Abstract
Previous studies have showed that the VQ motif–containing proteins in Arabidopsis thaliana and Oryza sativa play an important role in plant growth, development, and stress responses. However, little is known about the functions of the VQ genes in Brassica rapa (Chinese cabbage). In this study, we performed genome-wide identification, characterization, and expression analysis of the VQ genes in Chinese cabbage, especially under adverse environment. We identified 57 VQ genes and classified them into seven subgroups (I–VII), which were dispersedly distributed on chromosomes 1 to 10. The expansion of these genes mainly contributed to segmental and tandem duplication. Fifty-four VQ genes contained no introns and 50 VQ proteins were less than 300 amino acids in length. Quantitative real-time PCR showed that the VQ genes were differentially expressed in various tissues and during different abiotic stresses and plant hormone treatments. This study provides a comprehensive overview of Chinese cabbage VQ genes and will benefit the molecular breeding for resistance to stresses and disease, as well as further studies on the biological functions of the VQ proteins.
Keywords: VQ motif-containing proteins, Chinese cabbage, expression patterns
1. Introduction
To survive adverse environmental conditions, plants have evolved a wide range of complex mechanisms to respond to external stimuli [1]. Such responses are controlled by a complex network regulated by transcription factors (TFs) and other cofactors; though the cofactors do not bind DNA like TFs, they could interact with TFs to co-regulate plant transcriptional machinery in response to the surrounding environment [2].
Over the past several years, plant-specific VQ motif–containing proteins, which were named after the highly conserved amino acid sequence “FxxxVQxL/F/VTG”, were found in many monocotyledon and dicotyledon plants [3,4,5,6]. VQ motif–containing proteins can interact with the WRKY TFs [3] and play many important roles in plant growth and development. For example, the N-terminal peptide of AtVQ8, predicted to be a chloroplast targeting signal and mutation in this gene, results in pale-green and stunted-growth phenotypes [3]. IKU1, also named AtVQ14, is expressed in the endosperm during the early stages of seed development and directly regulates endosperm and seed growth [7]. AtVQ29 is expressed at a higher level in stem than root, rosette leaf, flower, and silique, and its over-expression reduces the hypocotyl growth under the far-red and low intensity of white light conditions [8]. In soybean, GmVQ1, -6 and -53 are highly expressed during seed development [5].
VQ proteins also have a vital function in resistance to abiotic and biotic stresses. AtVQ9 was reported to act antagonistically with WRKY8 to mediate responses to salt stress and decrease the DNA-binding activity of WRKY8 [9]. AtCaMBP25 (AtVQ15), a novel calmodulin-binding protein, functions as a negative regulator of plant responses to osmotic stress [10]. MKS1, another VQ motif protein (AtVQ21), acts as a substrate for MAP kinase 4 (MPK4) which functions as a regulator of pathogen defense responses, and MKS1 was also found to form complexes with WRKY25 and WRKY33 [11,12]. Nuclear-encoded sigma factor binding protein1, SIB1 and SIB2, which were renamed AtVQ23 and AtVQ16, respectively, can be rapidly and strongly induced by pathogens [13,14] and were found to recognize the C-terminal WRKY domain and stimulate the DNA-binding activity of WRKY33 [14]. Some AtVQ proteins were phosphorylated by mitogen-activated protein kinases (MAPKs), MPK3 and MPK6, and most of these MPK3/6-targeted VQ proteins interacted with WRKY TFs to regulate plant immune responses [15,16].
At present, 34, 39, 74, and 18 VQ genes were identified in Arabidopsis thaliana [3], Oryza sativa [4], Glycine max [5], and Vitis vinifera L. [6], respectively. Chinese cabbage (Brassica rapa L. ssp. pekinensis), an important vegetable crop known for its high nutritional value, is widely cultivated in Asia. However, to our knowledge, the VQ gene family from Chinese cabbage has not been characterized in detail.
In this study, we performed a genome-wide bioinformatics analysis of the VQ motif–containing proteins, including genome locations, evolutionary divergence, and gene structure. In addition, expression patterns of these genes were analyzed by quantitative real-time PCR (qRT-PCR) in different tissues and in response to abiotic stresses and hormone treatments. The detailed information provided in this study will facilitate further research on functional characterization of the VQ genes in Chinese cabbage.
2. Results
2.1. Identification and Sequence Analysis of VQ Genes in Chinese Cabbage
A total of 57 genes encoding highly conserved VQ motif–containing proteins were identified in Chinese cabbage and the sequences were downloaded from the Brassica database [17] (Tables S1 and S2). All the VQ genes were assigned specific names according to their A. thaliana orthologs [18] (Table 1), which were determined based on the instructions of Gramene [19]. If two or more Chinese cabbage genes had the same homologous gene in A. thaliana, one additional number was added after their specific name to distinguish them [20]. For instances, Bra007265, Bra014675, and Bra014674 were homologs of AtVQ23, so they were named BrVQ23-1, BrVQ23-2, and BrVQ23-3, respectively. Therefore, of the 57 BrVQs, 56 putative BrVQs were renamed based on the sequence similarity to 29 AtVQ proteins, and the remaining one (Bra006328), whose ortholog was AT5G14640 (shaggy-like kinase 13), was also identified as a BrVQ protein and renamed BrVQ35. Subsequent sequence analysis of these 57 BrVQ genes showed that the CDS ranges from 282bp to 1707bp and the predicted protein lengths vary in size from 93 to 568 amino acids (Table 1). The majority of the proteins (50/57; 87.7%) contain 300 amino acids or less, whereas two proteins (3.5%), BrVQ14-1 and BrVQ35, were more than 400 amino acids. This result was similar to previous studies in Arabidopsis [3] and rice [4], where 85.3% and 89.7% of VQ proteins contain less than 300 amino acids, respectively. Additionally, sequence analysis showed that the molecular weight of the BrVQ proteins ranged from 10.4 to 63.0 kDa and the theoretical isoelectric point (pI) from 4.67 to 10.53 (Table 1).
Table 1.
Properties of the Chinese cabbage VQ genes and proteins.
| Gene Name | Gene Locus | Chr. No. | Strand Direction | Location | CDS | Protein | ||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | Mol.Wt. (KDa) | pI | ||||||
