Abstract
The effects of methyl jasmonate (MeJA), an elicitor of plant defense mechanisms, on the biosynthesis of diosgenin, a steroidal saponin, were investigated in six fenugreek (Trigonella foenum-graecum) varieties (Gujarat Methi-2, Kasuri-1, Kasuri-2, Pusa Early Branching, Rajasthan Methi and Maharashtra Methi-5). Treatment with 0.01% MeJA increased diosgenin levels, in 12 days old seedlings, from 0.5%–0.9% to 1.1%–1.8%. In addition, MeJA upregulated the expression of two pivotal genes of the mevalonate pathway, the metabolic route leading to diosgenin: 3-hydroxy-3-methylglutaryl-CoA reductase (HMG) and sterol-3-β-glucosyl transferase (STRL). In particular, MeJA increased the expression of HMG and STRL genes by 3.2- and 22.2-fold, respectively, in the Gujarat Methi-2 variety, and by 25.4- and 28.4-fold, respectively, in the Kasuri-2 variety. Therefore, MeJA may be considered a promising elicitor for diosgenin production by fenugreek plants.
Keywords: elicitors, mevalonate pathway, steroidal saponins, 3-hydroxy-3-methylglutaryl-CoA reductase, sterol-3-β-glucosyl transferase, plant innate immunity
1. Introduction
Trigonella foenum-graecum (fenugreek) is a dicotyledonous plant belonging to the family of Fabaceae (subfamily Papilionaceae), widely used for a number of medicinal applications all over the world [1]. Fenugreek has been extensively used, in Indian Ayurveda and Chinese Traditional Medicine, as remedy for treatment of illnesses and conditions including epilepsy, paralysis, gout, dropsy, chronic cough, diabetes, piles, sinus, lung congestion, inflammation, infections, as well as in hair treatment, breast enhancement and for its aphrodisiac effects [2,3,4]. In addition, fenugreek plants possess anticancer, anti-fertility, anti-ageing, antimicrobial, anti-parasitic, galactagogue and hypocholesterolaemic effects [5,6,7,8,9,10,11,12,13,14,15]. Many bioactive phytochemicals have been reported in fenugreek, including saponins, a class of glycosylated triterpenes that show antimicrobial, antiviral and insecticidal activities [16]. These compounds represent a part of the plant defense mechanisms against biotic stress and, therefore, can also be classified as phytoprotectants [17,18]. Among steroidal saponins, diosgenin ((25R)-5-spirosten-3H-ol) (Pubchem ID 99474) is one of the most bioactive components of fenugreek which has been investigated due to its anticancer and anti-aging activities [19,20,21,22,23]. It is also used as contraceptive and to prevent cardiovascular diseases [24,25,26,27,28]. Notably, the synthesis of some steroidal drugs and hormones such as testosterone, glucocorticoids, norethisterone and progesterone may arise from diosgenin used as precursor [9].
Currently, certain wild species of Mexican yam (Dioscorea spp.) are used for diosgenin production, even if they need a long time to accumulate high diosgenin levels [29]. Therefore, fenugreek may represent a more suitable system for diosgenin production because of short growth cycle and low cost of production [30,31]. The maximum amount of diosgenin naturally present in fenugreek is found in young leaves and in mature seeds, with percentages ranging from 0.28% to 0.92%. However, to develop fenugreek as a source of diosgenin, it is pivotal to trigger the cascade of 11 key genes responsible for diosgenin biosynthesis identified in our previous study [32].
The aim of the present study is to increase the content of diosgenin in fenugreek by treatment with methyl jasmonate (MeJA), an elicitor able to trigger the plant defense mechanisms. MeJA was first isolated from the plant pathogenic fungus Lasiodiplodia theobromae as plant growth inhibitor and the role of jasmonic acid, in elicitor-induced signal transduction pathway, was first described by Gundlach et al. [33,34]. Then, MeJA, a plant growth regulator, was reported to increase the accumulation of secondary metabolites involved in plant resistance against pathogens [1,35,36,37,38,39,40]. It triggers a cascade of intracellular signals and also activates de novo transcription of genes as phenylalanine ammonia-lyase (PAL), the key enzyme of the phenylpropanoid pathway leading to accumulation of antimicrobial phytoalexins [41]. It is also known that the 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) is an important rate limiting and key regulatory enzyme for isoprenoid or mevalonate biosynthetic route in plants, that catalyzes the irreversible conversion of 3-hydroxy-3-methylglutaryl-CoA (HMG) to mevalonate. Therefore, expression of this enzyme increases the total sterol and cycloartenol accumulations by two and 100-fold, respectively [42]. Two pivotal genes of the metabolic pathway leading to diosgenin, 3-hydroxy-3-methylglutaryl-CoA reductase (HMG) and sterol-3-β-glucosyl transferase (STRL), were selected as target genes for the current study, in order to investigate their involvement in diosgenin biosythesis.
