studies aiming to identify potential miRNA-based therapies relies on the definition of the levels of the selected miRNA, the identification of crucial targets that support MM clone and/or MM-BMM, and finally the testing of mimics or antagomirs in vitro and in vivo to demonstrate their antitumor activity.
In the following section, we will review current available data on potential miRNA-based therapies directed towards different cellular effectors of MM microenvironment.
miRNA replacement
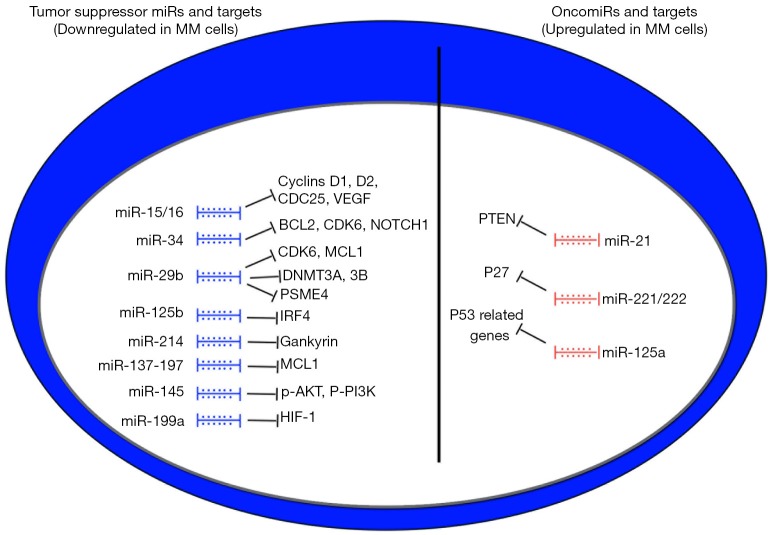
Pioneeristic studies (22) have demonstrated that replacement of down-regulated miRNAs could inhibit MM cell proliferation. MiR-15 and -16, which were initially identified as TS-miRNAs in chronic lymphocytic leukemia (CLL) (45), were found down-regulated in MM patients carrying ch13 deletion. Transfection of pre-miR-15 and -16 within MM cells inhibited DNA synthesis with about 60% reduction of proliferation. Cell cycle regulators as cyclin D1, D2 and CDC25A were suppressed by miR-15 and -16 transfection. MiR-15 and-16 restoration interfered also with NF-Kb pathway and reduced the formation of tumoral driven new vasculature (22). Indeed, it has been recently demonstrated that VEGF is a direct target of miR-15 and -16 (46) (Figure 1). Following preliminary reports of miR-34 replacement in a lung cancer model (47), this miRNA was evaluated also in MM. Di Martino et al. demonstrated that miR-34 expression is reduced in p53 mutated (p53 mut) MM cells and that enforced expression of miR-34 mimics in MM cells suppressed cell cycle regulators such as BCL2, CDK6 and NOTCH1, leading to the induction of apoptosis (48) (Figure 1). BCL2 direct targeting by miR-34 has been previously described in a lung cancer model (49). Treating SCID synth-hu mice bearing MM with lipid formulated miR-34 mimics exerted strong anti-tumor effects together with a survival prolongation as compared to controls (48,50-53). No toxicity was described with miR-34 treated mice.
Figure 1.
Dysregulated miRNAs in MM. Overview of the main dysregulated miRNAs and corresponding targets in MM cells. Blue colored miRNAs are down-regulated and exert tumor suppressor activity (TS-miRs), whereas red colored miRNAs are up-regulated and act as driver of MM cell proliferation and survival (oncomiRs). MM, multiple myeloma.