| BrVQ1-1 | Bra025998 | A06 | − | 6,588,703–6,588,999 | 297 | 98 | 10.81 | 4.75 |
| BrVQ1-2 | Bra016616 | A08 | + | 19,301,503–19,301,796 | 294 | 97 | 10.92 | 5.13 |
| BrVQ3-1 | Bra025892 | A06 | − | 8,759,573–8,760,265 | 693 | 230 | 25.07 | 8.67 |
| BrVQ3-2 | Bra012276 | A07 | + | 8,889,633–8,890,172 | 540 | 179 | 19.29 | 5.1 |
| BrVQ4 | Bra030082 | A07 | + | 6,712,561–6,713,295 | 735 | 244 | 26.74 | 9.66 |
| BrVQ5 | Bra035492 | A08 | − | 7,791,236–7,791,901 | 666 | 221 | 25.40 | 6.58 |
| BrVQ8 | Bra033934 | A02 | − | 108,00,121–10,800,534 | 414 | 137 | 15.36 | 10.19 |
| BrVQ9-1 | Bra035028 | A07 | − | 21,850,607–21,851,491 | 885 | 294 | 31.58 | 10.12 |
| BrVQ9-2 | Bra008356 | A02 | − | 14,998,946–14,999,815 | 870 | 289 | 31.19 | 10.39 |
| BrVQ10-1 | Bra008359 | A02 | + | 15,035,463–15,035,777 | 315 | 104 | 11.61 | 5.83 |
| BrVQ10-2 | Bra003642 | A07 | − | 14,203,152–14,203,469 | 318 | 105 | 11.69 | 5.01 |
| BrVQ10-3 | Bra035035 | A07 | + | 21,882,989–21,883,270 | 282 | 93 | 10.44 | 4.67 |
| BrVQ11-1 | Bra035182 | A07 | + | 22,479,008–22,479,511 | 504 | 167 | 18.96 | 7.96 |
| BrVQ11-2 | Bra003566 | A07 | − | 13,824,994–13,825,515 | 522 | 173 | 19.66 | 9.69 |
| BrVQ11-3 | Bra008473 | A02 | + | 15,858,716–15,859,210 | 495 | 164 | 18.79 | 8.74 |
| BrVQ12 | Bra039937 | A09 | + | 31,714,821–31,715,237 | 417 | 138 | 16.09 | 9.66 |
| BrVQ14-1 | Bra023004 | A03 | + | 8,183,495–8,184,808 | 1314 | 437 | 48.10 | 8.58 |
| BrVQ14-2 | Bra017329 | A04 | + | 15,346,657–15,347,685 | 1029 | 342 | 37.24 | 10.53 |
| BrVQ14-3 | Bra005358 | A05 | − | 5,012,814–5,014,478 | 1185 | 394 | 43.13 | 10.09 |
| BrVQ15 | Bra016956 | A04 | − | 17,404,492–17,405,181 | 690 | 229 | 24.84 | 7.87 |
| BrVQ16-1 | Bra000216 | A03 | + | 9,940,673–9,941,098 | 426 | 141 | 15.57 | 8.91 |
| BrVQ16-2 | Bra004604 | A05 | + | 1,001,763–1,002,185 | 423 | 140 | 15.29 | 4.89 |
| BrVQ18-1 | Bra004825 | A05 | + | 1,984,033–1,984,575 | 543 | 180 | 20.11 | 9.24 |
| BrVQ18-2 | Bra037658 | A04 | + | 18,293,805–18,294,347 | 543 | 180 | 19.98 | 9.33 |
| BrVQ19-1 | Bra027262 | A05 | + | 20,006,387–20,007,064 | 678 | 225 | 24.21 | 9.44 |
| BrVQ19-2 | Bra021096 | A01 | + | 23,997,911–23,998,561 | 651 | 216 | 23.49 | 9.33 |
| BrVQ20 | Bra037588 | A01 | + | 22,145,216–22,146,079 | 864 | 287 | 30.28 | 6.54 |
| BrVQ21-1 | Bra037569 | A01 | + | 22,005,523–22,006,176 | 654 | 217 | 22.73 | 6.05 |
| BrVQ21-2 | Bra022345 | A05 | + | 182,87,618–18,288,268 | 651 | 216 | 23.06 | 6.29 |
| BrVQ21-3 | Bra001716 | A03 | − | 18,018,055–18,018,699 | 645 | 214 | 22.95 | 6.22 |
| BrVQ22-1 | Bra023849 | A01 | + | 2,0411,389–20,411,979 | 591 | 196 | 20.61 | 9.69 |
| BrVQ22-2 | Bra041035 | Scaffold000402 | − | 11,806–12,390 | 585 | 194 | 20.40 | 9.89 |
| BrVQ23-1 | Bra007265 | A09 | − | 28,278,350–28,278,799 | 450 | 149 | 16.52 | 4.88 |
| BrVQ23-2 | Bra014675 | A04 | + | 2,158,610–2,159,095 | 486 | 161 | 18.10 | 5.1 |
| BrVQ23-3 | Bra014674 | A04 | + | 2,155,783–2,156,247 | 465 | 154 | 17.35 | 5.17 |
| BrVQ24-1 | Bra007279 | A09 | + | 28,339,520–28,340,206 | 687 | 228 | 23.79 | 6.59 |
| BrVQ24-2 | Bra014665 | A04 | − | 2,098,440–2,099,141 | 702 | 233 | 24.57 | 8.05 |
| BrVQ25-1 | Bra007373 | A09 | + | 28,879,454–28,879,999 | 546 | 181 | 20.12 | 6.19 |
| BrVQ25-2 | Bra014594 | A04 | − | 1,651,117–1,651,650 | 534 | 177 | 19.56 | 6.7 |
| BrVQ26-1 | Bra007505 | A09 | + | 29,533,410–29,533,874 | 465 | 154 | 17.58 | 7.02 |
| BrVQ26-2 | Bra014514 | A04 | − | 1,142,901–1,143,341 | 441 | 146 | 16.68 | 8 |
| BrVQ26-3 | Bra003400 | A07 | + | 13,026,327–13,029,056 | 846 | 281 | 32.87 | 7.16 |
| BrVQ27 | Bra039565 | A01 | − | 11,929,165–11,929,719 | 555 | 184 | 19.83 | 9.76 |
| BrVQ28 | Bra013438 | A01 | + | 5,666,359–5,666,979 | 621 | 206 | 23.15 | 5.37 |
| BrVQ29-1 | Bra010608 | A08 | + | 15,506,771–15,507,115 | 345 | 114 | 12.72 | 9.05 |
| BrVQ29-2 | Bra017849 | A03 | + | 30,916,676–30,917,020 | 345 | 114 | 12.84 | 9.05 |
| BrVQ30-1 | Bra010666 | A08 | + | 15,875,832–15,876,719 | 888 | 295 | 32.18 | 6.18 |
| BrVQ30-2 | Bra011838 | A01 | − | 282,249–283,121 | 873 | 290 | 31.74 | 7.94 |
| BrVQ31 | Bra005995 | A03 | + | 1,534,982–153,5506 | 525 | 174 | 19.11 | 9.37 |
| BrVQ32-1 | Bra022063 | A02 | + | 18,984,720–18,985,412 | 693 | 230 | 25.41 | 10.26 |
| BrVQ32-2 | Bra024996 | A06 | − | 24,596,158–24,596,844 | 687 | 228 | 25.12 | 9.99 |
| BrVQ33-1 | Bra003032 | A10 | + | 5,893,364–5,894,047 | 684 | 227 | 25.28 | 9.99 |
| BrVQ33-2 | Bra022675 | A02 | − | 8,093,063–8,093,788 | 726 | 241 | 26.93 | 9.75 |
| BrVQ34-1 | Bra024362 | A06 | + | 15,221,647–15,222,738 | 1092 | 363 | 39.11 | 5.76 |
| BrVQ34-2 | Bra037806 | A09 | + | 3,657,352–3,658,311 | 960 | 319 | 34.26 | 5.94 |
| BrVQ34-3 | Bra031876 | A02 | + | 27,356,488–27,357,465 | 978 | 325 | 35.03 | 6.65 |
| BrVQ35 | Bra006328 | A03 | − | 3,034,196–3,038,533 | 1707 | 568 | 62.99 | 8.89 |
2.2. Phylogenetic Tree, Gene Structure, and Conserved Domains Analysis in Chinese Cabbage
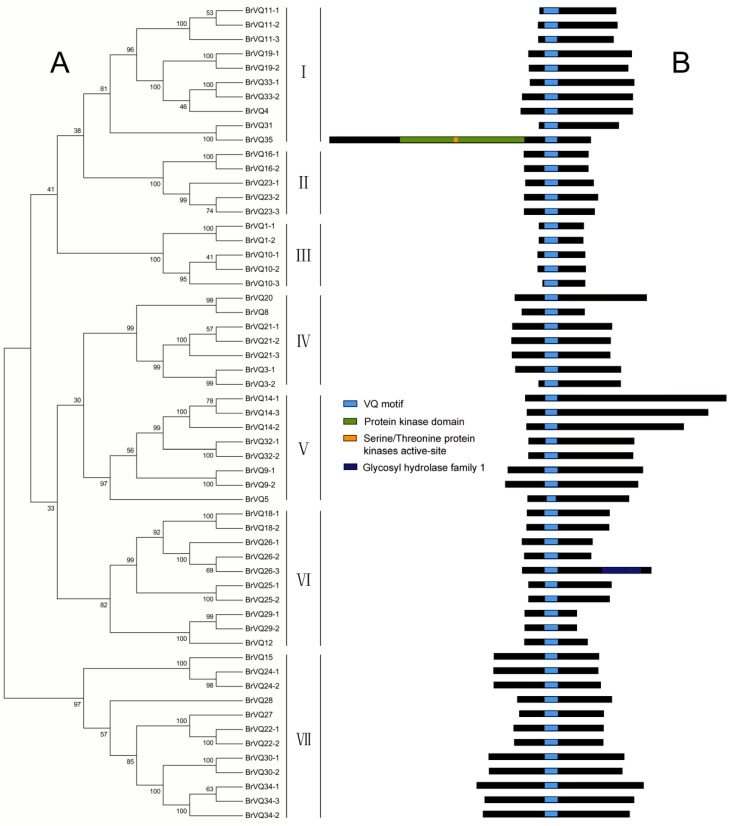
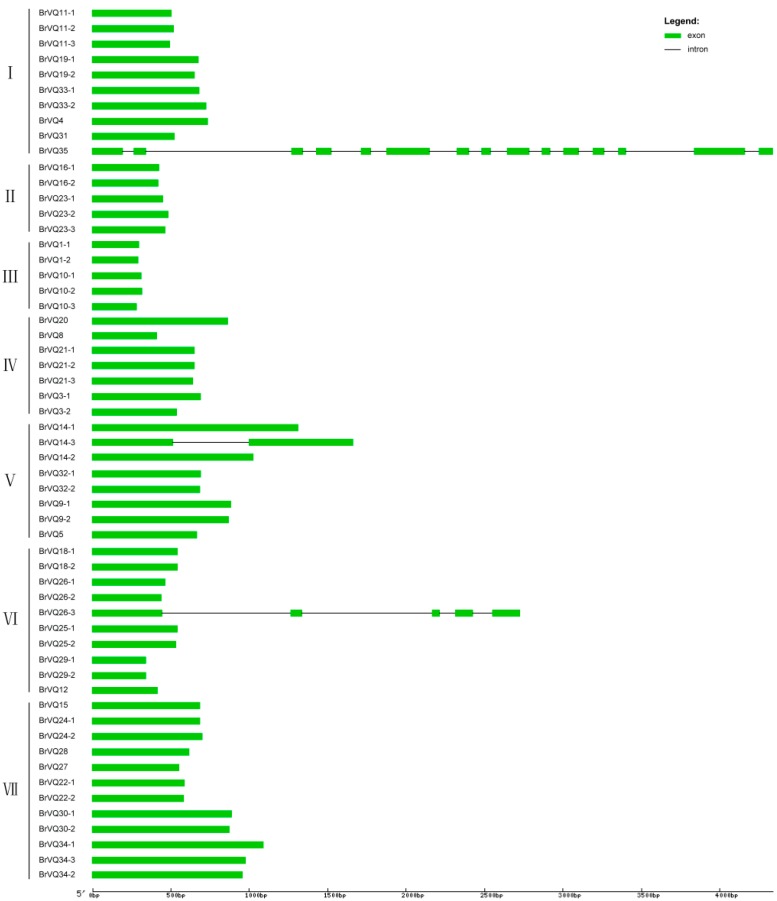
A phylogenetic tree was constructed on the basis of the full-length BrVQ protein sequences using the neighbor-joining method. The 57 BrVQ proteins can be divided into seven subgroups, with 10 proteins in subgroup I, five each in II and III, seven in IV, eight in V, 10 in VI, and 12 proteins in subgroup VII, respectively (Figure 1A). In addition, a large number of homologous genes have bootstrap value and alignment identity of more than 70% (Figure 1A, Table S3), implying that the putative BrVQ homologous genes have highly similar sequences. Moreover, the gene structure analysis showed that most BrVQ genes (54 genes; 94.7%) had no intron, whereas only three genes (BrVQ14-3, BrVQ26-3, and BrVQ35) contained one, four and 14 introns, respectively (Figure 2). The finding is consistent with a previous study [8] where the authors found as many as 30 genes in Arabidopsis and 37 genes in rice with no intron. We further performed motif analysis using the PlantsP database and found all BrVQ proteins contain a conserved VQ motif (Figure 1B). The same VQ motif (motif 1) was also detected in all BrVQ proteins when we did an independent analysis using the online tool MEME (Figure 3). By MEME analysis, we also found nine other motifs in the BrVQ proteins, including motif 2 in 19 BrVQ proteins, motif 3 in nine BrVQ proteins, motif 4 in five BrVQ proteins, motif 5 in seven BrVQ proteins, motif 6 in three BrVQ proteins, motif 7 in 10 BrVQ proteins, motif 8 in seven BrVQ proteins, motif 9 in four BrVQ proteins, and motif 10 in three BrVQ proteins, respectively (Figure 3).
