Abstract
Purpose
To examine the intra-examination repeatability of proton density fat fraction (PDFF) and T1 and T2 of liver water and fat as estimated by a novel multi-repetition time (TR)-echo time (TE) 1H MRS stimulated echo acquisition mode (STEAM) sequence that acquires 32 spectra for a range of TRs and TEs in single breath-hold.
Materials and Methods
Sixty-seven subjects undergoing liver MRI examinations at 3T had three multi-TR-TE sequences acquired consecutively in a single session. This sequence was designed to allow accurate estimation of T1 and T2 of both water and fat, as well as PDFF, in a single breath-hold. A standard long-TR, multi-TE sequence was also acquired to allow comparison of estimated PDFF. Regression and interclass correlation (ICC) analyses were performed.
Results
There was strong agreement between PDFF estimated by the multi-TR-TE and long-TR, multi-TE sequences (slope 0.997; intercept -0.03; R = 0.997). The multi-TR-TE sequence had high repeatability for estimating PDFF (ICC = 0.999), water T2 (ICC = 0.920), water T1 (ICC = 0.845) and fat T2 (ICC = 0.760), and moderate repeatability for estimating fat T1 (ICC = 0.556).
Conclusion
A novel multi-TR-TE sequence can estimate PDFF and water and fat T1 and T2 in a single breath-hold. Refinement may be needed to improve repeatability for fat T1 estimation.
Keywords: NASH, NAFLD, Magnetic Resonance Spectroscopy, Relaxation
INTRODUCTION
Proton magnetic resonance spectroscopy (1H MRS) is widely considered the most accurate non-invasive, quantitative method to assess liver fat (1-5). With appropriate parameter choice, 1H MRS can estimate proton density fat fraction (PDFF), an estimate of liver fat un-confounded by biological or technical factors (6-10). Accurate estimation of PDFF is required to detect and monitor non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) (7).
Currently, the most accurate 1H MRS technique to measure liver PDFF is a single-TR, multi-TE STEAM (Stimulated Echo Acquisition Mode) sequence in which multiple long-TR (≥ 3,000 ms), single-average spectra are collected at different TE values to provide a T1-independent, T2-corrected estimate of PDFF (11,12). The requirement to acquire multiple spectra in a single breath-hold, each with a long TR, limits the number of spectra that can be collected at different TE values. The limited number of TE values reduces estimation accuracy of liver fat T2, as the short TE range used to minimize J coupling effects is not well suited to accurately estimate fat T2, which is a significantly longer than liver water T2 (13). Also, as this sequence has TR far greater than the T1 of fat and water, it has lower signal-to-noise for the same acquisition time than sequences with lower TR and more averages (14). Lowering TR, however, confounds PDFF estimation by introducing subject-dependent T1 weighting, necessitating subject-specific water and fat T1 measurement (7).
While a single-breath-hold, long-TR, multi-TE spectroscopic sequence can simultaneously and accurately estimate PDFF and water T2, it does not also allow determination of water and fat T1. There are no commercially available single-breath-hold sequences to estimate simultaneously both T1 of water and fat. Previous studies have used separate single-TR, single-TE spectroscopic sequences, acquired over multiple breath-holds (15,16) to estimate water and fat T1. This allows T1 correction for short TR sequences, and perhaps also provides subject-specific T1 information that may help characterize disease status. However, breath-hold inconsistency may reduce estimation accuracy, while the requirement for multiple breath-holds may reduce practicality.
To permit simultaneous PDFF, and water and fat T1 and T2 estimation, we propose a novel, single-average, single-breath-hold, multi-spectra MRS sequence which in a single acquisition acquires both variable TR spectra at fixed TE, and variable TE spectra at fixed TR. The purpose of this study was to examine the intra-examination repeatability of PDFF and T1 and T2 of liver water estimated by this multi-TR-TE sequence. Additionally, the study examined agreement for PDFF estimated by this sequence and by a standard single-average, single-breath-hold, long-TR, multi-TE sequence.