The multifaceted effects of miRNA mimics are further underlined by recent achievements on miR-29b (7,54-57). This miRNA is down-regulated in MM PCs and its restoration leads to cell cycle arrest and induction of apoptosis by targeting CDK6 and MCL1 (56,58). Amodio et al. demonstrated that miR-29b transfection suppresses DNA methylation through down-regulation of DNMT-3A and -3B, thus controlling gene expression at an epigenetic level. The demethylating activity is further potentiated by 5-azacitidine, as expected. Several reports have demonstrated that miR-29b mimics potentiate the antitumor effects of proteasome inhibitors. Bortezomib up-regulates miR-29b by Sp1 dependent feedback loop (56), while miR-29b targets PSME4 reinforcing bortezomib dependent proteasome inhibition and aggregate formation (59). Indeed, down-regulation of PSME4 leads to reduced expression of PA200, a proteasome activator. Citotoxic activity of miR-29b is paralleled by inhibition of migration of malignant PCs due to the upregulation of SOCS-1, whose promoter is demethylated by miR-29b (55) (Figure 1).
The basic idea that down-regulated miRNAs in MM PCs can be replaced with the corresponding mimics as a therapeutic tool has been also confirmed in other recent reports. MiR-125b is consistently down-regulated in MM TC2 and TC3 groups (60). The main target of miR-125b is interferon regulatory factor 4 (IRF4) that triggers c-MYC and BLIMP-1 expression, promoting tumor growth and survival. Enforced expression of miR-125b induced apoptosis and autophagic related cell death. The effects obtained in vitro were recapitulated in a xenograft model of MM, where lipid formulated miR-125b mimics exerted potent antitumor activity. These data indicate that c-MYC targeting may be an attractive strategy for miR-based therapies.
miR-214 is suppressed in MM cells and its restoration promotes cell cycle arrest and apoptosis. The mechanism seems to be related to upregulation of p53, p21 and BAX via inhibition of the oncoprotein gankyrin (61).
Anti-MM effects have been also associated to miR-137 and miR-197 (62). The cytotoxic activity of these miRNAs has been mainly related to MCL1 targeting as for miR-29b. Tumor shrinkage by lipid-formulated delivery of both miRNAs was observed in a mouse model.
miR-145 is a putative tumor suppressor miRNA in human cancer as it can interfere with cell proliferation, migration and invasiveness (63,64). IRS1 is a main target of miR-145, leading to inhibition of AKT mediated signaling (64). Indeed, in MM, miR-145 mimics inhibited p-AKT and p-PI3K, impairing proliferation and survival of MM cells. in vivo experiments have shown tumor growth inhibition in a xenograft model (65) (Figure 1).
miRNA inhibition
The current view of up-regulated miRNAs as oncomiRs has led investigators to counteract their activity by selective inhibition. Among up-regulated miRNAs, miR-21 has been long recognized as an oncogenic driver for MM (20,25). Anti-miR-21 exposed MM cells either by lentiviral transfection and miR-21 inhibitors induced the upregulation of a crucial miR-21 target, PTEN. This molecule regulates the levels of PIP3, which is the main product of PI3K and promotes AKT recruitment and activation (66). As for other cancer, PTEN is involved in MM cell proliferation both in vitro and in vivo (67) (Figure 1). Another renown MM growth promoting miRNA is represented by the cluster miR-221/222 (24,68,69), which seems relevant in the pathogenesis of poor prognosis MM groups, such as those carrying the translocation t(4;14) and the TC groups 2, 3 and 4 (69). The main target of this cluster is the cell cycle regulator p27kip1 (Figure 1), although different pathways are likely involved in the antitumor activity as shown by GEP analysis. miR-221/222 inhibitors induced cell cycle arrest and tumor shrinkage in vivo as compared to controls. miR-221 performed better than miR-222 in terms of antineoplastic effects. Based on these premises and in order to translate these results in the clinical settings, a 13-mer locked nucleic acid (LNA) form of miR-221 inhibitor was recently generated in order to increase the stability of the molecule and prolong activity in vivo. This newly designed molecule demonstrated activity and safety profile in preclinical in vivo studies and represents a promising candidate as a therapeutic tool (24,69). This inhibitor is presently under advanced preclinical investigation for translational purposes.
MiR-155 behaves as an oncomiR in different human cancers (70). As demonstrated in a lymphoma model, oncogenic activity of miR-155 can be mainly attributed to SOCS1 and C/EBP targeting, that lead to STAT3 activation. The importance of STAT3 has been long recognized in MM (6,71). A recent report described the anti-MM activity of a miR-155 antagonism therapy. Anti-miR-155 treatment inhibited proliferation, migration and colony formation in vitro in 8,226 MM cells (72). Further studies will be needed to evaluate this approach in xenograft models.