Figure 1.
Phylogenetic tree and conserved domain analysis in Chinese cabbage. (A) The phylogenetic tree was determined in MEGA5 using the neighbor-joining method with 1000 bootstrap replicates. Based on the clustering of the VQ motif–containing proteins, we classified the proteins into seven groups from subgroup I–VII; (B) Domain was analyzed by searching the PlantsP database.
Figure 2.
Intron and exon structure of the VQ genes in Chinese cabbage. The majority of the BrVQ genes only have one exon, except BrVQ14-3, BrVQ26-3, and BrVQ35, which have one, four, and 14 introns, respectively.
Figure 3.
Motif analysis of the VQ proteins in Chinese cabbage. Distribution of the BrVQ conserved motifs in Chinese cabbage was analyzed by the online tool MEME.
2.3. Multiple Sequence Alignment and Motif Analysis
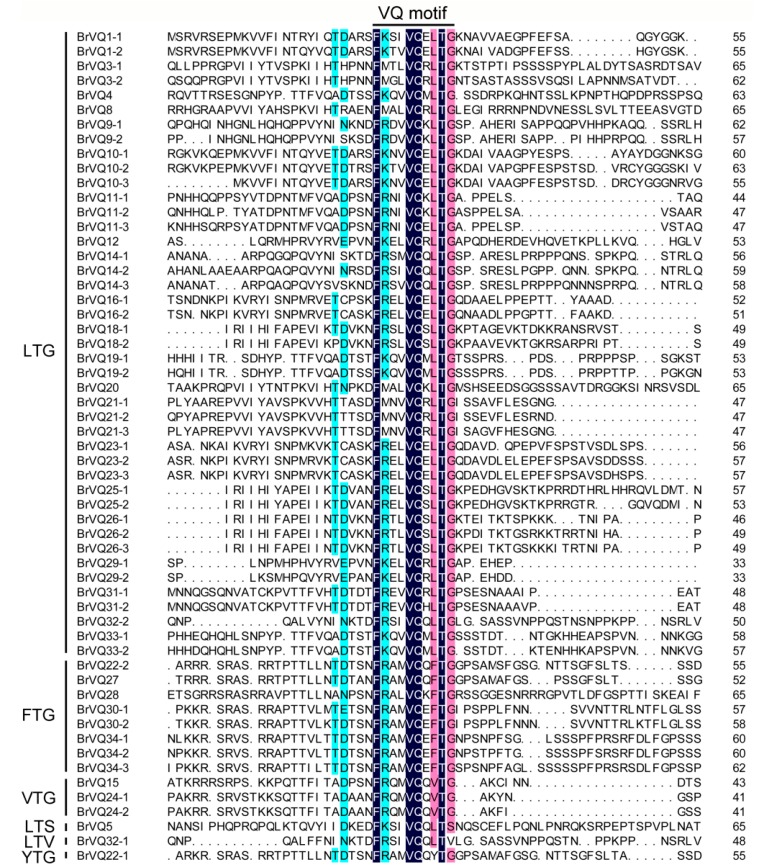
Multiple sequence alignment was constructed based on the types of B. rapa VQ domain proteins (Figure 4). In previous studies, six types of AtVQ proteins (LTG, LTS, LTD, FTG, VTG, YTG) [3] and four types of OsVQ proteins (ITG, LTG, VTG, FTG) [4] were identified. In our study, six types of VQ motifs, including FxxxVQxLTG (43/57), FxxxVQxFTG (8/57), FxxxVQxVTG (3/57), FxxxVQxLTS (1/57), FxxxVQxLTV (1/57), and FxxxVQxYTG (1/57), were identified in Chinese cabbage (Figure 4). Compared to the AtVQ and OsVQ proteins, no BrVQ protein contained the FxxxVQxLTD, FxxxVHxVTG, or FxxxVQxITG motifs, while a unique and conserved sequence “FxxxVQxLTV” was found in the BrVQ32-1 protein.
Figure 4.
Multiple sequence alignment of the VQ proteins in Chinese cabbage. The sequences were aligned using the DNAMAN software. The highly conserved motif is FxxxVQxLTG.
2.4. Chromosome Mapping and Syntenic Analysis of VQ Genes in B. rapa
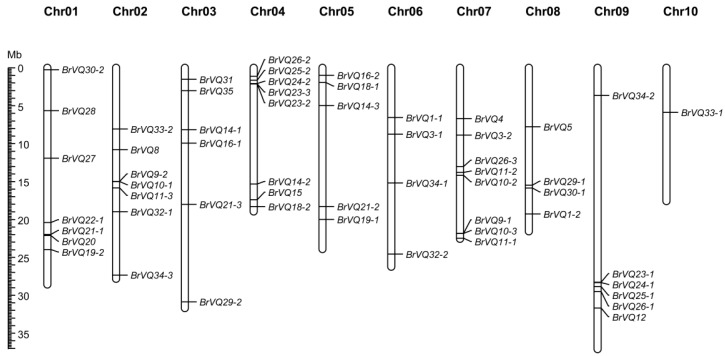
We mapped the physical locations of the BrVQ genes on 10 chromosomes of B.rapa except one gene, BrVQ22-2, which was located on Scaffold000402 (Table 1, Figure 5). Chromosomes 4 and 7 have the highest number of BrVQ genes (eight genes each). Chromosomes 1, 2, 3, 9, 5, 6, and 8 contain seven, seven, six, six, five, four, and four VQ genes, respectively, whereas chromosome 10 harbors the fewest (only one gene). To get a better understanding of the BrVQ gene evolution mechanism, we searched for the syntenic genes and possible BrVQ gene duplication events between A. thaliana and B.rapa with the BRAD program [17,21]. The results suggested that a total of 53 BrVQ genes derived from 13 blocks of seven tPCK (translocation Proto-Calepineae Karyotype) chromosomes of the ancestor, respectively, and were distributed on three subgenomes (LF, MF1, and MF2, which stands for less fractionized, more fractionized 1, and more fractionized 2, respectively), including 21 genes on LF, 23 genes on MF1, and 12 genes on MF2 (Table 2). A syntenic relationship between the 53 BrVQ genes and the 28 AtVQ genes was also detected. However, four genes (BrVQ3-1, BrVQ3-2, BrVQ22-2, and BrVQ35) in the B.rapa genome had no syntenic relationship with any A.thaliana VQ gene. On the other hand, six AtVQ genes (-2, -3, -6, -7, -13, and -17) were not in synteny with any B.rapa VQ genes. On average, for each AtVQ gene, there were one to three copies of a BrVQ gene. Seven BrVQ loci (the term “locus” instead of “gene” is used here according to the recommendations by Krishnamurthy et al. [22]) maintained three copies whereas other BrVQ loci maintained either a single copy (13 loci) or two copies (11 loci). Additionally, a total of 41 BrVQ genes were detected to have counterparts on segmental duplication, with every member of the segmentally duplicated genes dispersedly distributed on a different chromosome except for four genes (BrVQ10-2/10-3, BrVQ11-1/11-2) (Figure 5). Interestingly, one tandemly duplicated gene (BrVQ23-2/23-3) was found on chromosome 4 (Table 2, Figure 5) with a BrVQ23-2 protein sharing 83% similarity with BrVQ23-3 (Table S3).
Figure 5.
The distribution of the VQ genes of Chinese cabbage on 10 chromosomes. The chromosome number is indicated at the top of each chromosome. BrVQ gene numbers are shown on the right of each chromosome.