2. Results
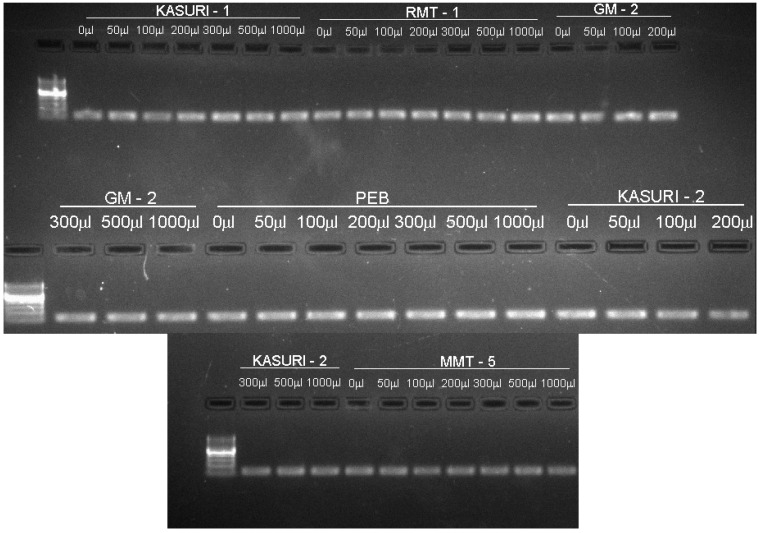
Total RNAs extracted from 36 treated and six control samples, analyzed with two peaks representing 28S and 18S ribosomal RNA along with a 5S ribosomal RNA peak, were observed on denaturing agarose gel electrophoresis and the RNA integrity number (RIN) value of all of them was found to be above six, which indicates that RNA can be used for further analyses. The size of the amplicons generated using housekeeping gene primers was 180 bp as confirmed in the agarose gel electrophoresis of all the 42 samples (Figure 1; Table 1). The amplicons generated for Kasuri-I and Kasuri-II variety were of low intensity, indicative of variation in the GC (guanine-cytosine) content of this plant.
Figure 1.
Electrophoretogram of amplified products generated for first strand complementary DNA (cDNA) from total RNA of methyl jasmonate (MeJA)-treated and control fenugreek (Trigonella foeum-graecum L.) plants using 18 housekeeping gene primers, analysed using 2% agarose gel electrophoresis.
Table 1.
Primer details of 3-hydroxy-3-methylglutaryl-CoA reductase (HMG), sterol-3-β-glucosyl transferase (STRL) and 18S housekeeping genes.
| Primer | Primer Sequence | Reference | PE ± SE * |
|---|---|---|---|
| 18S Ribosomal RNA | F primer: 5′→3′ GTCTCAACCATAAACGATGCCGACCA | Accession ID: KJ813746 | 2.0 ± 0.0 |
| R primer: ACCTGGTAAGTTTCCCCGTGTTGAGT | |||
| HMG | R primer: 5′→3′ ACTCAACACGGGGAAACTTACCAGGT | 171 bp Contig 19639-our transcriptome data | 2.1 ± 0.0 |
| F primer: 5′→3′ CACCCGCAAGATGAGATTTT | |||
| STRL | F primer: 5′→3′ TGGAAAATGAGGATGGGGTA | 144 bp Conting 10332-our transcriptome data | 2.0 ± 0.0 |
| R primer: 3′→5′TTGCATTGGATCAGGAACAA |
* PE ± SE: primer efficiency ± standard error.
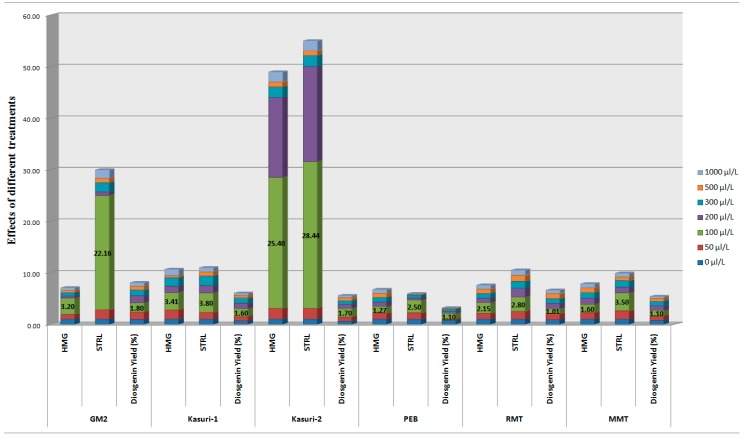
The expression of 3-hydroxy-3-methylglutaryl-CoA reductase (HMG) and sterol-3-β-glucosyl transferase (STRL) genes and diosgenin content of fenugreek seedlings are reported in Figure 2. Pattern of expression of both genes was found to be different among the fenugreek varieties, even if the highest expression was observed at 100 µL/L (0.01%) MeJA, in all varieties. Maximum expression of HMG gene was 3.2-fold in GM-2, 3.41-fold in Kasuri-1, 25.4-fold in Kasuri-2, 1.27-fold in PEB, 2.15 folds in RMT and 1.6 folds in MMT seedlings treated with 100 µL/L MeJA. The maximum expression of STRL gene was 22.16, 3.8, 28.44, 2.5, 2.8, and 3.5 folds in GM-2, Kasuri-1, Kasuri-2, PEB, RMT and MMT seedlings, respectively, after treatment with 100 µL/L MeJA. A two to three fold increase of diosgenin was recorded in all fenugreek varieties treated with 100 µL/L MeJA, in accordance with gene expression results (Figure 2). Notably, the eliciting effect of MeJA was shown to be dose-dependent on the expression of both target genes as well as on diosgenin levels, decreasing with increase of MeJA concentration up to 1000 µL/L (Table 2). Both results on gene expression and diosgenin content were statistically significant (p < 0.05) for 100 µL/L MeJA. However, the effects of MeJA treatments on seedling length, fresh weight and dry weight were not statistically significant (p > 0.05) (Table 3, Table 4 and Table 5).
Figure 2.
Effects of methyl jasmonate (MeJA) treatments on the expression of 3-hydroxy-3-methylglutaryl-CoA reductase (HMG) and sterol-3-β-glucosyl transferase (STRL) genes and diosgenin yield (%) in fenugreek (Trigonella foeum-graecum L.) plants (as per data reported in Supplementary Table S1).
Table 2.