Data from miR-34 studies (48) suggested that interference with p53 regulation is a promising strategy. MiR-125a is increased in MM cells leading to suppression of p53 pathway related genes (73). This effect is potentiated by MM cell adhesion to BMSCs. miR-125a inhibitors restored p53 related TS-miR-192 and -194 and impaired tumor cell proliferation and migration while promoting cell death (Figure 1). This study suggests the possibility to integrate miRNA inhibitory approaches with miRNA mimics to maximize antitumor effect.
miR-based therapies of multiple myeloma (MM) stem cell
An interesting innovative field of miRNA research regards the miRnome of the putative MM stem cell. MM stem cells have been identified as a small fraction of the malignant clone (side population, SP) that retains the unique feature of clonogenicity and capability to recapitulate the disease. The SP is usually defined as low Hoechst 33342 positive dye at cytofluorimetric analysis (74). By using this isolation method, Du et al. have shown that miR-451 is overexpressed in the SP as compared to main population (75). When using a miR-451 inhibitor, SP cells were more susceptible to drug treatments such as bortezomib and melphalan, likely due to down-regulation of MDR1 mRNA, which is involved in drug resistance. Furthermore, investigators observed that stemness feature of SP relies on the integrity of PI3K/AKT/mTOR pathway, which is sustained by down-regulation of a miR-451 direct target, TSC1 (tuberous sclerosis 1). MiR-451 antagonism disrupted the pathway as shown by the downstream effectors p-S6 and p-4EBP1. Finally, recent data have shown that miRNA-based approaches may help reverting standard chemotherapy drug resistance. Indeed miR-221/222 inhibition was able to overcome melphalan resistance in a MM model both in vitro and in vivo (76).
miR-based therapies towards multiple myeloma (MM) microenvironment
The BMM plays a central role for MM development and progression (77). MM cells rely on cell contact and soluble signals to survive and proliferate mainly deriving from BMSCs, while an immune permissive milieu is progressively established during the transition from MGUS to symptomatic MM, allowing the tumor cell clone expansion (7). In this context, malignant PCs trigger OCL-dependent bone resorption and suppress OBL-mediated bone apposition.
Several preclinical studies have investigated how miRNA modulation can affect cell components of the BMM to restore a more physiological microenvironment, independently from direct activity against the PC clone.
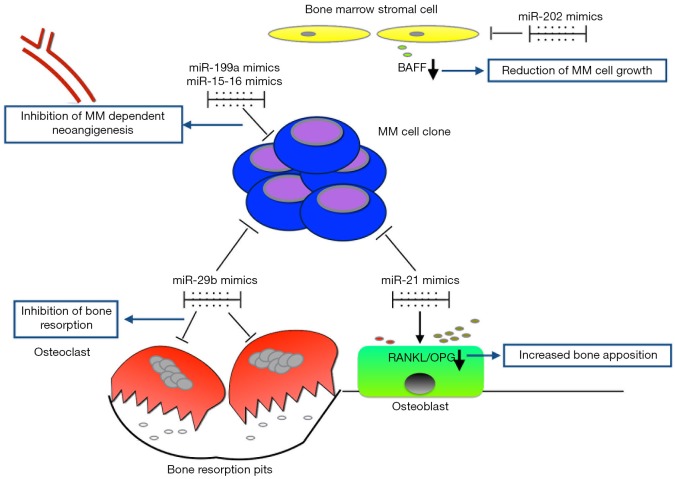
Based on the anti-MM and pro-OBL activities of miR-29b (54,56,57,78), Rossi et al. observed that this miRNA is progressively down-regulated during OCL differentiation. MiR-29b replacement within OCLs inhibited bone resorption activity even in the presence of MM cells. miR-29b treated OCLs are less responsive to RANK-L stimulation and present reduced levels of pro osteolytic enzymes. The main target of miR-29b turned to be c-FOS, which is crucial for OCL differentiation, final maturation and resorbing activity (79) (Figure 2).