Table 2.
Syntenic VQ genes between Arabidopsis and Chinese cabbage.
| tPCK Chr a | Block | Arabidopsis Gene | Chinese Cabbage Gene | ||
|---|---|---|---|---|---|
| LF b | MF1 c | MF2 c | |||
| tPCK1 | A | AtVQ1(AT1G17147) | BrVQ1-1 | BrVQ1-2 | – |
| – | – | – | BrVQ3-1 | – | – |
| – | – | – | – | – | BrVQ3-2 |
| tPCK1 | B | AtVQ4(AT1G28280) | – | – | BrVQ4 |
| tPCK1 | B | AtVQ5(AT1G32585) | – | BrVQ5 | – |
| tPCK6 | E | AtVQ8(AT1G68450) | – | BrVQ8 | – |
| tPCK6 | E | AtVQ9(AT1G78310) | BrVQ9-1 | BrVQ9-2 | – |
| tPCK6 | E | AtVQ10(AT1G78410) | BrVQ10-3 | BrVQ10-1 | BrVQ10-2 |
| tPCK6 | E | AtVQ11(AT1G80450) | BrVQ11-1 | BrVQ11-3 | BrVQ11-2 |
| tPCK3 | I | AtVQ12(AT2G22880) | BrVQ12 | – | – |
| tPCK3 | J | AtVQ14(AT2G35230) | BrVQ14-3 | BrVQ14-2 | BrVQ14-1 |
| tPCK3 | J | AtVQ15(AT2G41010) | – | BrVQ15 | – |
| tPCK3 | J | AtVQ16(AT2G41180) | BrVQ16-2 | – | BrVQ16-1 |
| tPCK3 | J | AtVQ18(AT2G44340) | BrVQ18-1 | BrVQ18-2 | – |
| tPCK2 | F | AtVQ19(AT3G15300) | BrVQ19-1 | BrVQ19-2 | – |
| tPCK2 | F | AtVQ20(AT3G18360) | – | BrVQ20 | – |
| tPCK2 | F | AtVQ21(AT3G18690) | BrVQ21-2 | BrVQ21-1 | BrVQ21-3 |
| tPCK2 | F | AtVQ22(AT3G22160) | – | BrVQ22-1 | – |
| tPCK6 | N | AtVQ23(AT3G56710) | BrVQ23-1 | BrVQ23-2/BrVQ23-3 | – |
| tPCK6 | N | AtVQ24(AT3G56880) | BrVQ24-1 | BrVQ24-2 | – |
| tPCK6 | N | AtVQ25(AT3G58000) | BrVQ25-1 | BrVQ25-2 | – |
| tPCK6 | N | AtVQ26(AT3G60090) | BrVQ26-1 | BrVQ26-2 | BrVQ26-3 |
| tPCK4 | T | AtVQ27(AT4G15120) | BrVQ27 | – | – |
| tPCK4 | U | AtVQ28(AT4G20000) | BrVQ28 | – | – |
| tPCK4 | U | AtVQ29(AT4G37710) | – | BrVQ29-2 | BrVQ29-1 |
| tPCK4 | U | AtVQ30(AT4G39720) | BrVQ30-2 | – | BrVQ30-1 |
| tPCK5 | R | AtVQ31(AT5G08480) | – | BrVQ31 | – |
| tPCK7 | V | AtVQ32(AT5G46780) | BrVQ32-2 | BrVQ32-1 | – |
| tPCK5 | Wb | AtVQ33(AT5G53830) | BrVQ33-1 | – | BrVQ33-2 |
| tPCK7 | X | AtVQ34(AT5G65170) | BrVQ34-1 | BrVQ34-3 | BrVQ34-2 |
| – | – | – | – | BrVQ35 | – |
2.5. Phylogenetic Tree of the VQ Domains in Arabidopsis, Rice and Chinese Cabbage
Using the neighbor-joining method, we constructed an un-rooted tree with 34, 57, and 39 VQ amino acid sequences in Arabidopsis, Chinese cabbage, and rice, respectively. The sequences were classified into eight groups (I–VIII) (Figure 6). The low bootstrap values in the tree are due to the divergent VQ sequence among the three species. This is not surprising, given that both A.thaliana and B. rapa belong to cruciferous plants, and the VQ genes in these two species were clustered together, while O. sativa VQ genes clustered by themselves. Moreover, the amino acid sequence of most VQ genes in A. thaliana and B. rapa revealed high similarity of more than 70% with each other (Figure 6, Table S3).
Figure 6.
Phylogenetic tree of the VQ motif–containing proteins from Arabidopsis, rice, and Chinese cabbage. The tree was determined using the neighbor-joining method with 1000 bootstrap replicates. Based on the clustering of the VQ motif–containing proteins, we classified proteins into eight different groups from Group I to Group VIII. Proteins from Arabidopsis, Chinese cabbage, and rice are denoted by green circles, blue squares, and yellow triangles, respectively.
2.6. Expression Pattern of the BrVQ Genes in Different Tissues
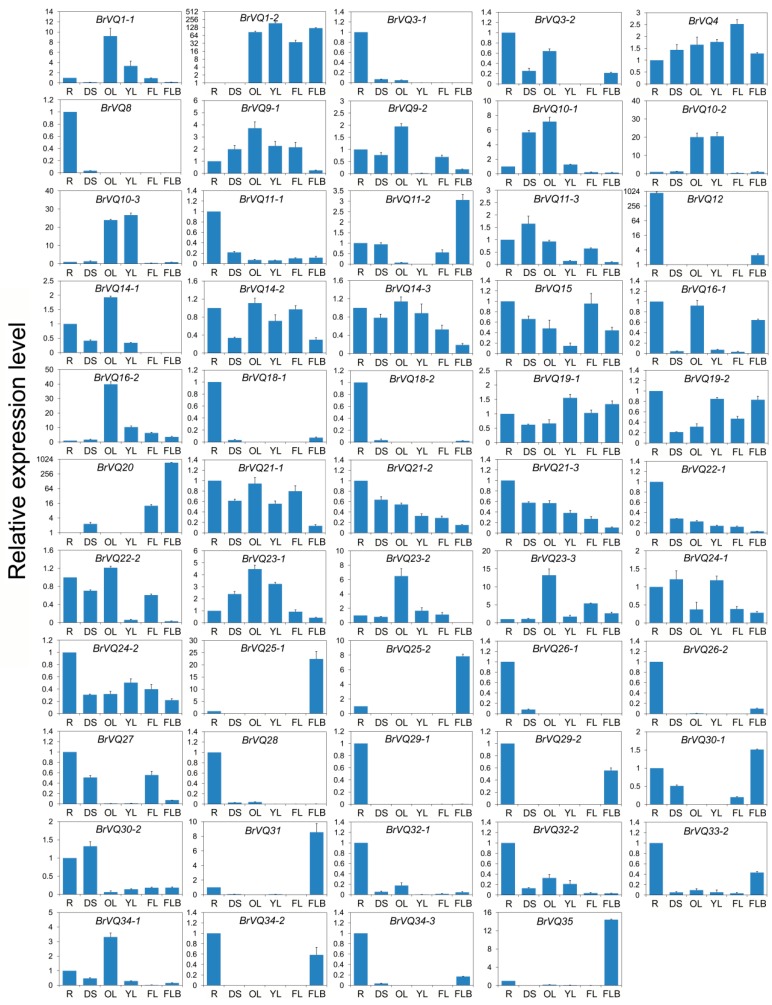
To explore the possible roles of the BrVQ genes in Chinese cabbage growth and development, we performed qRT-PCR expression analysis in six tissues, including root (R), dwarf stem (DS), old leaf (OL), young leaf (YL), flower (FL), and flower bud (FLB). Expression of 54 BrVQ genes were detected while the other three BrVQ genes (5, 26-3, 33-1) were either absent or poorly expressed. Expression patterns varied among the 54 BrVQ genes (Figure 7). For example, 26 BrVQ genes, including BrVQ3-1, 3-2, 8, 11-1, 12, 15, 16-1, 18-1, 18-2, 19-2, 21-1, 21-2, 21-3, 22-1, 24-2, 26-1, 26-2, 27, 28, 29-1, 29-2, 32-1, 32-2, 33-2, 34-2, and 34-3, showed higher expression levels in the R than in other tissues; 13 BrVQ genes (1-1, 9-1, 9-2, 10-1, 14-1, 14-2, 14-3, 16-2, 22-2, 23-1, 23-2, 23-3, and 34-1) were expressed more in the OL than in other tissues; seven BrVQ genes (11-2, 20, 25-1, 25-2, 30-1, 31, and 35) were expressed mainly in the FLB; four BrVQ genes (1-2, 10-2, 10-3, and 19-1) were expressed mainly in the YL; three BrVQ genes (11-3, 24-1, and 30-2) were expressed mainly in the DS and only the BrVQ4 gene was expressed in the FL.
Figure 7.
Expression analysis of the BrVQ genes in different tissues of Chinese cabbage. The surveyed tissues include root(R), dwarf stem (DS), old leaf (OL), young leaf (YL), flower (FL), and flower bud (FLB). Expression levels of the BrVQ genes were normalized to those of BrActin and shown relative to their expression in R, except for BrVQ1-2 and BrVQ12, whose expression level was relative to that in DS. The 2−ΔΔCt method was used to calculate the expression levels of target genes in different tissues. The expression levels of BrVQ1-2, BrVQ12, and BrVQ20 are shown by log2.