Effect of methyl jasmonate (MeJA) treatments on diosgenin content (%) in fenugreek (Trigonella foeum-graecum L.) varieties.
| MeJA Concentration | Fenugreek Varieties (Diosgenin Yield in %) | |||||
|---|---|---|---|---|---|---|
| GM2 | Kasuri-1 | Kasuri-2 | PEB | RMT | MMT | |
| 0 µL/L | 0.9 | 0.65 | 0.55 | 0.75 | 0.85 | 0.75 |
| 50 µL/L | 1.44 | 0.78 | 0.83 | 0.23 | 1.19 | 0.83 |
| 100 µL/L | 1.8 * | 1.6 * | 1.7 * | 1.1 * | 1.01 * | 1.1 * |
| 200 µL/L | 0.72 | 1.04 | 0.77 | 0.38 | 0.97 | 0.9 |
| 300 µL/L | 1.62 | 1.01 | 0.66 | 0.45 | 1.02 | 0.83 |
| 500 µL/L | 0.72 | 0.48 | 0.61 | 0.15 | 0.94 | 0.6 |
| 1000 µL/L | 1.36 | 0.46 | 0.44 | 0.11 | 0.68 | 0.38 |
* p < 0.05 significant by ANOVA with post-hoc Tukey HSD test.
Table 3.
Effect of methyl jasmonate (MeJA) treatments on the length (cm) of seedlings of different fenugreek (Trigonella foeum-graecum L.) varieties.
| Fenugreek Varieties | MeJA Treatment | ||||||
|---|---|---|---|---|---|---|---|
| 0 µL/L | 50 µL/L | 100 µL/L | 200 µL/L | 300 µL/L | 500 µL/L | 1000 µL/L | |
| GM-2 | 5.5 ± 2 * | 5.5 ± 3 | 5.2 ± 4 | 5.5 ± 3 | 5.8 ± 4 | 5.9 ± 2 | 5.8 ± 4 |
| K-1 | 4.8 ± 3 | 4.9 ± 2 | 4.3 ± 4 | 5.2 ± 2 | 5.1 ± 3 | 5 ± 2 | 4.8 ± 3 |
| K-2 | 4.6 ± 2 | 4.8 ± 3 | 4.2 ± 3 | 5 ± 2 | 4.9 ± 3 | 4.8 ± 2 | 4.9 ± 2 |
| PEB | 5.2 ± 3 | 5.2 ± 2 | 4.3 ± 3 | 4.9 ± 2 | 4.8 ± 2 | 4.9 ± 3 | 4.8 ± 2 |
| RMT | 7.2 ± 4 | 7.2 ± 3 | 6.4 ± 2 | 7 ± 3 | 9.8 ± 5 | 6.9 ± 3 | 7 ± 3 |
| MMT | 6.2 ± 3 | 6.2 ± 2 | 5.3± 4 | 6.2 ± 3 | 6 ± 2 | 5.9 ± 3 | 5.8 ± 4 |
* Mean value ± SD (n = 3); here p > 0.05 indicates data not significant.
Table 4.
Effect of methyl jasmonate (MeJA) treatments on the fresh weight (mg) of seedlings of different fenugreek (Trigonella foeum-graecum L.) varieties.
| Fenugreek Varieties | MeJA Treatment | ||||||
|---|---|---|---|---|---|---|---|
| 0 µL/L | 50 µL/L | 100 µL/L | 200 µL/L | 300 µL/L | 500 µL/L | 1000 µL/L | |
| GM-2 | 175 ± 1 * | 172 ± 4 | 180 ± 8 | 179 ± 0 | 178 ± 5 | 174 ± 6 | 173 ± 5 |
| K-1 | 145 ± 3 | 151 ± 2 | 152 ± 1 | 146 ± 5 | 147 ± 0 | 142 ± 2 | 140 ± 2 |
| K-2 | 140 ± 5 | 144 ± 8 | 148 ± 8 | 142 ± 7 | 142 ± 5 | 141 ± 2 | 140 ± 9 |
| PEB | 179 ± 5 | 175 ± 1 | 182 ± 3 | 173 ± 8 | 172 ± 8 | 170 ± 5 | 168 ± 5 |
| RMT | 190 ± 8 | 185 ± 2 | 192 ± 5 | 187 ± 0 | 184 ± 8 | 183 ± 5 | 180 ± 3 |
| MMT | 174 ± 6 | 176 ± 1 | 180 ± 7 | 178 ± 2 | 177 ± 5 | 175 ± 6 | 171 ± 3 |
* Mean value ± SD (n = 3); here p > 0.05 indicates data not significant.
Table 5.
Effect of methyl jasmonate (MeJA) treatments on the dry weight (mg) of seedlings of different fenugreek (Trigonella foeum-graecum L.) varieties.
| Fenugreek Varieties | MeJA Treatment | ||||||
|---|---|---|---|---|---|---|---|
| 0 µL/L | 50 µL/L | 100 µL/L | 200 µL/L | 300 µL/L | 500 µL/L | 1000 µL/L | |
| GM-2 | 130 ± 8 * | 128 ±1 | 134 ±9 | 131 ± 5 | 124 ± 5 | 122 ±5 | 120 ± 8 |
| K-1 | 85 ± 5 | 88 ± 3 | 92 ± 5 | 81 ± 5 | 83 ± 0 | 82 ± 8 | 79 ± 8 |
| K-2 | 80 ± 5 | 82 ± 5 | 85 ± 9 | 84 ± 8 | 83 ± 7 | 79 ± 8 | 78 ± 3 |
| PEB | 125 ± 5 | 124 ± 9 | 136 ± 8 | 134 ± 0 | 133 ± 8 | 128 ± 8 | 122 ± 2 |
| RMT | 135 ± 5 | 136 ± 0 | 142 ± 5 | 139 ± 8 | 138 ± 5 | 132 ± 7 | 130 ± 5 |
| MMT | 133 ± 9 | 134 ± 8 | 140 ± 5 | 138 ± 7 | 137 ± 1 | 136 ± 5 | 130 ± 2 |
* Mean value ± SD (n = 3); here p > 0.05 indicates data not significant.