Figure 2.
miRNA-targeted modulation of MM-related BMM. Examples of miR mimics (miR-29b, miR-202, miR-199a, miRs-15-16) and inhibitors (miR-21) are shown together with the main cellular targets of the BMM. miR29b promotes both MM growth and inhibition of bone resorption. miR-21 inhibitors restore the physiological RANKL/OPG ratio, favoring bone apposition. miR-202 mimics interfere with BMSC-MM cells cross talk through down-regulation of BAFF. miR-199a inhibits neoangiogenesis through suppression of HIF1, which is produced under hypoxic conditions by MM cells. miRs-15-16 inhibit neoangiogenesis by targeting VEGF. BAFF, b cell activating factor; MM, multiple myeloma; BMM, bone marrow microenvironment; OPG, osteoprotegerin; BMSC, BM stromal cell; RANKL, receptor activator nuclear factor Kappa-b ligand; VEGF, vascular endothelial growth factor; HIF1, hipoxia inducible factor 1.
As stated above, miR-21 is a critical oncomiR in MM and its expression is strictly related to BM IL-6 levels (25). Pitari et al. described that BMSCs up-regulate miR-21 in the presence of MM cells, thus contributing to malignant microenvironment. Interestingly, the authors found that miR-21 targets osteoprotegerin (OPG), a decoy receptor that neutralizes RANK-L (80). The RANK-L/OPG ratio is a critical factor to maintain bone homeostasis. In the presence of MM cells, this ratio is shifted towards RANK-L and consequent triggering of bone resorption (81). MiR-21 is likely involved in this process, as its levels are overall increased in MM milieu, contributing to the suppression of OPG. These premises paved the way to design miR-21 antagonistic strategies to revert OPG suppression and restore RANK-L/OPG to physiological levels. As expected, OPG production was re-established, while a consistent down-regulation of RANK-L was observed. To further explain this unpredicted effect of miR-21 inhibition, the authors investigated the putative molecular link between RANK-L expression and miR-21 activity. Indeed, protein inhibitor activated STAT3 (PIAS3) inhibits RANK-L expression in OBLs and it is suppressed in MM BMSCs (82). Moreover, Pitari et al. demonstrated that PIAS3 is a direct target of miR-21 and can be restored through miR-21 antagonism. The overall effect of miR-21 inhibition is both PIAS3 mediated RANK-L down-regulation and OPG restoration, leading to reduction of RANK-L/OPG ratio (80) (Figure 2).
Given the close dependence of PC clone on BMSCs support, these data suggest an alternative way to harness tumor cells. On this regard, miR-202 has been recently described as down-regulated in MM-related BMSCs and treatment with miR-202 mimics to restore its level within BMSCs, overcame growth promoting activity by reducing BCL-2 and BAFF levels (83) (Figure 2).
miRNA-based therapies can modulate the BMM also by directly targeting MM cells, without affecting proliferation rate or induction of apoptosis. Raimondi et al. explored the effects of miR-199a to halt MM related angiogenesis (84). Under hypoxic conditions, MM PCs trigger neoangiogenesis through upregulation of HIF-1a. HIF-1a is a predicted target of miR-199a, which is down-regulated in hypoxic MM cells. Treatment of MM cells with miR-199a mimics suppressed HIF-1a expression and consequent reduction of VEGF and IL-8. Taking into account also the concomitant down-regulation of bFGF, the overall effect of miR-199a treatment is the depression of MM related pro-angiogenic activity and BMSC dependent support for tumor cell proliferation and survival (Figure 2).
Taken together, these data strongly support the hypothesis that miRNA-based therapies can contribute to revert MM microenvironment through functional modulation of the BMM components such as OCL, OBLs and BMSCs. In many cases, it is possible to gain advantage from the pleiotropic effects of single miRNAs to target both MM cells and microenvironment effectors (e.g., miR-29b, miR-21 inhibitors).
The appropriate selection of such miRNAs will likely be favored by the introduction of ad hoc preclinical models that can recapitulate the disease at both MM cell and microenvironment level. The selected miRNAs will be the best candidates to be explored in next future clinical trials.
