Abstract
Introduction
Streptococcus agalactiae (group B streptococcus; GBS) is globally recognised as one of the leading causes of neonatal sepsis and meningitis. It also causes adverse pregnancy outcomes such as stillbirth and miscarriages. Incidence of invasive disease is increasing in non-pregnant adults with underlying medical conditions (e.g., diabetes mellitus). Epidemiological studies of GBS infections are based on capsular serotyping. Genotyping of the surface anchored protein genes is also becoming an important tool for GBS studies. Currently ten different GBS serotypes have been identified. This study was performed to determine the prevalence of GBS capsular types (CTs) and surface anchored protein genes in isolates from colonized pregnant women attending antenatal clinic, at Dr George Mukhari Academic Hospital, Garankuwa, Pretoria, South Africa.
Methods
The samples were collected over 11 months and cultured on selective media. GBS was identified using different morphological and biochemical tests. Capsular typing was done using latex agglutination test and conventional PCR. Multiplex PCR with specific primers was used to detect the surface anchored protein genes.
Results
Of the 413 pregnant women recruited, 128 (30.9%) were colonized with GBS. The capsular polysaccharide (CPS) typing test showed that CPS type III (29.7%) was the most prevalent capsular type followed by CPS type Ia (25.8%), II (15.6%), IV (8.6%), V (10.9%) and Ib (8.6%); 0.7% of the isolates were nontypeable. Multiplex PCR revealed that the surface proteins genes were possessed by all the capsular types: rib (44.5%), bca (24.7%), alp2/3 (17.9%), epsilon (8.6%) and alp4 (4.7%).
Conclusion
The common capsular types found in this study are Ia, III, and II. The most common protein genes identified were rib and bca, and the distribution of the surface protein genes among the isolates of different capsular types showed similar trends to the distribution reported from previous studies.
Keywords: Streptococcus agalactiae, pregnancy, serotypes, surface proteins
Introduction
Streptococcus agalactiae (group B streptococcus; GBS) has remained one of the leading causes of bacterial sepsis and meningitis in neonates for the past few decades.1 Non-pregnant adults with underlying diseases, can also be affected by group B streptococcal infections.1,2 More than 30% of pregnant women are colonized with GBS.3 Colonization in late pregnancy is a risk factor for poor pregnancy outcome because GBS has the ability to penetrate the intact amniotic membrane causing amnionitis thereby infecting the foetus in utero which could lead to miscarriage.3,4
Capsular polysaccharide (CPS) antigens in GBS have been studied extensively and are important antigenic markers. Virulence of clinical isolates with similar capsular composition may vary, suggesting that other bacterial factors in addition to the capsule are involved in the pathogenesis of GBS infection.5 There are ten antigenically distinct CPS types identified, namely Ia, Ib, II – IX.6 Capsular type distribution varies with geographical region and ethnic origin of the population.5 Types Ia, III and V are the most predominant types in most parts of the world except for Japan where types VI and VIII are more frequent.7,8
There are also surface–anchored protein antigens, in addition to capsular polysaccharides, that act as antigenic markers in GBS. These are: Cα, Cβ, the alpha-like proteins (Alps), namely alp1 (epsilon), alp2, alp3, alp4 and R4 (Rib).9 Some of these proteins contain large internal tandem repeats.10 Stable mosaic genes encode the surface proteins; these genes are created through modules’ recombination at one chromosomal locus.9 The size of each protein depends on the number of repeats in the corresponding gene. The number of repeats in Cα can undergo internal deletion as a way of dodging the host immune response.11 The surface-anchored protein antigens Cα and Rib are immunologically different, unlike alp2, alp3, epsilon and Cα, that cross-react during western blotting.12 These proteins are not necessarily immunologically the same based on cross-reactivity; cross-reacting can also be attributed to sharing similar epitopes or impurity of antibodies used.
Various laboratory techniques have been used to characterize antigens in GBS. These include immunological and molecular techniques. Serotyping kits have reduced the number of GBS nontypeable isolates by nearly 50%.13 PCR has been used successfully, for the detection of different types of GBS capsular polysaccharide antigen and surface anchored protein genes.10,14 To allow vaccine development, it is important to characterize antigenic markers which are able to induce protective immunity, especially in invasive GBS strains. The current study aimed to describe the distribution of capsular types and the surface anchored protein genes found in GBS isolates recovered from colonized pregnant women attending antenatal care at the Dr George Mukhari Academic Hospital, Garankuwa, Pretoria, South Africa.