MATERIALS AND METHODS
This was a prospective, single-site study approved by our Institutional Review Board and compliant with the Health Insurance Portability and Accountability Act. Adult human subjects were recruited from clinical NAFLD studies being conducted at our institution. These subjects either had biopsy proven NAFLD, or were at risk for NAFLD due to family history or obesity. Written informed consent was obtained from all subjects.
MRS Acquisition
Subjects were scanned at 3T (GE Signa EXCITE HDxt, GE Healthcare, Waukesha, WI) with an 8-channel torso array coil. Multi-planar localization images were acquired and a 20 × 20 × 20 mm voxel was selected within the right lobe of the liver that avoided liver edges as well as large biliary or vascular structures. 1H MR spectra were acquired using the STEAM sequence, which compared to the alternative technique of Point REsolved SpectroScopy (PRESS), allows a shorter minimum TE, minimizing J coupling effects (13). The mixing time (TM) for the STEAM sequence was fixed at a minimum value of 5 ms, also to minimize J coupling (17). There was no water saturation, and spatial saturation bands around the voxel were disabled to ensure a uniform spectral response across the frequency range of interest.
The timing of the multi-TR-TE sequence is shown in Table 1. The timing values are designed to provide a range of TR and TE to allow accurate estimation of T1 and T2 of both water and fat in a single breath-hold. A total of 32 spectra (including four pre-acquisitions excitations) are acquired in a 21 s breath-hold. Following the four pre-pulses, TR was altered between 150 - 2,000 ms to acquire 20 spectra with a fixed TE of 10 ms. Then, from spectrum 20 onwards, TR was fixed at 1,000 ms and TE was increased from 10 to 110 ms.
Table 1.
Sequence timing of the multi-TR-TE sequence
| Spect No. | P1 | P2 | P3 | P4 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|---|
| TR (ms) | - | 150 | 150 | 150 | 150 | 225 | 300 | 400 |
| τ (ms) | 10000 | 140 | 140 | 140 | 140 | 215 | 290 | 490 |
| TE (ms) | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Spect No. | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| TR (ms) | 600 | 900 | 2000 | 1500 | 700 | 450 | 325 | 250 |
| τ (ms) | 590 | 890 | 1990 | 1490 | 690 | 440 | 315 | 240 |
| TE (ms) | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Spect No. | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| TR (ms) | 175 | 200 | 275 | 350 | 500 | 800 | 1250 | 1000 |
| τ (ms) | 165 | 190 | 265 | 340 | 490 | 790 | 1240 | 990 |
| TE (ms) | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Spect No. | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| TR (ms) | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 |
| τ (ms) | 988 | 985 | 983 | 980 | 970 | 960 | 950 | 940 |
| TE (ms) | 15 | 20 | 25 | 30 | 50 | 70 | 90 | 110 |
Note: P1 to P4 are the preparation pulses. Spect No. = spectra number. TR = repetition time. TE = echo time. τ = time for recovery of longitudinal magnetization. P1 is also used to estimate T1, T2 and PDFF and is assumed to have an effective TR of 10,000 ms.
The current standard for MRS estimation of PDFF – a long-TR, multi-TE acquisition – was acquired for comparison to, and at the same location as the multi-TR-TE sequence (4,5,18). Five spectra and a single pre-acquisition excitation were acquired with a TR of 3,500 ms consecutively at progressively longer TEs of 10, 15, 20, 25 and 30 ms in a 21 s breath-hold. The long TR was required to minimize T1 effects, and the range of TE values was chosen to minimize J coupling effects (13). Thus both the multi-TR-TE and long-TR, multi-TE sequences had identical acquisition times.
To examine the repeatability of T1, T2 and PDFF estimated by the multi-TR-TE sequence, the sequence was repeated two more times with identical parameters for each subject. Thus a total of three co-localized multi-TR-TE spectra were acquired for each subject.