3. Discussion
The occurrence of diosgenin in plants is usually very low, even if its levels may be enhanced by treatment with elicitors or under stress conditions. A number of studies have provided evidence that some plant growth regulators, as ethylene and ethephon (an ethylene donor), may elicit the diosgenin accumulation in plants. Diosgenin content in fenugreek increased by 195% and 126% after treatment with 25 and 5 ppm ethephon, respectively [43,44]. MeJA was previously shown to be the most effective elicitor on diosgenin concentration in fenugreek seedlings, with a 10.5-fold increase at 100 µL/L MeJA, though varietal differences and gene expression were not investigated in this study [45]. In general, increase of secondary metabolite production can be mediated by the expression of key genes involved in specific metabolic routes [46,47].
Plants are sessile organisms continuously exposed to a plethora of biotic and abiotic stresses, and secondary metabolites are produced as part of the plant defense mechanisms [48]. A number of studies have proved that MeJA is a powerful inducer of secondary metabolites in various plants [49,50,51]. It enhanced the cell biomass and induced the phytochemical production in some plants of the Lamiaceae family, with positive or neutral effects on plant growth [52]. Earlier studies also reported that MeJA, originally isolated from fungal cell wall, up-regulates the expression of HMG and other genes involved in mevalonate pathway in some fungal species [53].
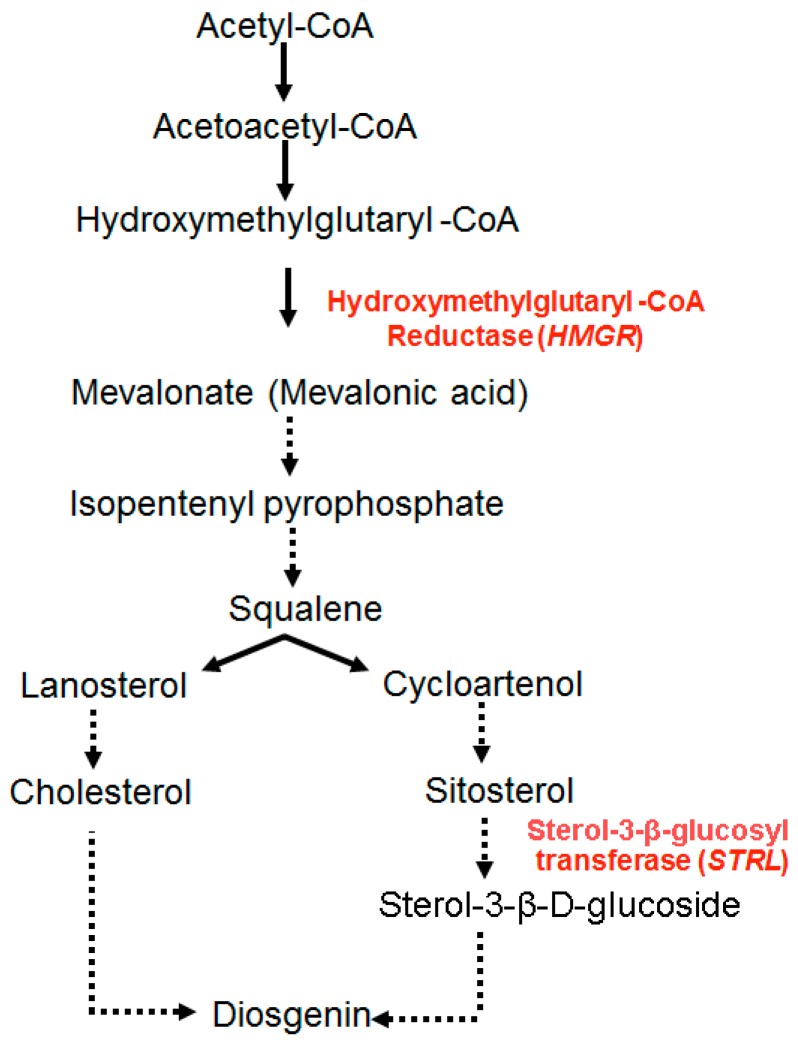
The expression of HMG gene is central to diosgenin biosynthesis, as the HMGR enzyme regulates the rate limiting step of the isoprenoid pathway and provides mevalonate, the precursor for the production of both primary and secondary metabolites (Figure 3). Ethylene treatment enhanced both HMG gene expression and diosgenin levels in in vitro cultures of Dioscorea zingiberensis [54]. Similarly, MeJA treatment stimulated HMG gene in Camptotheca acuminata [55].
Figure 3.
Proposed diosgenin biosynthetic pathway. 3-Hydroxy-3-methylglutaryl-CoA is converted to mevalonate by 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) enzyme. This is the major rate limiting step of isoprenoid pathway. Mevalonate is converted to isopentenyl pyrophosphate via series of multiple reactions. Then, diosgenin is produced from squalene by two routes: via cholesterol from lanosterol and via sitosterol from cycloartenol to sterol 3-β-d-glucoside. Finally, the latter is converted to diosgenin by sterol 3-β-glucosyl transferase (STRL) enzyme (dashed lines indicate multiple steps involved in the pathway; arrows indicate the flow of reaction in pathway).