The delivery of miR-based therapies: a matter of choice
The idea to use miRNAs as a therapeutic tool prompted investigators to find the most suitable delivery system to maximize antitumoral miRNA effects with the lowest toxicity profile. The simplest method to improve the stability of miRNA molecules in vivo is represented by the chemical modifications. This is the case of the LNA-miRNA. LNA miRNAs retain a phosphorothioate (PS) backbone, which confers resistance to enzymatic degradation together with increased biodistribution within tissues (85). The efficacy of this formulation has been investigated for the first time in human subjects in a recently published trial (86), where the LNA-miR-122 antagomir was successfully administered to HCV+ patients, leading to prolonged reduction of HCV RNA levels with a favorable toxicity profile. As stated above, LNA-miR-221 has been tested in a preclinical model of MM (69).
miRNA encapsulation in different vector types is also under intensive investigation. In preclinical models, lipid miRNA formulation has been preferred (47,48). Di Martino et al. tested the efficiency of stable nucleic acid lipid particles (SNALPs) to deliver miR-34a in vivo (52). SNALPs possess high serum stability, being an attractive option to translate miR based therapies in the clinics. According to these achievements, a liposomal carried form of miR-34a is currently under investigation in a phase I clinical trial in patients affected by several malignancies, including MM (clinicaltrials.gov: A multicenter phase I study of MRX34, miRNA miR-RX34 liposomal injection NCT01829971).
Conclusions
Since the discovery of miRNAs in Caenorhabditis elegans (87) in 1993, the comprehension of the role played by these molecules has impressively progressed. Scientists reasoned that miRNA networks have further raised the level of complexity of intra/intercellular pathways, shedding a new light in the pathophysiology of the diseases. This scenario is now rapidly evolving also in the light of new investigational approaches such as the integrative genomics (32,68). As for other cancer types, the description of MM miRnome is contributing to understand the behavior of MM cells and their milieu. Taking advantage from these findings, investigators are providing fascinating insights on the use of miRNAs as a promising therapeutic tool. miRNA therapy is fast becoming a clinical reality as trials have already been conducted in HCV+ patients (86) and are currently ongoing in different cancer types (clinicaltrials.gov: A multicenter phase I study of MRX34, MIRNA miR-RX34 liposomal injection NCT01829971). MM represents an attractive target for such kind of therapy for several reasons. MM is an incurable disease, whose outcome has significantly improved with innovative therapies that target MM cells and their BMM; the mid-long term OS of MM patients prompts the identification of therapies with low toxicity profile; the pathogenesis of the disease has been extensively studied and unveiled, paving the way to innovative molecularly targeted therapies. The availability of complex preclinical platforms (50,77,88-91) represent a robust base for predicting effective therapeutic strategies and to design clinical trials for miRNA-based treatments. The results of these studies will hopefully provide clinicians of a new and selective antitumor therapy with the lowest toxicity profile.
Acknowledgements
This work has been supported by the Italian Association for Cancer Research (AIRC), PI: PT. “Special Program Molecular Clinical Oncology- 5 per mille” n. 9980, 2010/15.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Kyle RA, Therneau TM, Rajkumar SV, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med 2002;346:564-9. [DOI] [PubMed] [Google Scholar]
- 2.Morabito F, Recchia AG, Mazzone C, et al. Targeted therapy of multiple myeloma: the changing paradigm at the beginning of the new millennium. Curr Cancer Drug Targets 2012;12:743-56. [DOI] [PubMed] [Google Scholar]
- 3.Basak GW, Carrier E. The search for multiple myeloma stem cells: the long and winding road. Biol Blood Marrow Transplant 2010;16:587-94. [DOI] [PubMed] [Google Scholar]
- 4.Tassone P, Tagliaferri P, Rossi M, et al. Genetics and molecular profiling of multiple myeloma: novel tools for clinical management? Eur J Cancer 2006;42:1530-8. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, Avet-Loiseau H, Oliva S, et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J Clin Oncol 2015;33:2863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi M, Di Martino MT, Morelli E, et al. Molecular targets for the treatment of multiple myeloma. Curr Cancer Drug Targets 2012;12:757-67. [DOI] [PubMed] [Google Scholar]
- 7.Rossi M, Botta C, Correale P, et al. Immunologic microenvironment and personalized treatment in multiple myeloma. Expert Opin Biol Ther 2013;13 Suppl 1:S83-93. [DOI] [PubMed] [Google Scholar]