MRS Analysis
An identical prior-knowledge-based analysis was used to fit spectra from both multi-TR-TE and long-TR, multi-TE sequences. The spectra from the individual channels were combined using singular value decomposition (19). A single experienced observer analyzed the spectra using the AMARES algorithm (20) included in the MRUI software package (21). The observed or measurable peaks were modeled by multiple Gaussian resonances. The 1.3 ppm peak was modeled by four Gaussians; the fat peaks at 2.1, and 0.9 ppm were each modeled by two Gaussians, while the 2.7 ppm peak was modeled by a single Gaussian. The frequency of the fat peaks was fixed relative to the main CH2 peak at 1.3 ppm. The composite water and fat peak in the 4 - 6 ppm range was modeled by five unconstrained Gaussians (i.e., the amplitude, linewidth and frequency of the peaks were all fitted freely) (8,18).
For both the multi-TR-TE and long-TR, multi-TE sequences, the results of the MRUI analysis were saved in a text file and analyzed using custom Matlab routines. In the multi-TR-TE spectra, the Matlab routine non-linearly minimized the difference between the measured peak area and the peak area given by the standard equation
to give the T1 and T2 and the T1-T2-corrected peak areas of fat and water. The recovery time for longitudinal magnetization is not TR but the time from the third 90° pulse of the STEAM sequence. Thus in this equation τ (as detailed in Table 1) is used rather than TR. While the second, third and fourth pre-acquisition excitations are discarded, the first pre-acquisition excitation is assigned a TR of 10,000 ms and included in the calculation. In the long-TR, multi-TE sequence, the Matlab routine used non-linear fitting to give the T2 and T2-corrected peak areas of fat and water assuming monoexponential T2 decay. No T1 correction was applied to the peak areas given by the long-TR, multi-TE sequence. PDFF was estimated as the ratio of fat signal to the sum of water and fat signals, corrected for fat subsumed by the ‘water’ peak from a previously-established standard liver spectrum (18).
For subjects with low levels of liver fat (PDFF < 5%), estimations of fat T2 from the long-TR, multi-TE sequence may be unreliable. The 5% value was derived observationally from previous studies involving this sequence, as below the 5% level non-physical estimates for fat T2 were observed. Thus, subjects with PDFF lower than 5% were excluded from statistical analyses that included fat T2 estimated by the long-TR, multi-TE sequence.
Statistical Analysis
PDFF, and water and fat T2 produced by the long-TR, multi-TE and multi-TR-TE sequences were compared using Bland-Altman plots. The Pearson-r correlation coefficient was also calculated. Paired t-tests were used to compare estimates produced by the two sequences. Intraclass correlation coefficient (ICC) was used to estimate the repeatability of the PDFF and water and fat T1 and T2 as estimated by the three multi-TR-TE acquisitions.
RESULTS
Sixty-seven adult subjects (26 male, 41 female; mean age 52.2 yrs) were recruited for this study between June 2012 and September 2014. Three of the 67 subjects had at least one multi-TR-TE spectral acquisition that proved unanalyzable and these subjects were excluded from the study. A further 13 subjects did not have a long-TR, multi-TE spectra and were thus excluded from multi-TR-TE vs. long-TR, multi-TE comparisons. Twenty-five subjects had PDFF < 5%, so these subjects were excluded from analyses involving the T2 of fat measured by the long-TR, multi-TE sequence.