Based on previous fenugreek transcriptome studies [33], we proposed a pathway for the production of diosgenin by combining mevalonate and sterol metabolic routes (Figure 3). We hypothesised that diosgenin may be formed from squalene 2,3-oxide in two ways, (i) from lanosterol, via the formation of cholesterol; and (ii) from cycloartenol via sitosterol. Mehrafarin et al. [56] tried to explain the biosynthesis of steroidal saponins from cholesterol, though all the steps were not completely defined. In the present study, the expression of the second target gene, STRL, was also elicited by MeJA treatment more than two fold in comparison with the HMG gene, thus supporting the proposed hypothesis of diosgenin biosynthesis from squalene via cycloartenol and sitosterol. The effects of MeJA on the increased expression of the two target genes of mevalonate pathway can be explained on the basis of transcriptional modulation of related genes as shown to be a common response to elicitor signals [57].
4. Materials and Methods
4.1. Fenugreek Seed Sterilization and Sowing
Seeds of six fenugreek (Trigonella foenum-graecum L.) varieties Gujarat Methi-2 (GM-2), Kasuri-1, Kasuri-2, Pusa Early Branching (PEB), Rajasthan Methi (RMT-1) and Maharashtra Methi-5 (MMT-5) were sterilized using sodium hypochlorite solution (4% W/V Merck, UN1791) for 10 min. Sterilized seeds were soaked in autoclaved water in petri-dishes for 48 h in dark for sprouting process. The germinated seeds were transferred to the pots filled with autoclaved soil from a farm near Ahmedabad (latitude 23°03′N and longitude 72°40′E). Then, in each plastic container (3.5 cm diameter at the bottom and 5.5 cm diameter at the top with 5.5 cm of depth), approximately 50 g of soil were transferred and 30 sprouted seeds transplanted; these were grown for 6 days in a sterile room with a light:dark cycle of 10:14 h at 25 °C temperature with the relative humidity around 36%–40% without any treatment.
4.2. Methyl Jasmonate Treatments
Methyl jasmonate (MeJA, Sigma Aldrich, St. Louis, MO, USA, reference number 392707, 95% pure) was dissolved in absolute ethanol and diluted in water to obtain 50, 100, 200, 300, 500 and 1000 µL/L (0.005%, 0.01%, 0.02%, 0.03%, 0.05% and 0.1%) solutions. Ethanol was used as control. One mL of each dilution was administered to each pot, in the morning and evening, 7 and 9 days after sprouted seed transplantation and pots were watered with 20 mL of water for uniform drenching of pot soil. A total of 36 experiments (6 fenugreek varieties 6× MeJA concentrations) and 6 control pots were grown in triplicate. Some seedlings harvested on the 12th day were washed thoroughly with autoclaved water to remove soil from the roots and stored in TMS RNA stabilizer (XGtms-100) solution at −80 °C for RNA isolation and gene expression studies, while others were kept 5 days in a hot air oven at 50 °C till complete dryness for diosgenin extraction.
4.3. Growth Parameter Measurement
Length and fresh weight of seedlings were recorded before diosgenin extraction; then they were dried for 5 days in a hot air oven at 50 °C to complete dryness and dry weight was registered.
4.4. Diosgenin Extraction and Analysis
Diosgenin extraction and determination were carried out according to Trivedi et al. [58] with modifications. Briefly, dried fenugreek seedlings were ground to fine powder using liquid nitrogen and re-fluxed with 50 mL of 2.5 M ethanol-sulfuric acid at 80 °C for 4 h. The solution was then filtered using Whatman filter paper no.1, diluted with 50 mL double distilled water and extracted with 50 mL of n-hexane 3 times, pooled and evaporated to dryness at room temperature. Dry residues were dissolved in 25 mL mobile phase (acetonitrile:water 90:10) and filtered through 0.22 µm filter before high performance liquid chromatography (HPLC) analysis. For HPLC analysis, Hypersil ODS C18, 5 µm, 250 × 4.6 mm (Thermo Scientific, Waltham, MA, USA) was used at flow rate of 1 mL per minute with mobile phase (acetonitrile:water 90:10), at 35 °C for 30 min. One mg per mL of diosgenin (D1634, SIGMA, Sigma Aldrich, St. Louis, MO, USA) was used as standard [58].
4.5. Primer Designing
Primers for the two target genes 3-hydroxy-3-methylglutaryl-CoA reductase (HMG) and sterol-3-β-glucosyl transferase (STRL) were designed as in the whole transcriptome data of Gujarat Methi-1 variety published by our lab [55]. Primer 3 plus software was used for primer designing and designed primers were synthesized in Primex facility at our lab. Primer efficiency of genes and their relative expressions were calculated using relative expression software tool [59]. Primer details are presented in Table 1.
4.6. RNA Extraction, Complementary DNA (cDNA) Preparation and Gene Expression
Fifty mg of frozen seedlings from all 42 pots were used for RNA isolation using Xcelgen plant RNA miniprep kit (XG6611-01) following the manufacturer’s instructions. The quantity of isolated total RNA was determined by absorbance ratios A260/280 and A260/230, on nanodrop spectrophotometer 8000 (Thermo Scientific), and quality of total RNA was analysed, on 1% denaturing agarose gel and Agilent Bioanalyzer 2100, using the RNA 6000 pico chip (Agilent Technologies, Santa Clara, Palo Alto, CA, USA). Then, 500 ng of total RNA was subjected to reverse transcription reaction for first strand cDNA synthesis using M-MLV reverse transcriptase enzyme (XG02032). For preparing first strand cDNA, 500 ng of total RNA with 0.5 µL RNase inhibitor were used with oligo dT primers. Reaction mixture used for cDNA synthesis contained 500–1000 ng of total RNA, 10 mM oligo-dt primer, 0.5 µL RNAse inhibitor and 10 mM dNTPs and the reaction was incubated at 65 °C for RNA denaturation. After incubation, 200 units of M-MLV enzyme, reverse trancriptase buffer and 50 mM dithiothreitol were added to the reaction and subjected to reverse transcription using three temperatures 25 °C for 10 min, 40 °C for 60 min and 70 °C for 10 min. First strand cDNA was quantified using nanodrop spectrophotometer and 100 ng were used for gene expression reaction. For standardization, polymerase chain reaction was set up in a gradient thermal cycler (Applied Biosystems Verity 96 well thermal cycler, 0.2 mL, Foster City, CA, USA) followed by analysis of the amplicons on 2% agarose gel. After standardization of the PCR conditions, the same conditions were used for gene expression assay using Light cycler 480II instrument (Roche lifesciences, Indianapolis, IN, USA). The gene expression reaction mixture contained 1X SYBR green master mix (Light cycler 480 SYBR green I master, 04707516001), 0.5 pM of each forward and reverse primers, 100 ng of first strand cDNA and 3 µL of nuclease free water (total volume 10 μL). All the samples were amplified using SYBR chemistry in triplicates using light cycler 480II instrument and CP values (crossing point (PCR-cycle) value) were recorded the same as Ct value (cycle threshold value); calculation of ΔΔCt was carried out using average Ct value and to analyze the gene expression pattern. Basic relative quantification method was used to analyze results as for studying physiological changes in the transcript level, as relative expression study is the most useful method [60]. Final results of the ΔΔCt value were described as fold change in the gene expression value.