- 8.Manni S, Brancalion A, Tubi LQ, et al. Protein kinase CK2 protects multiple myeloma cells from ER stress-induced apoptosis and from the cytotoxic effect of HSP90 inhibition through regulation of the unfolded protein response. Clin Cancer Res 2012;18:1888-900. [DOI] [PubMed] [Google Scholar]
- 9.Neri P, Ren L, Gratton K, et al. Bortezomib-induced "BRCAness" sensitizes multiple myeloma cells to PARP inhibitors. Blood 2011;118:6368-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neri P, Ren L, Azab AK, et al. Integrin beta7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood 2011;117:6202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marra M, Salzano G, Leonetti C, et al. Nanotechnologies to use bisphosphonates as potent anticancer agents: the effects of zoledronic acid encapsulated into liposomes. Nanomedicine 2011;7:955-64. [DOI] [PubMed] [Google Scholar]
- 12.Fulciniti M, Amin S, Nanjappa P, et al. Significant biological role of sp1 transactivation in multiple myeloma. Clin Cancer Res 2011;17:6500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulciniti M, Hideshima T, Vermot-Desroches C, et al. A high-affinity fully human anti-IL-6 mAb, 1339, for the treatment of multiple myeloma. Clin Cancer Res 2009;15:7144-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasui H, Hideshima T, Hamasaki M, et al. SDX-101, the R-enantiomer of etodolac, induces cytotoxicity, overcomes drug resistance, and enhances the activity of dexamethasone in multiple myeloma. Blood 2005;106:706-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamasaki M, Hideshima T, Tassone P, et al. Azaspirane (N-N-diethyl-8,8-dipropyl-2-azaspiro [4.5] decane-2-propanamine) inhibits human multiple myeloma cell growth in the bone marrow milieu in vitro and in vivo. Blood 2005;105:4470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misso G, Zappavigna S, Castellano M, et al. Emerging pathways as individualized therapeutic target of multiple myeloma. Expert Opin Biol Ther 2013;13 Suppl 1:S95-109. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [DOI] [PubMed] [Google Scholar]
- 18.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet 2013;14:535-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagliaferri P, Rossi M, Di Martino MT, et al. Promises and challenges of MicroRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets 2012;12:838-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A 2008;105:12885-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Chen L, Barlogie B, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A 2010;107:7904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roccaro AM, Sacco A, Thompson B, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009;113:6669-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionetti M, Biasiolo M, Agnelli L, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009;114:e20-6. [DOI] [PubMed] [Google Scholar]
- 24.Di Martino MT, Gulla A, Cantafio ME, et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 2013;4:242-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007;110:1330-3. [DOI] [PubMed] [Google Scholar]
- 26.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 2007;109:2276-84. [DOI] [PubMed] [Google Scholar]
- 27.Sarasquete ME, Gutierrez NC, Misiewicz-Krzeminska I, et al. Upregulation of Dicer is more frequent in monoclonal gammopathies of undetermined significance than in multiple myeloma patients and is associated with longer survival in symptomatic myeloma patients. Haematologica 2011;96:468-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaswal V, Lutherborrow M, Ma DD, et al. Identification of microRNA-mRNA modules using microarray data. BMC Genomics 2011;12:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biasiolo M, Forcato M, Possamai L, et al. Critical analysis of transcriptional and post-transcriptional regulatory networks in multiple myeloma. Pac Symp Biocomput 2010:397-408. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Li J, Tsykin A. Discovery of functional miRNA-mRNA regulatory modules with computational methods. J Biomed Inform 2009;42:685-91. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu W, Xu Y, et al. Identification of Subtype Specific miRNA-mRNA Functional Regulatory Modules in Matched miRNA-mRNA Expression Data: Multiple Myeloma as a Case. Biomed Res Int 2015;2015:501262. [DOI] [PMC free article] [PubMed]
- 32.Di Martino MT, Guzzi PH, Caracciolo D, et al. Integrated analysis of microRNAs, transcription factors and target genes expression discloses a specific molecular architecture of hyperdiploid multiple myeloma. Oncotarget 2015;6:19132-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi M, Amodio N, Di Martino MT, et al. MicroRNA and multiple myeloma: from laboratory findings to translational therapeutic approaches. Curr Pharm Biotechnol 2014;15:459-67. [DOI] [PubMed] [Google Scholar]