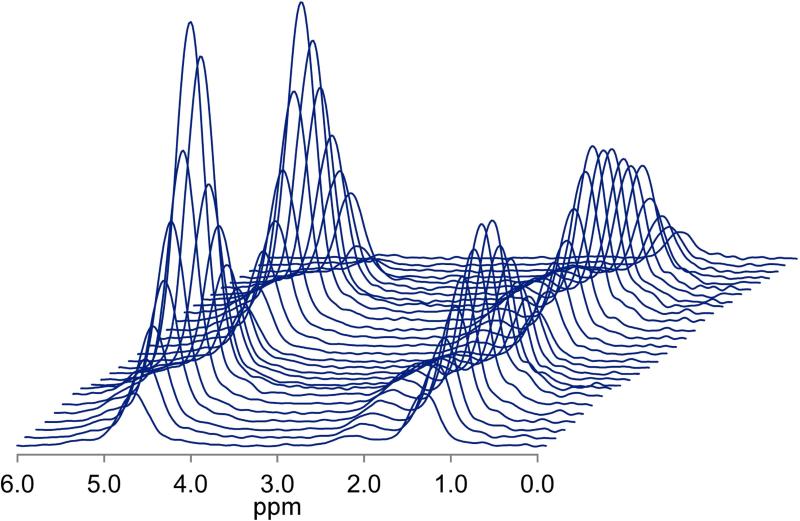
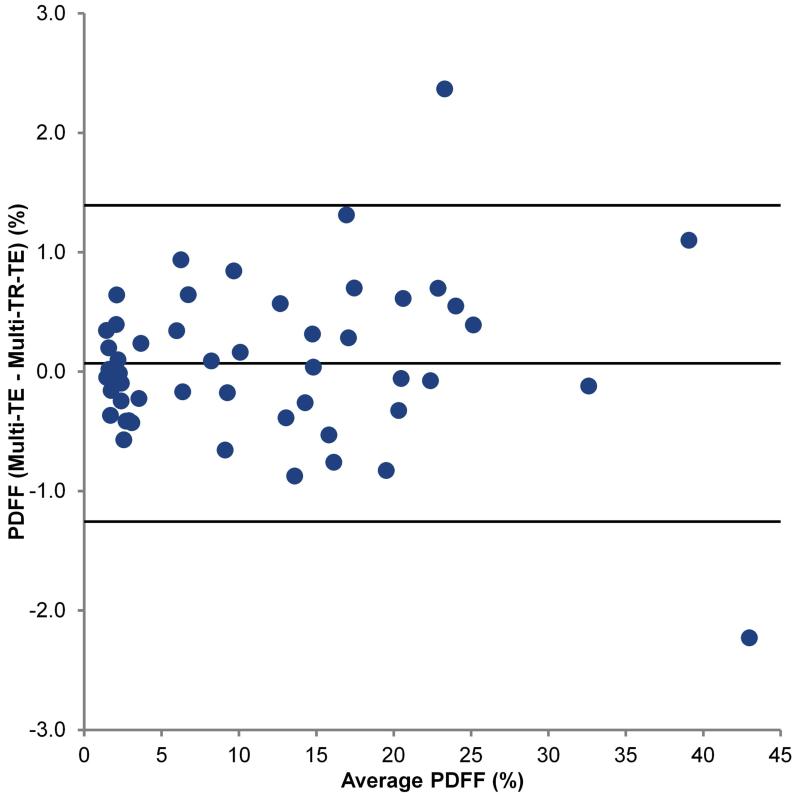
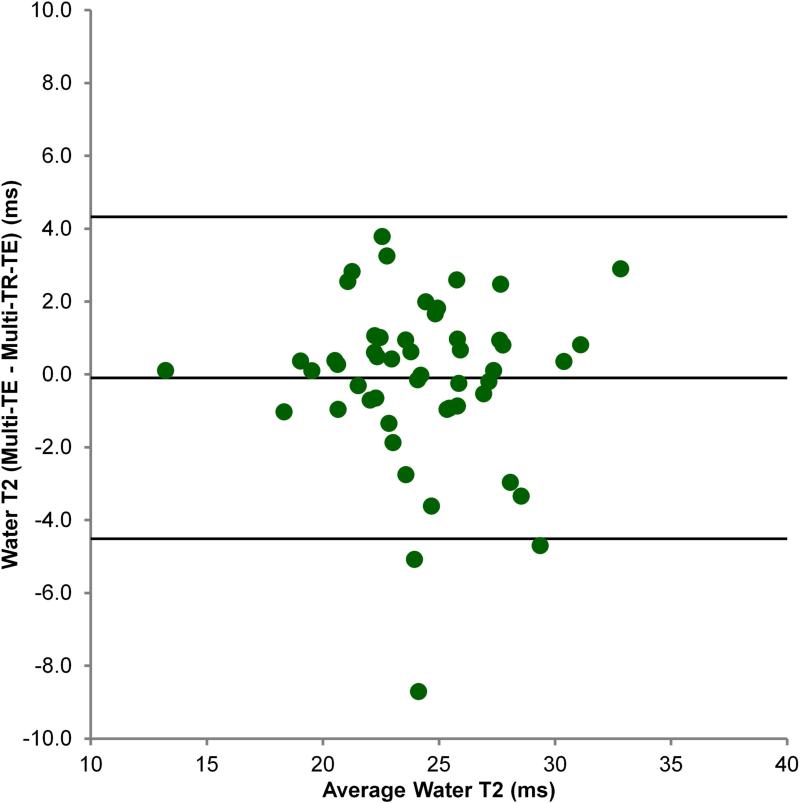
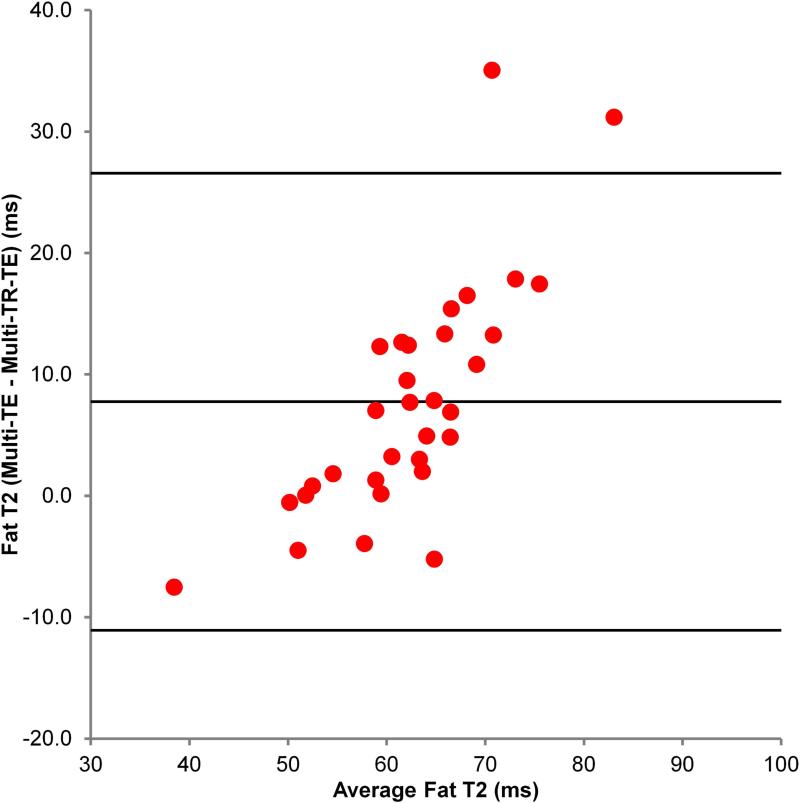
Figure 1 shows a typical multi-TR-TE acquisition. Table 2 shows the mean T1 and T2 values of fat and water, and PDFF as estimated by the multi-TR-TE and long-TR, multi-TE sequences. Figure 2 shows the Bland-Altman plot comparing mean PDFF estimated from the three multi-TR-TE sequences to that derived from the long-TR, multi-TE sequence, while Figures 3 and 4 show the same for water T2 and fat T2, respectively. PDFF values derived from the two sequences were strong correlated (Pearson-r = 0.997) and the mean difference between PDFF estimated by the long-TR, multi-TE and the multi-TR-TE sequence was just 0.07%. There was weaker correlation between water T2 estimated by the two methods (Pearson-r = 0.815), though the mean difference between the two sequences was just -0.1 ms. The poorest agreement was between estimates of fat T2 (Pearson-r = 0.711), with a mean difference of 7.7 ms, as expected since the TE range of the long-TR, multi-TE sequence is too short for accurate fat T2 estimation. The only significant difference in estimates from the two sequences was for fat T2.