4.7. Statistical Analysis
Statistical differences between control and treatments were assessed by one-way analysis of variance (ANOVA) with post-hoc Tukey HSD test. The p-value < 0.05 was considered significant.
5. Conclusions
In this study, our results showed that the 100 µL/L (0.01%) MeJA treatment increased diosgenin levels in seedlings of different fenugreek varieties, from 0.5%–0.9% (naturally occurring) to 1.1%–1.8%. In open-field conditions, it would require 1.24 µL/L MeJA which amounts, approximately, to $100/Ha crop area. The returns from increased diosgenin production might amount to around $100,000/Ha. MeJA elicited the expression of HMG and STRL genes, thus upregulating the mevalonate and sterol biosynthetic pathways and increasing diosgenin production from cycloartenol via the intermediate sitosterol. Therefore, MeJA may be considered a promising elicitor for diosgenin production by fenugreek plants.
Acknowledgments
We acknowledge the Management of Xcelris Labs Ltd., Ahmedabad, Gujarat, India, for financial support and encouragement during the study.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/12/26208/s1.
Author Contributions
Spandan Chaudhary has done literature survey, complete experimentation work and prepared the manuscript. Surendra K. Chikara and Mahesh C. Sharma has designed and guided for designing, experimentation and preparing the manuscript. Abhinav Chaudhary, Bakhtiyar Alam Syed and Pooja S. Chaudhary have helped in gene expression and diosgenin extraction and estimation studies. Aditya Mehta, Maulik Patel have done the statistical analysis of the data. Arpita Ghosh has analyzed the previous transcriptome data and has designed the primers for gene expression experiments. Marcello Iriti contributed to the study design, carried out extensive review and proofreading of the manuscript and finalized it for publication.
Conflicts of Interest
This study received the financial support of Xcelris Labs Ltd., Ahmedabad, India.
References
- 1.Acharya S.N., Thomas J.E., Basu S.K. Fenugreek, an alternative crop for semiarid regions of North America. Crop Sci. 2008;48:841–853. doi: 10.2135/cropsci2007.09.0519. [DOI] [Google Scholar]
- 2.Mullaicharam A.R., Geetali D., Uma M. Medicinal values of fenugreek—A review. Res. J. Pharm. Biol. Chem. Sci. 2013;4:1304–1313. [Google Scholar]
- 3.Leela N.K., Shafeekh K.M. Fenugreek. In: Parthasarathy V.A., Chempakam B., Zachariah T.J., editors. Chemistry of Spices. Biddles Ltd., CAB International; King’s Lynn, UK: 2008. pp. 242–259. [Google Scholar]
- 4.Gopu C.L., Gilda S.S., Paradkar A.R., Mahadik K.R. Development and validation of a densitometric TLC method for analysis of trigonelline and 4-hydroxyisoleucine in Fenugreek seeds. Acta Chromatogr. 2008;20:709–719. doi: 10.1556/AChrom.20.2008.4.15. [DOI] [Google Scholar]
- 5.Habori A., Raman M. Pharmacological Properties. In: Petropoulos G.A., editor. Fenugreek, the Genus Trigonella. Taylor and Francis; London, UK: 2002. pp. 163–182. [Google Scholar]
- 6.Tiran D. The use of fenugreek for breast feeding women. Complement. Ther. Nurs. Midwifery. 2003;9:155–156. doi: 10.1016/S1353-6117(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 7.Broca C., Breil V., Cruciani G.C., Manteghetti M., Rouault C., Derouet M., Rizkalla S., Pau B., Petit P., Ribes G., et al. The insulinotrophic agent 1D1101(4-hydroxyisoleucine) activates insulin signaling in rat. Am. J. Physiol. Endocrinol. Metab. 2004;287:E463–E471. doi: 10.1152/ajpendo.00163.2003. [DOI] [PubMed] [Google Scholar]
- 8.Devi B.A., Kamalakkannan N., Prince P.S. Supplementation of fenugreek leaves to diabetic rats-effect on carbohydrate metabolic enzymes in diabetic liver and kidney. Phytother. Res. 2003;17:1231–1233. doi: 10.1002/ptr.1357. [DOI] [PubMed] [Google Scholar]
- 9.Aasim M., Hussain N., Umer E.M., Zubair M., Hussain B.S., Saeed S., Rafique T.S., Sancak C. In vitro shoot regeneration of fenugreek (Trigonella foenum-graecum L.) using different cytokinins. Afr. J. Biotechnol. 2010;9:7174–7179. [Google Scholar]
- 10.Tahiliani P., Kar A. Mitigation of thyroxine induced hyperglycaemia by two plant extracts. Phytother. Res. 2003;17:294–296. doi: 10.1002/ptr.1121. [DOI] [PubMed] [Google Scholar]
- 11.Thakaran S., Salimuddin, Baquer N.Z. Oral administration of orthovanadate and Trigonella foenum-graecum seed powder restore the activities of mitochondrial enzymes in tissues of alloxan-induced diabetic rats. Mol. Cell. Biochem. 2003;247:45–53. doi: 10.1023/A:1024188600523. [DOI] [PubMed] [Google Scholar]
- 12.Suboh S.M., Bilto Y.Y., Aburjai T.A. Protective effects of selected medicinal plants against protein degradation, lipid peroxidation and deformability loss of oxidatively stressed human erythrocytes. Phytother. Res. 2004;18:280–284. doi: 10.1002/ptr.1380. [DOI] [PubMed] [Google Scholar]