Methods
Sample collection
The study population consisted of pregnant women with ages between 18-45 years, at the gestational period of 16 to 38 weeks, attending antenatal clinic, at Dr George Mukhari Academic Hospital, Garankuwa, Pretoria, South Africa. This area of Garankuwa is mainly made up of different black African tribal groups. The women had not received antibiotic treatment for at least two weeks prior to recruitment into the study and sample collection. Those who were regarded as critically ill enough to be admitted by the attending physician were also excluded from the study. Participants signed a written informed consent and confidentiality of their results was assured.
Specimens included rectal and vaginal swabs and were collected by a gynaecologist, over the course of eleven months, from 413 pregnant women. The accurately labelled swabs were placed in Amies transport medium (Rochelle Chemical, Pretoria, South Africa), in a cooler box containing ice packs, and transported to the laboratory at the Department of Microbiological Pathology, University of Limpopo, Medunsa campus within 2-4 hours of collection. Specimens were cultured onto selective media, 5% sheep blood, Columbia colistin and nalidixic acid (CNA) agar (DMP-National Health Laboratory Service, Pretoria, South Africa). Details of the microbiological processing have been described previously.15
Capsular typing
Serological capsular typing was performed with the latex agglutination test kit (Staten Serum Institut, Copenhagen, Denmark) using specific antisera against type Ia, Ib, II-IX to detect the capsular polysaccharide antigens as described by Slotved et al., 2003.13
Molecular typing
Conventional PCR method was used for the molecular capsular typing of GBS isolates. Firstly, DNA was extracted with genomic DNA isolation kit (Norgen Biotek, Thorold, Canada) following the manufacturer’s instructions. The specific primers for typing capsular types I to V used are shown in Table 1 and were synthesized locally (Davies Diagnostics, Randburg, South Africa).16,17 A final volume of 50 µL consisting of 2.5 µL of dNTP, 2 µL of 10x PCR buffer, 3 µL of MgCl, 33.2 µL of dH2O, 0.3 µL of AmpliTag Gold, 2 µL of forward primer, 2 µL of reverse primer and 5 µL of DNA template were used as the master-mix. PCR was done using conditions described before on a Gene Amp® PCR system 9700 version 3.05 for the amplification.10
Table 1. Oligonucleotide primers used for capsular typing.
| Capsular type | Primer | Expected amplicon size | Sequence |
|---|---|---|---|
| Ia | IacpsHS1-R | 354 | 5'-GGC CTG CTGGGA TTA ATG AAT ATA GTT CCA GGT TTG C-3' |
| Ia and III | CpsIA-F | - | 5'-GTA TAA CTT CTA TCA ATG GAT GAG TCT GTT GTAGTACGG-3' |
| Ib | IbcpsIS-F | 523 | 5'-GAT AAT AGT GGA GAA ATT TGT GAT AAT TTA TCT CAA AAA GAC G-3' |
| Ib | IbcpsIA1-R | - | 5'-CCT GAT TCA TTG CAG AAG TCT TTA CGA TGC GAT AGG TG-3' |
| III | IIIcpsHS-F | 641 | 5'-GAA TAC TAT TGG TCT GTA TGT TGG TTT TAT TAG CAT CGC-3' |
| II | cpsIIK-F | 526 | 5'-CCA ACG GCA ATA AAA TAC AT-3' |
| II | cpsIIK-R | - | 5'-GCATTGAGATTAGAGTAGTC-3' |
| IV | IVcpsHS1-F | 379 | 5'-CCC AAG TAT AGT TAT GAA TAT TAG TTG GAT GGT TTT TGG-3' |
| IV | VcpsMA-R | - | 5'-GGG TCA ATT GTA TCGTCGCTG TCA ACA AAA CCA ATC AAA TC-3' |
| V | VcpsHS2-F | 374 | 5'-CCC AGT GTG GTA ATG AAT ATT AGT TGG CTA GTT TTT GG-3' |
| V | VcpsMA-R | - | 5'-CCC CCC ATA AGT ATA AATAATATC CAA TCT TGC ATA GTC AG-3' |
Amplified products were detected on 2% agarose gel. A photographic copy of the positive amplicons (bands) on the gel was taken with a Gel DocTM EZ Imager (BioRad, Hercules, CA, USA) as shown in Figure 1.