Figure 1.
Typical multi-TR-TE acquisition from a 62 yr-old female with a PDFF of 27.6%. The four pre-acquisitions excitations are not shown.
Table 2.
Water and fat T1 and T2, and PDFF provided by the multi-TR-TE and long-TR, multi-TE sequences
| Multi-TR-TE | Long-TR, Multi-TE | ||||
|---|---|---|---|---|---|
| n | Mean (SD) | n | Mean (SD) | p | |
| Water T1 (ms) | 64 | 822 (123) | - | - | - |
| Fat T1 (ms) | 64 | 312 (48) | - | - | - |
| Water T2 (ms) | 64 | 24.1 (4.5) | 51 | 24.1 (3.6) | > 0.05 (n = 51) |
| Fat T2 (ms) | 64 | 53.4 (11.7) | 32 | 66.3 (12.5) | < 0.001 (n = 32) |
| PDFF(%) | 64 | 11.5 (10.6) | 51 | 11.7 (10.2) | > 0.05 (n = 51) |
Note: SD - standard deviation
Figure 2.
Bland-Altman plot comparing PDFF estimated by the multi-TR-TE and long-TR, multi-TE sequences.
Figure 3.
Bland-Altman plot comparing water T2 estimated by the multi-TR-TE and long-TR, multi-TE sequences.
Figure 4.
Bland-Altman plot comparing fat T2 estimated by the multi-TR-TE and long-TR, multi-TE sequences.
The multi-TR-TE sequence showed high repeatability when estimating PDFF (ICC = 0.999), water T2 (ICC = 0.920) and water T1 (ICC = 0.845). There was good repeatability for fat T2 (ICC = 0.760), but there was only moderate repeatability for fat T1 (ICC = 0.556).
DISCUSSION
This study showed that by acquiring single-average spectra at multiple TRs and TEs in a single acquisition, PDFF, and water and fat T1 and T2 can be estimated in a single breath-hold. PDFF estimated by the multi-TR-TE sequence agrees with that of the long-TR, multi-TE sequence. The multi-TR-TE sequence showed high ICC values for PDFF and for water T1 and T2, though ICC values were lower for fat T1 and T2 indicating lower repeatability for these quantities.
The multi-TR-TE sequence was designed to take the same time as the standard long-TR, multi-TE sequence. Thus for the same acquisition time, in addition to PDFF and water T2, water T1, and fat T1 and T2 can also be estimated which cannot be provided by the long-TR, multi-TE sequence. We showed excellent agreement for PDFF derived from these two sequences. We also showed agreement between water T2 estimated by the multi-TR-TE and the long-TR, multi-TE sequences.
Agreement for fat T2 estimation was poor. This is an expected result given the short TE range of the multi-TE sequence. The multi-TR-TE sequence is expected to provide a better estimate of water T2 than the multi-TE sequence, as it acquires more spectra over a larger range of TEs. We speculate that the multi-TR-TE sequence provides a less noisy estimate of PDFF than the long-TR, multi-TE sequence, as the long TR required by the multi-TE sequence sacrifices signal-to-noise to minimize T1 weighting, a compromise not required by the multi-TR-TE sequence. However, as we did not acquire repeat acquisitions of the long-TR, multi-TE sequence, we cannot conclude this definitively.
The multi-TR-TE sequence showed high repeatability (ICC > 0.8) for PDFF, and for water T1 and T2. Estimates of fat T1 and T2 were comparatively less repeatable. This may simply be due to the lower signal of fat. In the liver, fat signal is usually smaller than water signal, and this may be reflected in the lower ICC of fat T1 and T2.