- 13.Thompson Coon J.S., Ernst E. Herbs for serum cholesterol reduction: A systematic view. J. Fam. Pract. 2003;52:468–478. [PubMed] [Google Scholar]
- 14.Venkatesan N., Devaraj S.N., Devraj H. Increased binding of LDL and VLDL to apo B, E receptors of hepatic plasma membrane of rats treated with Fibranat. Eur. J. Nutr. 2003;42:262–271. doi: 10.1007/s00394-003-0420-8. [DOI] [PubMed] [Google Scholar]
- 15.Devasena T., Menon V.P. Fenugreek affects the activity of β-glucuronidase and mucinase in the colon. Phytother. Res. 2003;17:1088–1091. doi: 10.1002/ptr.1331. [DOI] [PubMed] [Google Scholar]
- 16.Harsha K., Preeti P., Manish K.P., Harendra K., Amit C.K., Devi D.J. Foaming glycosides: A review. IOSR J. Pharm. 2012;2:23–28. [Google Scholar]
- 17.Freeman B.C., Beatiie G.A. An overview of plant defenses against pathogens and herbivores. Plant Health Instruct. 2008 doi: 10.1094/PHI-I-2008-0226-01. [DOI] [Google Scholar]
- 18.Gonzalez L.R., Mitchell G., Gattuso M., Diarra M.S., Malouin F., Bouarab K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009;10:3400–3419. doi: 10.3390/ijms10083400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal M., Agarwal M., Jain S.C. In vitro regulation of bioactive compounds in Trigonella species by mutagenic treatments. J. Plant Sci. 2015;3:40–44. [Google Scholar]
- 20.Lee J., Jung K., Kim Y.S., Park D. Diosgenin inhibits melanogenesis through the activation of phosphatidylinositol-3-kinase pathway (PI3K) signaling. Life Sci. 2007;81:249–254. doi: 10.1016/j.lfs.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Tada Y., Kanda N., Haratake A., Tobiishi M., Uchiwa H., Watanabe S. Novel effects of diosgenin on skin aging. Steroids. 2009;74:504–511. doi: 10.1016/j.steroids.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Yan L.L., Zhang Y.J., Gao W.Y., Man S.L., Wang Y. In vitro and in vivo anticancer activity of steroid saponins of Paris polyphylla var. yunnanensis. Exp. Oncol. 2009;31:27–32. [PubMed] [Google Scholar]
- 23.Gong G., Qin Y., Huang W., Zhous S., Wu X., Yang X., Zhao Y., Li D. Protective effects of diosgenin in the hyperlipidemic rat model and in human vascular endothelial cells against hydrogen peroxide-induced apoptosis. Chem.-Biol. Interact. 2010;184:366–375. doi: 10.1016/j.cbi.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Katy G.D., Nadja A.C., Kristhea G.P. Mechanisms involved in the vasodilator effect induced by diosgenin in rat superior mesenteric artery. Eur. J. Pharmacol. 2007;574:172–178. doi: 10.1016/j.ejphar.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Qin T.C., Zhang Y.D., Zhang J.Z. Distribution and utilization of diosgenin resources in Hubei province. Biol. Resour. 1997;13:200–202. [Google Scholar]
- 26.Liu P., Wu X.Y., Yue D.Y. Comprehensive utilization of diosgenin resources. Resour. Econ. Compr. Util. 1993;12:47–49. [Google Scholar]
- 27.Aradhana M., Rao A.C., Kale R.K. Diosgenin a growth stimulator of mammary gland of ovariectomized mouse. Indian J. Exp. Biol. 1992;30:367–370. [PubMed] [Google Scholar]
- 28.Patel K., Gadewar M., Tahilyani V., Patel D.K. A review on pharmacological and analytical aspects of diosgenin: A concise report. Nat. Prod. Bioprospect. 2012;2:46–52. doi: 10.1007/s13659-012-0014-3. [DOI] [Google Scholar]
- 29.Rosser A. The day of the yam. Nurs. Times. 1985;81:47–48. [Google Scholar]
- 30.Hardman R. Pharmaceutical products from plant steroids. Trop. Sci. 1969;11:196–222. [Google Scholar]
- 31.Petropoulos G.A. Ph.D. Thesis. Bath University; Bath, UK: 1973. Agronomic, Genetic and Chemical Studies of Trigonella foenum-graecum L. [Google Scholar]
- 32.Kanak V., Arpita G., Vinay K., Spandan C., Navin S., Kalpesh K., Tanushree T., Surendra K.C. De novo transcriptome sequencing in Trigonella foenum-graecum L. to identify genes involved in the biosynthesis of diosgenin. Plant Genome. 2013;6:1–11. [Google Scholar]
- 33.Paul E.S., Wenpei S., Stephen H.H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sembdner G., Parthier B. The biochemistry and the physiological and molecular actions of jasmonates. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:569–589. doi: 10.1146/annurev.pp.44.060193.003033. [DOI] [Google Scholar]
- 35.Aldridge D.C., Galt S., Giles D., Turner W.B. Metabolites of Lasiodiplodia theobromae. J. Chem. Soc. 1971:1623–1627. doi: 10.1039/j39710001623. [DOI] [Google Scholar]
- 36.Goodrich-Tanrikulu M., Mahoney N.E., Rodriguez S.B. The plant growth regulator methyl jasmonate inhibits aflatoxin production by Aspergillus flavus. Microbiology. 1995;141:2831–2837. doi: 10.1099/13500872-141-11-2831. [DOI] [PubMed] [Google Scholar]