Additionally, some paralogs showed similar expression patterns in different tissues. For example, BrVQ10-2/10-3 had higher expression levels in both OL and YL. BrVQ21-2/21-3 showed similar expression tendency in all tissues. On the contrary, some exhibited different expression patterns in different tissues, including BrVQ22-1/22-2, BrVQ24-1/24-2, and BrVQ30-1/30-2.
2.7. Expression Analysis of the BrVQ Genes under Abiotic Stresses
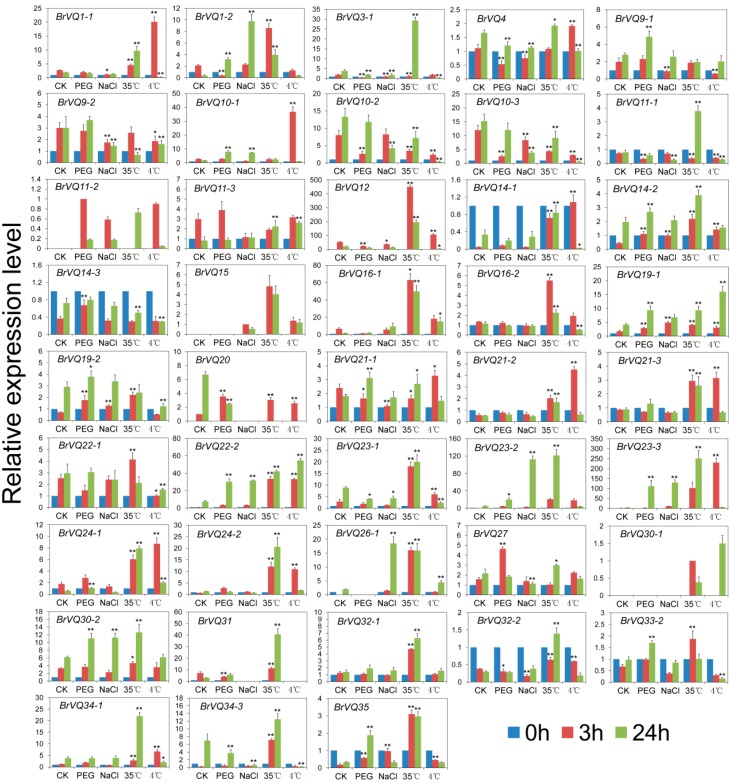
To further understand the possible roles of the BrVQ genes in response to abiotic stresses, we investigated their expression levels under the most common osmotic (polyethylene glycol, PEG6000), salt (NaCl), heat (35 °C), and cold (4 °C) stress. Altogether, 43 BrVQ genes displayed differential expression compared to the untreated control after at least one stressor treatment (Figure 8), while the expression patterns of the other 14 genes were not detectable. For example, after PEG6000 treatment, four (14-2, 19-2, 20, and 27) and six genes (1-2, 10-1, 19-1, 22-2, 23-2, and 23-3) were up-regulated whereas seven (1-2, 3-1, 4, 10-2, 10-3, 11-1, and 12) and two genes (20 and 23-1) were down-regulated more than two-fold compared to the untreated control at 3 h and 24 h of treatment, respectively. BrVQ35 was up-regulated more than two-fold at both 3 h and 24 h. During salt stress, four genes (14-2, 19-1, 19-2, and 35) and five genes (1-2, 22-2, 23-2, 23-3, and 26-1) were significantly induced at 3 and 24 h. On the contrary, five genes (1-1, 3-1, 9-2, 10-3, and 23-1) were down-regulated at 3 h and their expression levels were further decreased two-fold at 24 h with the exception of BrVQ1-1. For 35°C treatment, 27 genes were rapidly up-regulated at 3 h. Among these genes, 19 genes (1-2, 12, 16-1, 16-2, 21-2, 21-3, 22-2, 23-2, 23-3, 24-1, 24-2, 26-1, 30-2, 31, 32-1, 32-2, 34-1, 34-3, and 35) showed the highest expression levels at 3 h and 24 h. In contrast, three genes (9-2, 10-2, and 10-3) showed a trend of down-regulation from 3 h to 24 h. Interestingly, BrVQ3-1 and BrVQ11-1 were initially down-regulated about two-fold at 3 h but up-regulated more than four-fold at 24 h. In the case of low temperature treatment, five genes (10-1, 14-2, 21-2, 21-3, and 34-1) at 3 h and two genes (19-1 and 26-1) at 24 h exhibited at least a two-fold increase in expression compared to the untreated control, while two genes (9-1 and 22-1) at 3 h and four genes (3-1, 14-3, 19-2, and 34-3) at 24 h showed the opposite expression trend. Interestingly, four genes (1-1, 12, 14-1, and 23-1) were up-regulated at 3 h and down-regulated at 24 h more than two-fold. Two genes (22-2 and 24-1) and two genes (10-2 and 10-3) were induced and inhibited more than two-fold at both 3 h and 24 h, respectively. Four genes (22-2, 23-2, 23-3, and 35) were induced to a different extent after osmotic, salt, heat, and cold treatments, while two genes (10-2 and 10-3) were inhibited under those conditions.
Figure 8.
Expression analysis of the BrVQ genes under abiotic stresses. Three-week-old plants were treated with 20% (w/v) PEG6000, 20 mmol/L NaCl, 35 °C, and 4 °C for 0, 3, and 24 h before the mature leaves were harvested. Expression of the BrVQ genes were normalized to those of BrActin and shown relative to the expression of CK at 0 h, except for four BrVQ genes (11-2, 15, 20, 30-1), whose expression levels were related to the expression of CK treated with PEG, NaCl, CK, 35 °C at 3 h, respectively. The 2−ΔΔCt method was used to calculate the expression of target genes in different tissues. * indicated that the expression level is significantly different from the value of the control (* p < 0.05, ** p < 0.01).
2.8. Expression Analysis of the BrVQ Genes under Phytohormone Treatment
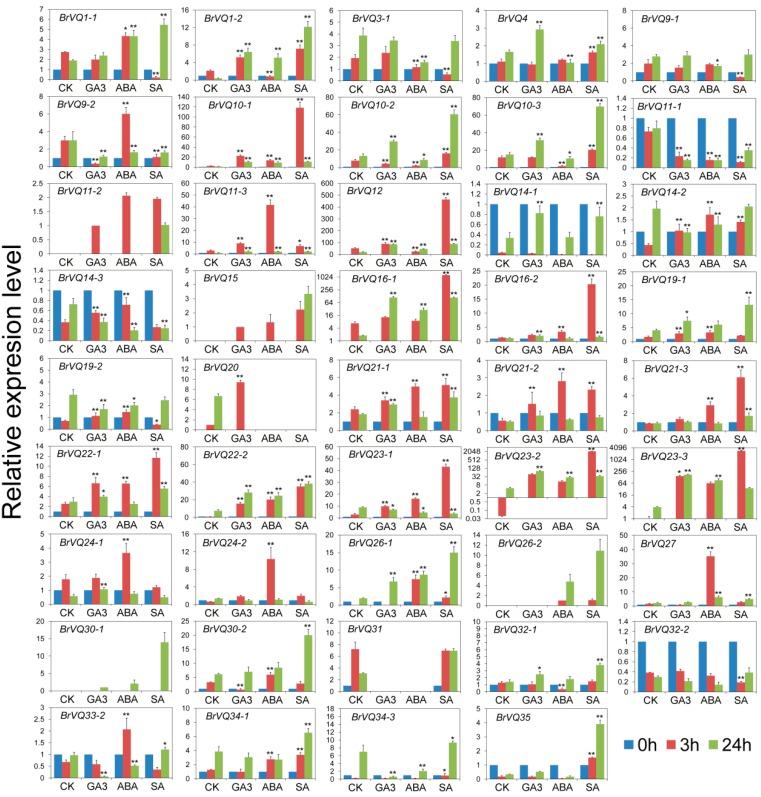
More and more studies have demonstrated that plant hormones play important roles in plant growth and defense signaling [23,24,25]. To understand the expression response of the BrVQ genes to various plant hormones, we carried out Gibberellin A3 (GA3), abscisic Acid (ABA) or salicylic acid (SA) treatments in Chinese cabbage plants. We detected the expression of a total of 44 BrVQ genes (Figure 9). Upon GA3 treatment, nine genes (1-2, 10-1, 10-3, 11-3, 12, 21-1, 22-1, 22-2, and 23-3) were up-regulated at 3 h and 24 h, and two genes (9-2 and 11-1) were down-regulated. Additionally, some genes showed significant differential expression at one time-point compared to the untreated controls. For example, at 3 h, BrVQ30-2 was down-regulated more than four-fold while BrVQ21-2 was up-regulated at least two-fold. At 24 h, BrVQ33-2 and 34-3 were down-regulated while BrVQ23-2 was up-regulated at least 10-fold. In the case of ABA treatment, about 25 BrVQ genes were induced at 3 h, and some genes (16-1, 19-1, 22-2, 23-2, 23-3, and 26-1) maintained the up-regulated trend at 24 h, while several other genes (9-2, 14-2, 23-1, and 33-2) showed a down-regulated trend at 24 h. Apart from these, six genes (1-2, 10-2, 10-3, 11-1, 12, and 32-1) showed down-expression of more than two-fold at 3h, and then, three genes (10-2, 10-3, and 11-1) kept this tendency up to 24 h, while the other two genes (1-2 and 12) changed to an up-regulation tendency at 24 h. For SA treatment, the expression levels of 17 genes were higher than the untreated control at 3 h and 24 h, including 1-2, 4, 10-1, 10-2, 10-3, 11-3, 12, 21-1, 21-3, 22-1, 22-2, 23-2, 26-1, 27, 34-1, 34-3, and 35. BrVQ23-2 and BrVQ23-3, in particular, were up-regulated more than one thousand-fold at 3h. BrVQ9-2 and BrVQ11-1 were down-regulated two-fold at 3 h and 24 h. The expression of BrVQ23-1 showed a trend of initial increase at 3 h, followed by a decrease at 24 h, compared to the untreated control; however, BrVQ1-1 showed the opposite expression trend.