- 34.Rossi M, Amodio N, Di Martino MT, et al. From target therapy to miRNA therapeutics of human multiple myeloma: theoretical and technological issues in the evolving scenario. Curr Drug Targets 2013;14:1144-9. [DOI] [PubMed] [Google Scholar]
- 35.Amodio N, Di Martino MT, Neri A, et al. Non-coding RNA: a novel opportunity for the personalized treatment of multiple myeloma. Expert Opin Biol Ther 2013;13 Suppl 1:S125-37. [DOI] [PubMed] [Google Scholar]
- 36.Lionetti M, Agnelli L, Lombardi L, et al. MicroRNAs in the pathobiology of multiple myeloma. Curr Cancer Drug Targets 2012;12:823-37. [DOI] [PubMed] [Google Scholar]
- 37.Di Martino MT, Amodio N, Tassone P, et al. Functional Analysis of microRNA in Multiple Myeloma. Methods Mol Biol 2015. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 38.Duchaine TF, Slack FJ. RNA interference and micro RNA-oriented therapy in cancer: rationales, promises, and challenges. Curr Oncol 2009;16:61-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martz CA, Ottina KA, Singleton KR, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal 2014;7:ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter PS, Sarosiek KA, Lin KH, et al. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci Signal 2014;7:ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J. Signaling pathways in HPV-associated cancers and therapeutic implications. Rev Med Virol 2015;25 Suppl 1:24-53. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Zhang XD, Proud C. Dissecting the signaling pathways that mediate cancer in PTEN and LKB1 double-knockout mice. Sci Signal 2015;8:pe1. [DOI] [PubMed] [Google Scholar]
- 43.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 2009;106:3207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou CD, Zhao WM, Wang XN, et al. MicroRNA-107: a novel promoter of tumor progression that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Oncotarget 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A 2008;105:5166-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun CY, She XM, Qin Y, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013;34:426-35. [DOI] [PubMed] [Google Scholar]
- 47.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010;70:5923-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Martino MT, Leone E, Amodio N, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res 2012;18:6260-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res 2012;72:5576-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calimeri T, Battista E, Conforti F, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia 2011;25:707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scognamiglio I, Di Martino MT, Campani V, et al. Transferrin-conjugated SNALPs encapsulating 2'-O-methylated miR-34a for the treatment of multiple myeloma. Biomed Res Int 2014;2014:217365. [DOI] [PMC free article] [PubMed]
- 52.Di Martino MT, Campani V, Misso G, et al. In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS One 2014;9:e90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misso G, Di Martino MT, De Rosa G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 2014;3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amodio N, Rossi M, Raimondi L, et al. miR-29s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015;6:12837-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amodio N, Bellizzi D, Leotta M, et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle 2013;12:3650-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amodio N, Di Martino MT, Foresta U, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis 2012;3:e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amodio N, Leotta M, Bellizzi D, et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget 2012;3:1246-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang YK, Wang H, Leng Y, et al. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun 2011;414:233-9. [DOI] [PubMed] [Google Scholar]
- 59.Jagannathan S, Vad N, Vallabhapurapu S, et al. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia 2015;29:727-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morelli E, Leone E, Cantafio ME, et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia 2015;29:2173-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misiewicz-Krzeminska I, Sarasquete ME, Quwaider D, et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 2013;98:640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Li F, Saha MN, et al. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin Cancer Res 2015;21:2399-411. [DOI] [PubMed] [Google Scholar]