Figure 1. Gel image of the conventional PCR for detection of capsular type genes.
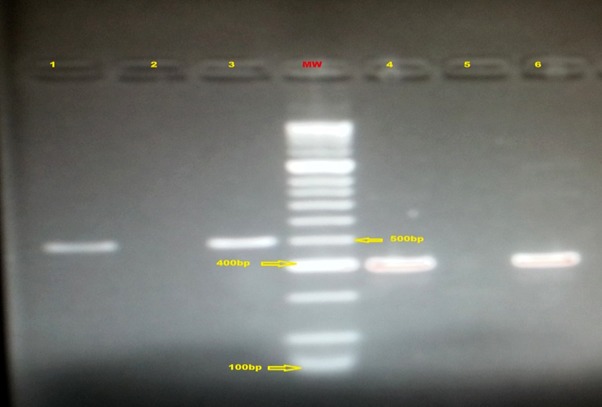
Capsular type Ib (lane 1), capsular type II (lane 3), capsular type V (lane 4) and capsular type IV (lane 6). Lane MW is the molecular weight marker (HyperLadderTM 100bp Bioline Reagent Limited, London, United Kingdom) with lane 2 and 5 as the negative controls.
Subtypes of GBS isolates
Multiplex PCR method was used to detect the surface protein genes bca, rib, epsilon, alp2/3 and alp4 in 128 GBS isolates, using primer nucleotide threads corresponding to the distinctive threads as a reverse primer (Integrated DNA Technologies, Pretoria, South Africa) and a nucleotide thread, that is common to all the surface protein genes, as universal forward primer14 (Table 2).
Table 2. Primers for detecting GBS surface protein genes.
| Surface protein gene | Primer | Expected amplicon size | Sequence |
|---|---|---|---|
| Universal | Universal-F | - | 5'-TGA TAC TTC ACA GAC GAA ACA ACG-3' |
| bca | Alpha-C-R | 398 | 5'-TAC ATG TGG TAG TCC ATC TTC ACC-3' |
| rib | Rib-R | 295 | 5'-CAT ACT GAG CTT TTA AAT CAG GTG A-3' |
| epsilon | Epsilon-R | 200 | 5'-CCA GAT ACA TTT TTTACT AAA GCG G-3' |
| alp2/3 | Alp2/3-R | 334 | 5'-CAC TCG GAT TAC TAT AAT ATT TAG CAC-3' |
| alp4 | Alp4-R | 110 | 5'-TTA ATT TGC ACC GGA TTA ACA CCA C-3' |
Ethical considerations
Permission to conduct the study was sought and granted from the authorities, including MEDUNSA Research and Ethics Committees, Dr George Mukhari Academic Hospital Management and UNISA, College of Agriculture and Environmental Health Sciences, Research and Higher Degrees Ethics Committee.
Results
Of the 413 pregnant women recruited in this study, GBS was identified from 128 (30.9%) participants. All the participants were from the same ethnicity but from three different locations (urban, rural-urban and rural areas) which are within and around George Mukhari Academic Hospital, Garankuwa. These were located in communities which differed in the availability of socio-economic and health services. Most isolates were recovered from participants residing in the urban area (113/128), followed by rural-urban (8/128), and rural (4); 3 women did not mention their location. Meaningful comparison of serotype distribution data from the areas was not useful as most of the isolates came from urban women.
Capsular type distribution
Capsular typing was done on 100 (78.1%) of the 128 isolates, obtained from colonized mothers with Strep B Latex typing kit. A total of 7/100 (7%) isolates were serologically nontypeable. Table 3 shows all the capsular types obtained by latex agglutination method. Capsular type III was the most common serotype constituting 26%, followed by Ia at almost the same proportion. The least common were type Ib and the nontypeables at 7%.