The long-TR, multi-TE sequence uses a short TE range which minimizes J coupling, but is unable to estimate fat T2 accurately. The multi-TR-TE sequence collects more spectra at longer TEs to allow fat T2 estimation but this may introduce systematic errors due to J coupling (13,17). The strong agreement between multi-TR-TE and long-TR, multi-TE estimates of PDFF suggests J coupling is not a major source of confounding for the multi-TR-TE sequence. This may be because most spectra are collected in the low TE range in which J coupling effects are small; only four spectra have TE > 30 ms, where J coupling effects are meaningful.
J coupled peaks may also be affected by TM (17). Although it is possible to estimate T1 by altering TM, we choose not to follow this approach to ensure that the fat T1 estimates were not systematically affected by J coupling.
Repeatability of fat T1 estimation was only modest. One plausible explanation is that the minimum TR of the multi-TR-TE sequence, 150 ms, was too long to permit accurate estimation fat T1. It is possible that shortening the minimum TR will improve fat T1 estimation repeatability. However, a TR of 150 ms was the minimum TR achievable by the sequence design used in this study.
The fat spectrum is complex with multiple peaks each with their own T1 and T2. However, there is insufficient spectral resolution in the liver in vivo consistently to resolve the individual fat peaks and to allow estimation of T1 and T2 in a breath-hold sequence. Thus, T1 and T2 values estimated for fat are in effect a weighted average of the T1 and T2 values of the individual fat peaks in the 0 - 3 ppm range. Further, water peak relaxation values include small contributions from fat. While it is possible to correct the contribution to PDFF for these peaks using a previously defined spectrum (18), it was not possible to correct the water T1 and T2 values. However, even at a PDFF of 40%, signals from fat peaks included in the water peak only contribute 6% of the total water peak signal.
While the multi-TR-TE and long-TR, multi-TE sequences have identical acquisition times, the time required to analyze spectra acquired with the multi-TR-TE sequence is greater. Although MRUI analyzes all spectra acquired from either acquisition in a single batch, the 32 spectra resulting from the multi-TR-TE sequence take longer to pre-process, analyze, and check compared to the six spectra from the long-TR, multi-TE sequence.
As the multi-TR-TE sequence is the first sequence that attempts to estimate T1 and T2 of both water and fat, and PDFF in a single sequence, the optimal methodology to acquire that sequence was not known at the start of this study. At that time, available data on the range of liver fat and water relaxation values was limited. It is possible that different sequence timing from that used here, or other methods of estimating T1 such as inversion recovery or changing TM may provide more accurate and repeatable estimates of T1, T2 and PDFF. The data collected in this manuscript and from other sources will help inform future sequence design allowing possibly more optimal selection of sequence parameters to obtain better estimates of these parameters.
The purpose of this study was to examine the repeatability of the multi-TR-TE sequence; it did not aim to prove its accuracy definitively. We lacked histological comparison in this study, though the long-TR, multi-TE sequence is generally regarded as the non-invasive standard for PDFF estimation. There is no biological measure that would directly allow comparison of the T1 and T2 of fat and water. Comparison of sequences allowed us to demonstrate agreement for water T2, but not for fat T2, since the long-TR, multi-TE sequence does not accurately estimate fat T2. No comparison inversion recovery sequences were carried on subjects to estimate T1; such comparison sequences would have required multiple breath-holds, reducing their accuracy, and many of the technical issues (poor shim, incorrect RF pulse calibration, etc.) which affect our multi-TR-TE sequence would also affect inversion-recovery-based methods of measuring T1.
In conclusion, this study showed that in a single breath-hold, water and fat T1 and T2, and PDFF can be estimated repeatably in a single breath-hold using the multi-TR-TE sequence with no extra acquisition time cost compared to the long-TR, multi-TE sequence. Previous studies have required multiple acquisitions over many breath-holds to provide estimates for the same data that can now be acquired in a single breath-hold. By acquiring this data more rapidly, it is hoped that the multi-TR-TE sequence will provide new opportunities to characterize and monitor chronic liver disease.