- 63.Bandopadhyay M, Banerjee A, Sarkar N, et al. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer 2014;14:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Hu C, Cheng J, et al. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun 2014;446:1255-60. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Yan W, Bai Y, et al. Synthetic miR-145 mimic inhibits multiple myeloma cell growth in vitro and in vivo. Oncol Rep 2015;33:448-56. [DOI] [PubMed] [Google Scholar]
- 66.Carnero A, Blanco-Aparicio C, Renner O, et al. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 2008;8:187-98. [DOI] [PubMed] [Google Scholar]
- 67.Leone E, Morelli E, Di Martino MT, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res 2013;19:2096-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calura E, Bisognin A, Manzoni M, et al. Disentangling the microRNA regulatory milieu in multiple myeloma: integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Di Martino MT, Gulla A, Gallo Cantafio ME, et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One 2014;9:e89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Czyzyk-Krzeska MF, Zhang X. MiR-155 at the heart of oncogenic pathways. Oncogene 2014;33:677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bommert K, Bargou RC, Stuhmer T. Signalling and survival pathways in multiple myeloma. Eur J Cancer 2006;42:1574-80. [DOI] [PubMed] [Google Scholar]
- 72.Feng M, Luo X, Gu C, et al. Seed targeting with tiny anti-miR-155 inhibits malignant progression of multiple myeloma cells. J Drug Target 2015;23:59-66. [DOI] [PubMed] [Google Scholar]
- 73.Leotta M, Biamonte L, Raimondi L, et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol 2014;229:2106-16. [DOI] [PubMed] [Google Scholar]
- 74.Goodell MA, Brose K, Paradis G, et al. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med 1996;183:1797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du J, Liu S, He J, et al. MicroRNA-451 regulates stemness of side population cells via PI3K/Akt/mTOR signaling pathway in multiple myeloma. Oncotarget 2015;6:14993-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gulla A, Di Martino MT, Gallo Cantafio ME, et al. A 13 mer LNA-i-miR-221 inhibitor restores drug-sensitivity in melphalan-refractory multiple myeloma cells. Clin Cancer Res 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tassone P, Tagliaferri P, Rossi M, et al. Challenging the current approaches to multiple myeloma-related bone disease: from bisphosphonates to target therapy. Curr Cancer Drug Targets 2009;9:854-70. [DOI] [PubMed] [Google Scholar]
- 78.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci 2014;71:4747-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossi M, Pitari MR, Amodio N, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol 2013;228:1506-15. [DOI] [PubMed] [Google Scholar]
- 80.Pitari MR, Rossi M, Amodio N, et al. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bone marrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget 2015;6:27343-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giuliani N, Bataille R, Mancini C, et al. Myeloma cells induce imbalance in the osteoprotegerin/osteoprotegerin ligand system in the human bone marrow environment. Blood 2001;98:3527-33. [DOI] [PubMed] [Google Scholar]
- 82.Hikata T, Takaishi H, Takito J, et al. PIAS3 negatively regulates RANKL-mediated osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblasts. Blood 2009;113:2202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen X, Guo Y, Yu J, et al. miRNA-202 in bone marrow stromal cells affects the growth and adhesion of multiple myeloma cells by regulating B cell-activating factor. Clin Exp Med 2015. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 84.Raimondi L, Amodio N, Di Martino MT, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget 2014;5:3039-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geary RS, Yu RZ, Levin AA. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr Opin Investig Drugs 2001;2:562-73. [PubMed] [Google Scholar]
- 86.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013;368:1685-94. [DOI] [PubMed] [Google Scholar]
- 87.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [DOI] [PubMed] [Google Scholar]
- 88.Tassone P, Neri P, Burger R, et al. Mouse models as a translational platform for the development of new therapeutic agents in multiple myeloma. Curr Cancer Drug Targets 2012;12:814-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tassone P, Neri P, Kutok JL, et al. A SCID-hu in vivo model of human Waldenstrom macroglobulinemia. Blood 2005;106:1341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tassone P, Neri P, Carrasco DR, et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood 2005;106:713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu D, Wang Z, Zhao JJ, et al. The Cyclophilin A-CD147 complex promotes the proliferation and homing of multiple myeloma cells. Nat Med 2015;21:572-80. [DOI] [PMC free article] [PubMed] [Google Scholar]