Table 3. Distribution of GBS capsular types detected using the two methods.
| Serotypes | Latex, no. (%) | PCR, no. (%) | Total, no. (%) |
|---|---|---|---|
| Ia | 25 (25.0) | 8 (22.9) | 33 (25.8) |
| Ib | 7 (6.0) | 4 (11.4) | 11 (8.6) |
| II | 18 (18.0) | 2 (5.7) | 20 (15.6) |
| III | 26 (26.0) | 12 (34.3) | 38 (29.7) |
| V | 9 (9.0) | 5 (14.3) | 14 (10.9) |
| IV | 8 (8.0) | 3 (8.5) | 11 (8.6) |
| Nontypeable | 7 (7.0) | 1 (2.8) | |
| Total | 100 | 35 | 128 |
Molecular typing
Conventional PCR was designed with the intention to type all isolates and compare with the latex agglutination typing results. However reagents were inadequate and only available to type 28/128 (21.9%) of the isolates, in addition to seven nontypeable isolates making a total of 35 isolates. Due to the limitation of funding, the intended comparison of PCR typing and latex agglutination typing could not be achieved for the isolates as was planned originally. Serotype III occurred more often, with the rate of 34.3% and the least frequent was type II. One isolate remained nontypeable. Table 3 shows the distribution of the GBS capsular gene of the 35 isolates, using this molecular method.
The overall distribution of GBS capsular types found in the current study is shown in Table 3. A total of 93/100 GBS isolates were serologically typeable by latex agglutination and 34/35 isolates were typeable by conventional PCR method. The overall distribution of the capsular types mainly consisted of type III with the least being type Ib and type IV at 8.6%. No strain of type VI, VII, VIII and IX was found using the latex agglutination test.
A typical gel image of the electrophoresis of the conventional PCR used in typing the GBS isolates is shown in Figure 1. It shows the banding pattern of capsular type Ib, II, V and IV. Lane MW is the 100 base pairs molecular weight marker used in the study, which helped in the interpretation of the amplified products. This was the PCR assay performed with specific capsular type primers on the 28 GBS isolates and the 7 serologically nontypeable strains.
Antigenic subtypes of GBS detected
Surface-anchored protein genes were investigated in the 128 GBS isolates. Multiplex PCR method was used to detect genes bca, rib, epsilon, alp2/3 and alp4. Table 4 shows the rate of detection of the genes encoding these proteins. Isolates of various serotypes could be segregated into various subtypes. The gene rib (44.5%) and bca (24.2%) were detected more often than the other surface protein genes. Figure 2 shows the image of gel electrophoresis of multiplex PCR amplified products. Analysis of the amplicon size was used for the determination of the identity of the genes for these GBS surface proteins. Lane MW is the 50 base pairs molecular weight maker. Alp4 protein gene encoding the alp4 protein was detected using the amplicon size of 110, in lanes 1, 2, 8, 9 and 13. Epsilon gene was detected by the presence of amplicon size of 200.
Table 4. Distribution of the protein genes found among GBS serotypes.
| Surface protein gene | Serotypes/number of surface protein genes possessed | |||||||
|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | NT | Total | |
| alp4 | 2 | 0 | 2 | 0 | 0 | 2 | 0 | 6 |
| epsilon | 9 | 0 | 0 | 0 | 0 | 1 | 0 | 10 |
| alp2/3 | 2 | 2 | 8 | 2 | 1 | 8 | 0 | 23 |
| bca | 19 | 6 | 2 | 0 | 4 | 0 | 0 | 31 |
| rib | 0 | 2 | 6 | 36 | 8 | 5 | 0 | 57 |
| Total | 32 | 10 | 18 | 38 | 13 | 16 | 1 | 128 |
Figure 2. Gel image of the multiplex PCR.
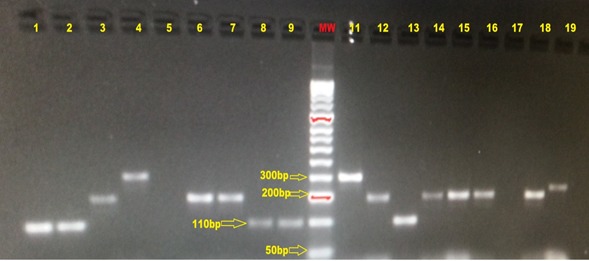
Alp4 (lanes 1, 2, 8, 9, and 13), Epsilon (lanes 3, 6, 7, 12, 14, 15, 16 and 18), Alp3 (lane 19), Rib (lane11), Alp2/3 (lane 4), Lane 5 and 17 are the negative controls. MW is the molecular weight marker (HyperLadderTM 50bp Bioline Reagent Limited, London, United Kingdom)
The rib gene was the most frequently detected surface protein gene. It was found in all the serotypes isolated, except for serotype Ia. A total of 36 isolates with the rib gene were detected mainly from serotype III but none in type Ia. Isolates with the alpha C gene were found mainly in serotype Ia isolates. Alp2/3 was detected in all the serotypes. Epsilon was found mainly in serotype Ia. Alp4 was detected in CPS types Ia, II and V as shown in Table 4. No protein was detected from the single isolate which remained nontypeable by PCR. The nontypeable isolate did not possess any protein gene.