Figure 9.
Expression analysis of the BrVQ genes under phytohormones. Three-week-old plants were treated with 200 μM GA3, 100 μM ABA, and 200 μM SA for 0, 3, and 24 h before the mature leaves were harvested. Expression of the BrVQ genes were normalized to those of BrActin and shown relative to the expression of CK at 0 h, except for five BrVQ genes (11-2, 15, 20, 26-2, 30-1), whose expression levels were related to the expression of CK treated with GA3, GA3, CK, ABA at 3 h, and GA3 at 24 h, respectively. The 2−ΔΔ Ct method was used to calculate the expression of target genes in different tissues. The expression levels of three BrVQ genes (16-1, 23-2, 23-3) are shown by log2. * indicated that the expression level is significantly different from the value of the control (* p < 0.05, ** p < 0.01).
2.9. Comparison of the Expression Patterns of BrVQ Genes and Their Orthologs in Arabidopsis
To further compare the expression patterns of BrVQ genes and their orthologs in A.thaliana, the expression patterns of AtVQ genes were extracted from Genevestigator [26,27]. We found that the expression patterns in different tissues between AtVQ and BrVQ genes were similar except for some individual genes (Table S4, Figure 7); for instance, the transcriptional expressions of BrVQ8, BrVQ12, BrVQ28, and BrVQ29 were mainly detected in the roots, while AtVQ8, AtVQ12, AtVQ28, and AtVQ29 genes could be detected in the roots, stem, leaf, and flower. Additionally, most of the orthologs of the VQ genes between B. rapa and A. thaliana also have similar expression patterns under abiotic and hormone treatment. For example, AtVQ4/BrVQ4 and -10 were induced by cold and SA treatment, AtVQ11/BrVQ11 were induced by cold and ABA treatment, AtVQ12/BrVQ12, -14, and -21 were induced by heat, cold, and SA, AtVQ16/BrVQ16 were induced by salt and SA treatment (Table S5, Figure 8 and Figure 9). In summary, after comparing the expression patterns of BrVQ genes and their orthologs in A. thaliana, we found most, but not all of them, displayed similar expression trends in some tissues and in their responses to various stresses and hormone stimulus. These results further explained the speculation that VQ genes may have similar functions in some aspect in two different species; however, the different expression patterns of VQ ortholog genes between these two species should also be noticed.
3. Discussion
Previous studies showed that VQ genes play an essential role in plant growth, development, and response to adverse environment [3,7,8,9,10,11,12,13,14]. However, there is little information on the characterization of VQ motif–containing proteins in B. rapa. Therefore, the comprehensive analysis of BrVQ genes and their expression patterns under various abiotic and hormone treatments could be beneficial to further understanding the mechanisms of influencing plant growth and development as well as applying them to Chinese cabbage molecular breeding.
3.1. VQ Gene Duplication in Chinese Cabbage
Genome duplication plays an important role in expanding genome content and diversifying gene function because a duplication event can evolve into genes with new functions [28]. After genome duplication, processes such as nonfunctionalization (duplicated genes are silenced), subfunctionalization (function is partitioned between the new paralogs), and neofunctionalization (duplicated genes gain new functions) generally take place so that genes are either lost or fixed [22,29,30]. Brassica rapa is a mesopolyploid crop that has undergone the whole genome triplication (WGT) event since its divergence from Arabidopsis thaliana [31]. Since there are 34 VQ genes in the A. thaliana genome, the predicted VQ genes could number more than 100 in the B. rapa genome. However, in this study, we found that only 57 VQ genes were retained in the B. rapa genome, suggesting that there was extensive gene loss during genome duplication [32,33]. Similar cases were also reported in other B. rapa gene families, such as MADS-box [31], mitogen-activated protein kinase (MAPK) [34], WRKY [35], etc. Even so, the WGT event has indeed expanded the quantity of the B. rapa gene family members. We found 53 BrVQ genes showed a syntenic relationship with 28 AtVQ genes, implying that the numbers of duplicated BrVQ genes contain only approximately twice as many as that of the AtVQ genes. This result is consistent with a previous study which showed the triplicated B. rapa genome contains only approximately twice the numbers in that of A. thaliana because of genome shrinkage and differential loss of duplicated genes [36]. The expansion of the BrVQ gene family seems to mainly depend on the segmental duplication (41/57 genes; 71.9%), while one tandem duplicated pair (2/57 genes, 3.5%) may play a minor role. A similar phenomenon was reported in the B. rapa expansin superfamily which contains high segmental duplication (68%) and low tandem duplication (6.3%) [22]. Similarly, in the A. thaliana genome, a large proportion of gene families also fall into the low tandem and high segmental duplication class [37]. Besides these, gene duplication can generate gene functional redundancy, and these duplicate genes could develop divergent patterns of gene expression for stably maintaining through subfunctionalization [30,38]. For example, the two duplicated MADS-box genes, BdMADS2 and BdMADS4 from Brachypodium distachyon, displayed different expression patterns in all floral organs, and ectopic expression these two genes in Arabidopsis caused different phenotypic effects between them [39]. In our study, some paralogs showed different expression patterns, such as BrVQ22-1/22-2, BrVQ24-1/24-2, and BrVQ30-1/30-2 in different tissues, and BrVQ10-1/10-2, BrVQ11-1/11-3, BrVQ14-1/14-3, and BrVQ32-1/32-2 in various abiotic and hormone treatments, suggesting these BrVQ paralogs might be maintained through subfunctionalization. Additionally, a similar report was found in the duplicate NAC TF pairs from [40] and AGAMOUS (AG) genes from Thalictrum thalictroides [41].
3.2. Function of the VQ Proteins in Plant Growth and Development
Accumulating evidence has demonstrated that the transcription of the VQ genes is regulated by various endogenous and environmental signals, consistent with their diverse roles in plant growth and development [42]. The IKU1/AtVQ14 gene is expressed preferentially in the early endosperm. AtVQ14 mutation reduces endosperm growth and produces small seeds, suggesting the AtVQ14 gene may be involved in seed development [7]. The loss-of-function mutation of AtVQ14 results in a decrease of expression of IKU2 (encoding a leucine-rich repeat kinase) and MINI3 (encoding a WRKY family protein), both of which play an important role in seed development [43]. AtVQ29 is expressed at a higher level in the stem than the root, rosette leaf, flower, and silique. Over-expression of AtVQ29 causes hyposensitivity of hypocotyl growth to far-red and low-light conditions while its loss-of-function mutants display decreased hypocotyl elongation under the low intensity of far-red and white light, implying the VQ protein maybe involved in regulating plant seedling photomorphogenesis [8]. Moreover, over-expression of AtVQ29 also substantially delays the blossom of the transgenic plant compared to the wild type [3]. AtVQ8 was located in plastid. The recessive loss-of-function AtVQ8 mutants exhibit pale-green and stunted-growth phenotypes throughout the entire life cycle, suggesting a predominant role in chloroplast development or photosystem assembly [3]. Besides these, over-expression of AtVQ17, AtVQ18, and AtVQ22 also causes highly stunted growth [3]. In this study, we assessed the expression levels of the BrVQ genes in six Chinese cabbage tissues (Figure 7). The result showed that the majority of the genes were differentially expressed in the tissues that we analyzed. The BrVQ genes were expressed mainly in specific organs and tissues, suggesting that they may play important roles in the growth and development of these organs or tissues. Previous studies have shown that Gibberellin acid (GA3) has various regulation functions in high plants, such as simulating early seed development and organ growth and controlling fertilization time [44]. Thus, we also examined the expression profiles of the BrVQ genes in response to exogenous GA3 (Figure 9). The result showed expression of half the BrVQ genes was induced and some genes were up-regulated by more than two-fold. Besides these, similar expression patterns of BrVQ genes and their orthologs in A. thaliana were detected in different tissues. Taken together, these results indicate that the VQ gene family is extensively involved in plant growth and development.