Acknowledgments
Grant Support: Funding provided by: Atlantic Philanthropies, Inc, the John A. Hartford Foundation, the Association of Specialty Professors, and the American Gastroenterological Association and NIH grants K23-DK090303; R01-DK088925-05 and 5R01-DK075128-04
REFERENCES
- 1.Hu HH, Kan HE. Quantitative proton MR techniques for measuring fat. NMR Biomed. 2013;26:1609–1629. doi: 10.1002/nbm.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longo R, Pollesello P, Ricci C, et al. Proton MR spectroscopy in quantitative in vivo determination of fat content in human liver steatosis. J Magn Reson Imaging. 1995;5:281–285. doi: 10.1002/jmri.1880050311. [DOI] [PubMed] [Google Scholar]
- 3.Szczepaniak LS, Nurenberg P, Leonard D, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288:E462–E468. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 4.Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology. 2011;258:749–759. doi: 10.1148/radiol.10100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meisamy S, Hines CD, Hamilton G, et al. Quantification of hepatic steatosis with T13 independent, T23corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology. 2011;258:767–775. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging. 2012;36:1011–1014. doi: 10.1002/jmri.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeder SB, Cruite I, Hamilton G, Sirlin CB. Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy. J Magn Reson Imaging. 2011;34:729–749. doi: 10.1002/jmri.22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ligabue G, Besutti G, Scaglioni R, Stentarelli C, Guaraldi G. MR quantitative biomarkers of non-alcoholic fatty liver disease: technical evolutions and future trends. Quant Imaging Med Surg. 2013;3:192–195. doi: 10.3978/j.issn.2223-4292.2013.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le TA, Chen J, Changchien C, et al. Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial. Hepatology. 2012;56:922–932. doi: 10.1002/hep.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noureddin M, Lam J, Peterson MR, et al. Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology. 2013;58:1930–1940. doi: 10.1002/hep.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineda N, Sharma P, Xu Q, Hu X, Vos M, Martin DR. Measurement of hepatic lipid: high-speed T23corrected multiecho acquisition at 1H MR spectroscopy-a rapid and accurate technique. Radiology. 2009;252:568–576. doi: 10.1148/radiol.2523082084. [DOI] [PubMed] [Google Scholar]
- 12.Yokoo T, Bydder M, Hamilton G, et al. Nonalcoholic Fatty Liver Disease: Diagnostic and fat-grading accuracy of low-flip-angle multiecho gradient-recalled-echo MR Imaging at 1.5 T. Radiology. 2009;251:67–76. doi: 10.1148/radiol.2511080666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton G, Middleton MS, Bydder M, et al. The effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantification. J Magn Reson Imaging. 2009;30:145–152. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambarota G, Tanner M, van der Graaf M, Mulkern RV, Newbould RD. 1H-MRS of hepatic fat using short TR at 3T: SNR optimization and fast T2 relaxometry. MAGMA. 2011;24:339–345. doi: 10.1007/s10334-011-0278-3. [DOI] [PubMed] [Google Scholar]
- 15.Ren J, Dimitrov I, Sherry AD, Malloy CR. Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res. 2008;49:2055–2062. doi: 10.1194/jlr.D800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomsen C, Becker U, Winkler K, Christoffersen P, Jensen M, Henriksen O. Quantification of liver fat using magnetic resonance spectroscopy. Magn Reson Imaging. 1994;12:487–495. doi: 10.1016/0730-725x(94)92543-7. [DOI] [PubMed] [Google Scholar]
- 17.de Graaf RA, Rothman DL. In vivo detection and quantification of scalar coupled 1H NMR resonances. Concepts in Magnetic Resonance. 2001;13:32–76. [Google Scholar]
- 18.Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat 1H MR spectrum. NMR Biomed. 2011;24:784–790. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bydder M, Hamilton G, Yokoo T, Sirlin CB. Optimal phased-array combination for spectroscopy. Magn Reson Imaging. 2008;26:847–850. doi: 10.1016/j.mri.2008.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 21.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12:141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]