Discussion
The colonization rate in our study was similar to the rate previously reported in other Southern African countries: South Africa and Zimbabwe, but differed from the colonization rate reported in some developed countries.18-20 Incidences as low as 0.3-5% has been reported in East Asian countries, while African countries such as Zimbabwe have reported greater than 60% colonization rate in pregnant patients.4,21
Ethnicity could have accounted for these differences. For instance, African American heritage is considered a risk factor for early-onset sepsis in neonates born to colonized mothers.22
To determine the capsular types and subtypes of GBS strains isolated from colonized pregnant women at DGMH, we used two methods, serotype-specific latex agglutination method and PCR based approach. The most common method for identifying capsular type is with serotype-specific latex agglutination, but this method has been regarded as expensive when used on large patient populations.23 The method however resulted in a high number of isolates which are nontypable (Table 3) as compared to molecular typing methods. The percentage of nontypeable isolates reported in our study was lower than that reported in studies primarily using the serotype-specific latex agglutination method.18,23 The reason is that molecular typing is more sensitive than serological typing.
In comparing the capsular type distribution, the present study reported a similar frequency to previously reported studies from Zimbabwe, Malawi and South Africa.18,24-26 This suggests that capsular type prevalence has remained stable in Africa over the past decades. However, intra-country regional differences of communities’ socio-demographic characteristics may affect GBS capsular distribution. Serotypic distribution shifts have been previously reported among different regions.21
The common capsular types found in this study are Ia, Ib, II, III, IV and V. Serotypes VI, VII, VIII and IX were not detected using the latex agglutination test. The distributions agree with the reports from other African countries where the above serotypes were found in clinical isolates.4,18,19 The two serotypes that were predominant among the colonizing isolates were type III and type Ia. Globally higher rates of capsular types Ia, Ib, IV, and V have been reported.20,26
All the GBS surface protein genes that we searched for in the current study, namely alp4, epsilon, alp2/3, bca and rib, were detected among the GBS isolates. These surface protein genes were found in association with six serotypes that were identified earlier. However it is important to note that there were other surface protein genes described before, which were not included in the current study, such as alp1, sar5 and bac genes which encode for alp1, R5 and the Cβ; proteins respectively. These were excluded partly because their estimated frequency is known from previous studies done from this region and considering the limited financial resources for the current study it was not practically possible to include all of them.27,28
In the present study, the most common gene identified was rib (44.5%) followed by bca (24%), alp2/3 (18%) epsilon (8.6%) and alp4 (4.7%). This result is similar to the data previously reported from Norway.[37,29] The distribution of the GBS surface proteins among the isolates of different capsular types showed similarities to the distribution elsewhere despite use of immunological techniques by the earlier researchers.26 Rib, the commonly detected surface protein gene was found in capsular types III, IV, II, V and Ib. The bca gene encoding Cα protein was detected in capsular types Ia, Ib, II and IV. Epsilon and bca genes were identified in 8.6% and 24.2% respectively in serotype Ia. This result is in agreement with the findings reported by Mavenyengwa et al., in 2008, although they reported a higher rate of alp1 gene and a lower rate of bca gene in serotype Ia. All the capsular types identified expressed alp2/3 protein antigen. This result suggests that, if this surface protein antigen has some protective immunogenic properties, then its inclusion in a vaccine would cover infection by many capsular types.27
Some studies have reported a significant relationship between capsular types and several surface protein genes.10,14 For example, Creti et al. noted a relationship between serotypes Ia, Ib, and II with Alpha-C protein and capsular type III with Rib and serotype V and VIII with Alp3, but it was not absolute.14 Furthermore another study reported associations between epsilon/alp gene with serotype Ia, bca with serotype Ib and II, rib with serotype III and alp3 with serotype V.29 The immunological significance which might have an impact on vaccine development needs to be explored further.