3.3. Function of the VQ Proteins in Abiotic and Biotic Resistance
Drought, salt, heat, cold, and pathogen stresses are the main factors of reducing crop production. Many studies have been carried out to understand how tolerance to these stresses is regulated in plants. In the present study, we found that the majority of the BrVQ genes were induced by PEG6000 and NaCl treatments (Figure 8). Similar results were also found for the VQ genes in Oryza sativa and Vitis vinifera L. For instance, 22 OsVQ genes [4] and 18 VvVQ genes [6] were up-regulated by drought stress. However, studies also showed that the up-regulation of some VQ genes may have a negative effect on abiotic stress resistance. For example, AtVQ9 expression is strongly induced by salt stress; however, the over-expression of AtVQ9 rendered plants hypersensitive to salt stress [9]. AtVQ15 was induced by dehydration and high salinity, whereas its over-expression lines exhibited an increased sensitivity to both salt and mannitol stresses during seed germination and seedling growth. On the contrary, the antisense lines were significantly more tolerant to these stresses [10]. We also found that the BrVQ genes were more responsive to heat and cold stresses, where the numbers of induced genes were more than those under PEG and NaCl stresses.
There are various stress perception and signaling pathways, some of which are special, but others may cross-talk [45]. Abscisic acid (ABA) is an important phytohormone and plays a critical role in the response to various abiotic stress signals [46]. Stress-responsive genes could be regulated by either the ABA-dependent or ABA-independent signaling pathway [45,47,48] and the ABA-dependent signaling pathway plays an important role in stress-responsive genes under osmotic stress [49]. The majority of the BrVQ genes either up- or down-regulated more than two-fold upon ABA stress compared to the untreated control (Figure 9). Similar results were also observed in the OsVQ genes upon ABA treatment [4]. In combined analysis of the expression patterns of BrVQ genes (Figure 8 and Figure 9), we found that some BrVQ genes displayed similar expression tendency under abiotic stress and ABA treatment. For example, BrVQ1-2 was down-regulated at 3 h and followed by an increase at 24 h under PEG and ABA treatments. BrVQ3-1 showed a down-regulation trend at 3 and 24 h under PEG, NaCl, and ABA treatments. The expression of BrVQ22-2 exhibited similar accumulation trends during abiotic stresses and ABA treatment but BrVQ10-2/10-3 showed the opposite tendency. These results suggested these BrVQ genes might be involved in stress response and ABA signaling. Salicylic acid (SA) is an important signal molecule that accumulates under abiotic and biotic stress [50]. We found that, after SA treatments (Figure 9), the expression of many BrVQ genes were up- or down-regulated compared to the untreated control. Similar results were observed in previous studies for other VQ genes. For example, 34 AtVQ genes are induced and differentially expressed in different tissues in response to SA treatment and pathogen infection [3]; 27 OsVQ genes are induced by at least one pathogen infection [4]. Sixteen VvVQ genes were induced by SA treatment [6]. AtVQ21/MKS1, as substrate of MPK4, is required for SA-dependent resistance in the mpk4 mutants, and over-expression of AtVQ21 in the wild-type plants is sufficient to activate SA-dependent resistance [11]. The AtVQ23/SIB1 loss-of-function mutants compromised the induction of some defense-related genes by pathogen infection and SA treatments. However, over-expression lines increased the expression of defense-related genes upon pathogen infection and SA treatment [13]. Interestingly, BrVQ21-1/21-2/21-3 and BrVQ23-1/23-2/23-3 were up-regulated to a different extent after SA treatment, suggesting they might have similar regulation to that of AtVQ21 and AtVQ23, respectively. Furthermore, we found that the responses of VQ genes in Chinese cabbage and Arabidopsis in the expression levels toward some abiotic stresses and hormone treatments were similar. Taken together, these results revealed that some VQ members could be actively involved in regulating plant responses to various abiotic and biotic stresses.
4. Experimental Section
4.1. Plant Materials, Growth Conditions, and Stress Treatments
Chinese cabbage cultivar “Guangdongzao” was used for all experiments. Plant seeds were sown in a glass Petri dish containing two wet filter papers. After germination, seedlings were transferred into pots (five seedlings in one pot) containing a growth medium with vermiculite and peat (3:1) and grown in a greenhouse at 20 ± 2 °C with a photoperiod of 16 h light and 8 h dark. Three-week-old seedlings were used for the abiotic and hormone treatments. For salinity and osmotic treatments, plant samples were irrigated with 200 mM NaCl and 20% (w/v) polyethylene glycol (PEG6000), respectively, until the solution flowed out from the bottom of the pot. For high and low temperature treatments, plant samples which were grown in the green house were transferred to an incubator at 35 °C or a refrigerating chamber at 4 °C, respectively. For planthormone treatments, we sprayed plant leaves with 200 μM Gibberellin A3 (GA3), 100 μM abscisic acid (ABA), and 200 μM salicylic acid (SA) solutions, respectively, until drops began to fall from the leaves. Then, the fully opened leaves of seedlings were harvested after 0, 3, and 24 h of the above abiotic and hormone treatments. For analysis of VQ gene expression in different tissues, plant organs were harvested after the plants bloomed; plant organs were harvested, including root (R), dwarf stem (DS), old leaf (OL), young leaf (YL), flower (FL), and flower bud (FLB), in three biological replicates for RNA preparation. All harvested samples were immediately frozen in liquid nitrogen and stored at −80 °C until use.
4.2. Sequence Retrieval
The VQ motif sequences are listed in the Pfam Database under the motif ID “PF05678” [51]. Chinese cabbage VQ motif–containing proteins were identified by using the local BLASTP in the Brassica database [17,32]. To confirm the presence of the VQ domain, the web tools from the Interpro program [52] and the SMART program [53] were used on the VQ proteins in B. rapa. The coding sequences (CDS) and amino acid sequences of the B. rapa VQ genes were downloaded from the Brassica database [17,32]. Thirty-four AtVQ gene and protein sequences from A. thaliana were retrieved from The Arabidopsis Information Resource (TAIR) [3,54]. Thirty-nine OsVQ gene and protein sequences from O. sativa were retrieved from the Rice Genome Annotation Project (RGAP) [4,55]. The homology searches between A. thaliana and B. rapa were performed by using the Gramene database [19] and the Basic Local Alignment Search Tool (BLAST) [56].
4.3. Identification and Analysis of the VQ Genes and Proteins in Chinese Cabbage
The physical locations of the BrVQ genes on the Chinese cabbage chromosomes were mapped by using Mapchart 2.2 (Plant Research International, Wageningen, The Netherlands). The BrVQ amino acid sequences were aligned by the software DNAMAN 6.0.40 (Lynnon Biosoft, Quebec, QC, Canada). Intron/exon structure analysis was performed by using the Gene Structure Display Server (GSDS) [57]. The protein size, molecular weight (Mw), and theoretical isoelectric point (pI) were computed by using the ProtParam tool [58]. Phylogenetic tree was constructed with MEGA5 [59] on the basis of alignment with the full-length VQ protein sequences using the neighbor-joining method [60] with 1000 bootstrap replicates [61]. The distribution of the conserved motifs and domains were detected using the MEME suite [62] and the PlantsP database [20,63]. A MEME search was carried out with the following parameters: optimum motif width ≥6 and ≤300 and the maximum number of motifs set at 10.
4.4. RNA Isolation and qRT-PCR
Total RNA was isolated from each sample using a Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNA synthesis was carried out using a PrimeScript™ RT reagent kit with a gDNA Eraser (Takara, Dalian, China). Quantitative real-time PCR (qRT-PCR) was performed using a SYBR Green Master mix (Takara, Dalian, China) on an IQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The qRT-PCR primers for the BrVQ genes and actin gene are listed in Table S6. The actin gene was used as a constitutive expression control in the qRT-PCR experiments. Reactions were set up in a total volume of 20 μL containing 10μL of SYBR Green Master mix, 0.4 μL of each primer (10 µM), 7.2 μL of double-distilled water, and 2 μL of cDNA template. The PCR cycling conditions comprised an initial polymerase activation step of 95 °C for 1 min, followed by 45 cycles of 95 °C for 15 s, and 60 °C for 70 s. After each PCR run, a dissociation curve was plotted to confirm the specificity of the product and to avoid the production of primer dimers. Three replicates of each sample were conducted to calculate the average Ct values. The relative expression level was calculated by the comparative 2−ΔΔCt method [64]. Three biological replicates were carried out and the significance was determined with SPSS software (SPSS 17.0, IBM, Chicago, IL, USA) (p < 0.05).
5. Conclusions
We identified 57 BrVQ proteins by genome-wide identification, characterization, and expression analysis. Phylogenetic relationship analysis indicated that the VQ family in Chinese cabbage closely resembled that of Arabidopsis. Due to genome shrinkage and differential loss of duplicated genes during the WGT events, the number of the BrVQ genes is approximately twice of that of the AtVQ genes. The BrVQ genes were differentially expressed in six tissues as well as when the plants were exposed to various abiotic stresses and hormone stimulus. We found some similar expression patterns existing in BrVQ genes and their orthologs in Arabidopsis. Although some AtVQ genes have been functionally analyzed, other members of VQ remain to be further studied due to VQ gene structure diversification with the exception of the VQ motif. This information will provide a solid foundation for further functional studies of Chinese cabbage VQ proteins, and will be useful to better understanding the roles that VQ genes play in plant growth and development as well as mediating the cross-talk between abiotic stresses and hormone signaling.