- 37.Aerts R.J., Gisi D., Carolis E., Luca V., Baumann T.W. Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J. 1994;5:635–643. doi: 10.1111/j.1365-313X.1994.00635.x. [DOI] [Google Scholar]
- 38.Wasternack C., Parthier B. Jasmonate-signalled gene expression. Trends Plant Sci. 1997;2:302–307. doi: 10.1016/S1360-1385(97)89952-9. [DOI] [Google Scholar]
- 39.Morrissey J.P., Osbourn A.E. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 1999;63:708–724. doi: 10.1128/mmbr.63.3.708-724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Wang X., Li Z., Kong L., Liu G., Fu J., Wang A. Molecular cloning, tissue expression and protein structure prediction of the porcine 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) gene. Gene. 2012;495:170–177. doi: 10.1016/j.gene.2011.12.051. [DOI] [PubMed] [Google Scholar]
- 41.Heidrun G., Martin J.M., Tonim K., Meinhart H.Z. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joseph C., Fred W., Jeanne P., Rick C., Court S. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plant. Plant Physiol. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oncina R., Del R.J.A., Gomez P., Ortuno A. Effect of ethylene on diosgenin accumulation in callus cultures of Trigonella foenum-graecum L. Food Chem. 2002;76:475–479. doi: 10.1016/S0308-8146(01)00309-0. [DOI] [Google Scholar]
- 44.Gomez P., Ortuno A., Del R.J.A. Ultrastructural changes and diosgenin content in cell suspensions of Trigonella foenum-graecum L. by ethylene treatment. Plant Growth Regul. 2004;44:93–99. doi: 10.1023/B:GROW.0000049415.62026.c3. [DOI] [Google Scholar]
- 45.Debjani D., Bratati D. Elicitation of diosgenin production in Trigonella foenum-graecum L. seedlings by heavy metals and signaling molecules. Acta Physiol. Plant. 2011;33:1585–1590. [Google Scholar]
- 46.Romeo R., Teresa S., Christopher B., Tajalli K. Elicitation of plants and microbial cell systems. Biotechnol. Appl. Biochem. 2003;37:91–102. doi: 10.1042/ba20020118. [DOI] [PubMed] [Google Scholar]
- 47.Namdeo A.G. Plant cell elicitation for production of secondary metabolites: A review. Pharmacogn. Rev. 2007;1:69–79. [Google Scholar]
- 48.Iriti M., Faoro F. Chemical diversity and defence metabolism: How plants cope with pathogens and ozone pollution. Int. J. Mol. Sci. 2009;10:3371–3399. doi: 10.3390/ijms10083371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H.J., Chen F., Wang X., Rajapakse N.C. Effect of methyl jasmonate on secondary metabolites of sweet basil (Ocimum basilicum L.) J. Agric. Food Chem. 2006;54:2327–2332. doi: 10.1021/jf051979g. [DOI] [PubMed] [Google Scholar]
- 50.Keinanen M., Oldham N., Baldwin I.T. Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J. Agric. Food Chem. 2001;49:3553–3558. doi: 10.1021/jf010200+. [DOI] [PubMed] [Google Scholar]
- 51.Dixon R.A., Paiva N.L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rebecca M., Deepa S.P. Effect of methyl jasmonate and chitosan on growth characteristics of Ocimum basilicum L., Ocimum sanctum L. and Ocimum gratissimum L. cell suspension cultures. Afr. J. Biotechnol. 2012;11:4759–4766. doi: 10.5897/AJB11.3183. [DOI] [Google Scholar]
- 53.Ren A., Qin L., Shi L., Dong X., Mu da S., Li Y.X., Zhao M.W. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour. Technol. 2010;101:6785–6790. doi: 10.1016/j.biortech.2010.03.118. [DOI] [PubMed] [Google Scholar]
- 54.Diarra S.T., Ji H., Jianbo W., Jiaru L. Ethylene treatment improves diosgenin accumulation in in vitro cultures of Dioscorea zingiberensis via up-regulation of CAS and HMGR gene expression. Electron. J. Biotechnol. 2013 doi: 10.2225/vol16-issue5-fulltext-9. [DOI] [Google Scholar]
- 55.Burnett R.J., Maldonado-Mendoza I.E., McKnight T.D., Nessler C.L. Expression of a 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Camptotheca acuminata is differentially regulated by wounding and methyl jasmonate. Plant Physiol. 1993;103:41–48. doi: 10.1104/pp.103.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehrafarin A., Qaderi A., Rezazadeh S., Naghdi Badi H., Noormohammadi G.H., Zand E. Bioengineering of important secondary metabolites and metabolic pathways in fenugreek (Trigonella foenum-graecum L.) J. Med. Plants. 2010;9:1–18. [Google Scholar]
- 57.Zhao J., Davis L.C., Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Trivedi P.D., Pundarikakshudu K., Rathnam S., Shah K.S. A validated quantitative thin-layer chromatographic method for estimation of diosgenin in various plant samples, extract, and market formulation. J. AOAC Int. 2007;90:358–363. [PubMed] [Google Scholar]
- 59.Michael W.P., Graham W.H., Leo D. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.