Conclusion
The common capsular types found in this study are Ia, III, and II. The most common surface protein genes identified were rib and bca, and the distribution of the surface protein genes among the isolates of different capsular types showed similarities to the distribution elsewhere. Based on the combinations of capsular types and surface protein genes found in the present study, future vaccine formulations based on capsular types Ia, III, and II and surface protein genes rib, bca, and epsilon would potentially have a broad coverage among female patients from South Africa.
Acknowledgment
We express gratitude to the staff at the Department of Microbiological Pathology, University of Limpopo – MEDUNSA campus – especially Ms B. De Villiers and Dr M. Le Roux. We sincerely thank the staff of the National Health Laboratory Service (NHLS) – DGMAH – Tertiary Laboratory, especially Ms M. Makhado for all her technical support.
Footnotes
Authors’ contributions statement: All authors contributed equally to study design, implementation, analysis and write-up of the manuscript. All authors read and approved the final manuscript.
Conflicts of interest: All authors – none to declare.
Funding: The study was funded by National Research Fund (NRF), VLIR/UL partnership program and UNISA Research directorate.
Presented in part at the 19th Lancefield International Symposium on streptococci and streptococcal diseases, Buenos Aires, Argentina, 2014.
References
- 1.Edwards MS, Baker CJ. Group B streptococcal infections in elderly adults. Clin Infect Dis. 2005;6:839–47. doi: 10.1086/432804. [DOI] [PubMed] [Google Scholar]
- 2.Skoff TH, Farley MM, Petit S, et al. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin Infect Dis. 2009;49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 3.Hansen SM, Uldbjerg N, Kilian M, Sørensen UB. Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J Clin Microbiol. 2004;42:83–9. doi: 10.1128/JCM.42.1.83-89.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavenyengwa RT, Afset JE, Schei B, et al. Group B Streptococcus colonization during pregnancy and maternal-fetal transmission in Zimbabwe. Acta Obstet Gynecol Scand. 2010;89:250–5. doi: 10.3109/00016340903398029. [DOI] [PubMed] [Google Scholar]
- 5.Spellerberg B. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2000;2:1733–42. doi: 10.1016/s1286-4579(00)01328-9. [DOI] [PubMed] [Google Scholar]
- 6.Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. Serotype IX, a proposed new Streptococcus agalactiae serotype. J Clin Microbiol. 2007;45:2929–36. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murayama SY, Seki C, Sakata H, et al. Capsular type and antibiotic resistance in Streptococcus agalactiae isolates from patients, ranging from newborns to the elderly, with invasive infections. Antimicrob Agents Chemother. 2009;53:2650–3. doi: 10.1128/AAC.01716-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z, Kong F, Zeng X, Gidding HF, Morgan J, Gilbert GL. Distribution of genotypes and antibiotic resistance genes among invasive Streptococcus agalactiae (group B streptococcus) isolates from Australasian patients belonging to different age groups. Clin Microbiol Infect. 2008;14:260–7. doi: 10.1111/j.1469-0691.2007.01914.x. [DOI] [PubMed] [Google Scholar]
- 9.Creti R, Michel J, Orefici LG. Genetic variability of the locus encoding alpha-C like proteins in Streptococcus agalactiae . In: Martin DR, Tagg J, editors. Streptococci and streptococcal diseases: Entering the new millennium. Proceedings of the XIV Lancefield international symposium on streptococci and streptococcal diseases. Securacopy; Porirua, New Zealand: 2000. pp. 397–9. [Google Scholar]