Acknowledgments
This work was supported by the National High Technology Research and Development Program, China (2012AA100103009); the Young Talents Training Program of Shandong Academy of Agricultural Science, China (NKYSCS-01); Modern Agricultural Industrial Technology System Funding of Shandong Province, China (SDAIT-02-022-04); National Natural Science Foundation of China (31471884); the China agriculture research system (CARS-25) and the Project for Cultivation of Major Achievements in Science and Technology in SAAS, China (2015CGPY09).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/12/26127/s1.
Author Contributions
Fengde Wang and Jianwei Gao conceived and designed the experiments. Gaoyuan Zhang, Fengde Wang, Jingjuan Li, and Qian Ding performed the experiments. Gaoyuan Zhang, Yihui Zhang and Huayin Li analyzed the data. Gaoyuan Zhang, Jiannong Zhang and Jianwei Gao drafted the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y., Zhou Y., Yang Y., Chi Y.J., Zhou J., Chen J.Y., Wang F., Fan B., Shi K., Zhou Y.H., et al. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012;159:810–825. doi: 10.1104/pp.112.196816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D.Y., Kwon S.I., Choi C., Lee H., Ahn I., Park S.R., Bae S.C., Lee S.C., Hwang D.J. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene. 2013;529:208–214. doi: 10.1016/j.gene.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Zhang H., Sun G., Jin Y., Qiu L. Identification of active VQ motif-containing genes and the expression patterns under low nitrogen treatment in soybean. Gene. 2014;543:237–243. doi: 10.1016/j.gene.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Wang M., Vannozzi A., Wang G., Zhong Y., Corso M., Cavallini E., Cheng Z.M. A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 2015;6:417. doi: 10.3389/fpls.2015.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang A., Garcia D., Zhang H., Feng K., Chaudhury A., Berger F., Peacock W.J., Dennis E.S., Luo M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010;63:670–679. doi: 10.1111/j.1365-313X.2010.04271.x. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Jing Y., Li J., Xu G., Lin R. Arabidopsis VQ-motif-containing-protein 29 represses seedling de-etiolation by interacting with PIF1. Plant Physiol. 2014;164:2068–2080. doi: 10.1104/pp.113.234492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y., Chen L., Wang H., Zhang L., Wang F., Yu D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013;74:730–745. doi: 10.1111/tpj.12159. [DOI] [PubMed] [Google Scholar]
- 10.Perruc E., Charpenteau M., Ramirez B.C., Jauneau A., Galaud J.P., Ranjeva R., Ranty B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 2004;38:410–420. doi: 10.1111/j.1365-313X.2004.02062.x. [DOI] [PubMed] [Google Scholar]
- 11.Andreasson E., Jenkins T., Brodersen P., Thorgrimsen S., Petersen N.H., Zhu S., Qiu J.L., Micheelsen P., Rocher A., Petersen M., et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu J.L., Fiil B.K., Petersen K., Nielsen H.B., Botanga C.J., Thorgrimsen S., Palma K., Suarez-Rodriquez M.C., Sandbech-Clausen S., Lichota J., et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008;27:2214–2221. doi: 10.1038/emboj.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Y.D., Li W., Guo D., Dong J., Zhang Q., Fu Y., Ren D., Peng M., Xia Y. The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ. 2010;33:828–839. doi: 10.1111/j.1365-3040.2009.02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Z., Li Y., Wang F., Cheng Y., Fan B., Yu J.Q., Chen Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell. 2011;23:3824–3841. doi: 10.1105/tpc.111.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecher P., Eschen-Lippold L., Herklotz S., Kuhle K., Naumann K., Bethke G., Uhrig J., Weyhe M., Scheel D., Lee J. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of “VQ-motif”-containing proteins to regulate immune responses. New Phytol. 2014;203:592–606. doi: 10.1111/nph.12817. [DOI] [PubMed] [Google Scholar]
- 16.Weyhe M., Eschen-Lipplod L., Pecher P., Scheel D., Lee J. Ménage à trois: The complex relationships between mitogen-activated protein kinases, WRKY transcription factors, and VQ-motif-containing proteins. Plant Signal. Behav. 2014;9:e29519. doi: 10.4161/psb.29519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. BRAD, The genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohanta T.K., Arora P.K., Mohanta N., Parida P., Bae H. Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genom. 2015;16:58. doi: 10.1186/s12864-015-1244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C., Jaiswal P., Hebbard C., Avraham S., Buckler E.S., Casstevens T., Hurwitz B., McCouch S., Ni J., Pujar A., et al. Gramene: A growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Pan C., Wang Y., Ye L., Wu J., Chen L., Zou T., Lu G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015;16:386. doi: 10.1186/s12864-015-1621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F., Wu J., Fang L., Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012;3:198. doi: 10.3389/fpls.2012.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy P., Hong J.K., Kim J.A., Jeong M.J., Lee Y.H., Lee S.L. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol. Genet. Genom. 2015;290:521–530. doi: 10.1007/s00438-014-0935-0. [DOI] [PubMed] [Google Scholar]
- 23.Bari R., Jones J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 24.Denancé N., Sánchez-Vallet A., Goffner D., Molina A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013;4:155. doi: 10.3389/fpls.2013.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santner A., Calderon-Villalobos L.I., Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Zhang M., Kong L., Lv Y., Zou M., Lu G., Cao J., Yu X. Genome-wide identification, phylogeny, duplication and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis) DNA Res. 2014;21:379–396. doi: 10.1093/dnares/dsu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker M.S., Baute G.J., Liu S.L. Duplications and turnover in plant genomes. Plant Genome Divers. 2012;1:155–169. [Google Scholar]
- 30.Zhang J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 31.Saha G., Park J.I., Jung H.J., Ahmed N.U., Kayum M.A., Chung M.Y., Hur Y., Cho Y.G., Watanabe M., Nou I.S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015;16:178. doi: 10.1186/s12864-015-1349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun JH., Bancroft I., Cheng F., et al. The Brassica rapa Genome Sequencing Project Consortium: The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 33.Cheng F., Mandáková T., Wu J., Xie Q., Lysak M.A., Wang X. Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell. 2013;25:1541–1554. doi: 10.1105/tpc.113.110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu K., Guo W., Lu J., Yu H., Qu C., Tang Z., Li J., Chai Y., Liang Y. Genome-wide survey and expression profile analysis of the mitogen-activated protein kinase (MAPK) gene family in Brassica rapa. PLoS ONE. 2015;10:e0132051. doi: 10.1371/journal.pone.0132051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang J., Wang F., Hou X.L., Wang Z., Huang Z.N. Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014;32:781–795. [Google Scholar]
- 36.Mun J.H., Kwon S.J., Yang T.J., Seol Y.J., Jin M., Kim J.A., Lim M.H., Kim J.S., Baek S., Choi B.S., et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duarte J.M., Cui L., Wall P.K., Zhang Q., Zhang X., Leebens-Mack J., Ma H., Altman N., dePamphilis C.W. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 39.Wei B., Liu D., Guo J., Leseberg C.H., Zhang X., Mao L. Functional divergence of two duplicated D-lineage MADS-box genes BdMADS2 and BdMADS4 from Brachypodium distachyon. J. Plant Physiol. 2013;170:424–431. doi: 10.1016/j.jplph.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Shang H., Li W., Zou C., Yuan Y. Analyses of the NAC transcription factor gene family in Gossypium raimondii UIbr.: Chromosomal location, structure, phylogeny, and expression patterns. J. Integr. Plant Biol. 2013;55:663–676. doi: 10.1111/jipb.12085. [DOI] [PubMed] [Google Scholar]
- 41.Galimba K.D., diStilio V.S. Sub-functionalization to ovule development following duplication of a floral organ identity gene. Dev. Biol. 2015;405:158–172. doi: 10.1016/j.ydbio.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 42.Jing Y., Lin R. VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol. 2015;169:371–378. doi: 10.1104/pp.15.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. MINISEED3(MINI3), A WRKY family gene, and HAIKU2(IKU2), A leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedden P., Thomas S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 45.Chinnusamy V., Schumaker K., Zhu J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 46.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi-Shinozaki K., Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong L., Lee H., Ishitani M., Zhu J.K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- 50.Fragnière C., Serrano M., Abou-Mansour E., Métraux J.P., L’Haridon F. Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 2011;585:1847–1852. doi: 10.1016/j.febslet.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 51.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Interpro. [(accessed on 11 November 2015)]. Available online: http://www.ebi.ac.uk/interpro/
- 53.SMART. [(accessed on 11 November 2015)]. Available online: http://smart.embl.de/
- 54.TAIR. [(accessed on 11 November 2015)]. Available online: http://www.Arabidopsis.org/
- 55.RGAP. [(accessed on 11 November 2015)]. Available online: http://rice.plantbiology.msu.edu.
- 56.BLAST. [(accessed on 11 November 2015)]; Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 57.GSDS. [(accessed on 11 November 2015)]. Available online: http://gsds.cbi.pku.edu.cn/
- 58.ProtParam. [(accessed on 11 November 2015)]. Available online: http://web.expasy.org/protparam/
- 59.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 61.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 62.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gribskov M., Fana F., Harper J., Hope D.A., Harmon A.C., Smith D.W., Tax F.E., Zhang G. PlantsP: A functional genomics database for plant phosphorylation. Nucleic Acids Res. 2001;29:111–113. doi: 10.1093/nar/29.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2011;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.