- 10.Kong F, Gowan S, Martin D, James G, Gilbert GL. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J Clin Microbiol. 2002;40:620–6. doi: 10.1128/JCM.40.2.620-626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madoff LC, Michel JL, Gong EW, Kling DE, Kasper DL. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci U S A. 1996;93:4131–6. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lachenauer CS, Madoff LC. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect Immun. 1996;64:4255–60. doi: 10.1128/iai.64.10.4255-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slotved HC, Elliott J, Thompson T, Konradsen HB. Latex assay for serotyping of group B streptococcus isolates. J Clin Microbiol. 2003;41:4445–7. doi: 10.1128/JCM.41.9.4445-4447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creti R, Fabretti F, Orefici G, von Hunolstein C. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J Clin Microbiol. 2004;42:1326–9. doi: 10.1128/JCM.42.3.1326-1329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolukaoto JY, Monyama CM, Chukwu MO, et al. Antibiotic resistance of Streptococcus agalactiae isolated from pregnant women in Garankuwa, South Africa. BMC Res Notes. 2015;8:364. doi: 10.1186/s13104-015-1328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borchardt SM, Foxman B, Chaffin DO, et al. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J Clin Microbiol. 2004;42:146–50. doi: 10.1128/JCM.42.1.146-150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong F, Ma L, Gilbert GL. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J Med Microbiol. 2005;54:1133–8. doi: 10.1099/jmm.0.46244-0. [DOI] [PubMed] [Google Scholar]
- 18.Madzivhandila M, Adrian PV, Cutland CL, Kuwanda L, Schrag SJ, Madhi SA. Serotype distribution and invasive potential of group B streptococcus isolates causing disease in infants and colonizing maternal-newborn dyads. PLoS One. 2011;6:e17861. doi: 10.1371/journal.pone.0017861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyo SR, Mudzori J, Tswana SA, Maeland JA. Prevalence, capsular type distribution, anthropometric and obstetric factors of group B Streptococcus (Streptococcus agalactiae) colonization in pregnancy. Cent Afr J Med. 2000;46:115–20. doi: 10.4314/cajm.v46i5.8533. [DOI] [PubMed] [Google Scholar]
- 20.Ippolito DL, James WA, Tinnemore D, et al. Group B streptococcus serotype prevalence in reproductive-age women at a tertiary care military medical center relative to global serotype distribution. BMC Infect Dis. 2010;10:336. doi: 10.1186/1471-2334-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BK, Song YR, Kim MY, et al. Epidemiology of group B streptococcus in Korean pregnant women. Epidemiol Infect. 2010;138:292–8. doi: 10.1017/S0950268809990859. [DOI] [PubMed] [Google Scholar]
- 22.Phares CR, Lynfield R, Farley MM, et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA. 2008;299:2056–65. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 23.Elliott JA, Thompson TA, Facklam RR, Slotved HC. Increased sensitivity of a latex agglutination method for serotyping group B streptococcus. J Clin Microbiol. 2004;42:3907. doi: 10.1128/JCM.42.8.3907.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray KJ, Bennett SL, French N, Phiri AJ, Graham SM. Invasive group B streptococcal infection in infants, Malawi. Emerg Infect Dis. 2007;13:223–9. doi: 10.3201/eid1302.060680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhi SA, Radebe K, Crewe-Brown H, et al. High burden of invasive Streptococcus agalactiae disease in South African infants. Ann Trop Paediatr. 2003;23:15–23. doi: 10.1179/000349803125002814. [DOI] [PubMed] [Google Scholar]
- 26.Moyo SR, Maeland JA, Bergh K. Typing of human isolates of Streptococcus agalactiae (group B streptococcus, GBS) strain from Zimbabwe. J Med Microbiol. 2002;51:595–600. doi: 10.1099/0022-1317-51-7-595. [DOI] [PubMed] [Google Scholar]
- 27.Mavenyengwa RT, Maeland JA, Moyo SR. Distinctive features of surface-anchored proteins of Streptococcus agalactiae strains from Zimbabwe revealed by PCR and dot blotting. Clin Vaccine Immunol. 2008;15:1420–24. doi: 10.1128/CVI.00112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavenyengwa RT, Maeland JA, Moyo SR. Serotype markers in a Streptococcus agalactiae strain collection from Zimbabwe. Indian J Med Microbiol. 2010;28:313–9. doi: 10.4103/0255-0857.71819. [DOI] [PubMed] [Google Scholar]
- 29.Persson E, Berg S, Trollfors B, et al. Serotypes and clinical manifestations of invasive group B streptococcal infections in western Sweden 1998–2001. Clin Microbiol Infect. 2004;10:791–6. doi: 10.1111/j.1469-0691.2004.00931.x. [DOI] [PubMed] [Google Scholar]


