Abstract
Purpose
To identify computer extracted in vivo dynamic contrast enhanced (DCE) magnetic resonance imaging (MRI) markers associated with quantitative histomorphometric (QH) characteristics of microvessels and Gleason scores (GS) in prostate cancer.
Materials and Methods
This study considered retrospective data from 23 biopsy confirmed prostate cancer patients who underwent 3 Tesla multiparametric MRI prior to radical prostatectomy (RP). Representative slices from RP specimens were stained with vascular marker CD31. Tumor extent was mapped from RP sections onto DCE MRI using nonlinear registration methods. 77 microvessel QH features and 18 DCE MRI kinetic features were extracted and evaluated for their ability to distinguish low from intermediate and high GS. The effect of temporal sampling on kinetic features was assessed and correlations between those robust to temporal resolution and microvessel features discriminative of GS were examined.
Results
A total of 12 microvessel architectural features were discriminative of low and intermediate/high grade tumors with AUC > 0.7. These features were most highly correlated with mean washout gradient (WG) (max rho = −0.62). Independent analysis revealed WG to be moderately robust to temporal resolution (ICC=0.63) and WG variance, which was poorly correlated with microvessel features, to be predictive of low grade tumors (AUC=0.77). Enhancement ratio was the most robust (ICC = 0.96) and discriminative (AUC = 0.78) kinetic feature but was moderately correlated with microvessel features (max rho = −0.52).
Conclusion
Computer extracted features of prostate DCE MRI appear to be correlated with microvessel architecture and may be discriminative of low vs. intermediate and high GS.
Keywords: Imaging biomarkers, prostate cancer, quantitative histomorphometry, microvessel architecture, DCE MRI, Gleason grades
INTRODUCTION
Overdiagnosis and overtreatment of prostate cancer [1,2] has led to the growing interest in adopting conservative disease management strategies such as active surveillance [3]. However, success of such strategies rely on the ability to selectively recruit patients with low-risk tumors, where low-risk refers to low likelihood of death on account of prostate cancer [4]. Although pathology is the clinical standard for prostate cancer risk assessment, with Gleason scores and microvessel density (MVD) being some of the most well established surrogates of outcome [5–7], pathological criteria for in vivo risk determination is significantly limited by biopsy sampling errors [8,9].
Multiparametric (MP) magnetic resonance imaging (MRI) has emerged as a promising modality for prostate cancer diagnosis given its high sensitivity and specificity for detection of clinically significant prostate cancer in vivo [5]. Among the MP-MRI sequences, dynamic contrast enhanced (DCE) MRI has shown promise in its ability to assess tumor aggressiveness [10,11]. Growing tumors comprising large number of leaky microvessels are expected to show increased enhancement as well as faster contrast uptake and washout as compared to indolent tumors. Previous studies have shown associations between various kinetic and pharmacokinetic features and Gleason scores. Most recently, Vos et al. [11] found statistics of wash-in, washout rate as well as pharmacokinetic parameters, Ktrans and Kep, to be significantly different in patients with low and high Gleason scores. However, Gleason grade for an average prostate cancer patient spans a narrow range between 6 and 7, where scores ≤ 6 are considered low risk, scores of 7 are considered intermediate risk and scores > 7 are considered high risk [12]. Provided that active surveillance is recommended for low risk prostate cancer [13,14], it is necessary to identify markers that distinguish low from both intermediate and high risk tumors. In this work, we therefore consider a patient population with Gleason scores between 6 and 8, the range where each increment in score results in a change in risk categorization, and seek DCE MRI markers capable of distinguishing low risk (Gleason score ≤ 6) from intermediate and high risk (Gleason score > 6) prostate cancer within this population.
However, roughly 49% of the prostate cancer population comprises of moderately differentiated, intermediate grade tumors [15] which can have up to three-fold differences in the risk of mortality [16]. Therefore, Gleason score, and consequently its MRI correlate, is likely to be insufficient in determining candidates for active surveillance. Bostwick et al. [17] found that combining Gleason scores and preoperative PSA with microvessel density improved their ability to predict extraprostatic extension, a surrogate of outcome. In this work, we therefore additionally explore correlations between quantitative DCE MRI characteristics and microvessel attributes to identify in vivo markers of prostate cancer risk.
Microvessel morphology [18] and microvessel density (MVD) are known to be associated with prostate cancer risk [5][6]. MVD is pathologically determined as the number of microvessels per unit area within hotspots, or regions of high MVD within the tumor. Determination of the location and size of these hotspots is subjective due to lack of standard, established criteria for identifying microvessel hotspots [19]. As such, a recent attempt at identifying imaging correlates of MVD to discover in vivo DCE MRI markers of prostate cancer presence relied on computing MVD within local neighborhood around each pixel [20]. Tretiakova et al. [19] qualitatively demonstrated that differences in spatial distribution of microvessels has contributed to the perceived differences in MVD between tumor and benign tissues despite the similarities in total microvessel counts in the two tissue types. Measures of microvessel arrangement may therefore serve as more direct and objective alternatives to MVD. As depicted with representative image patches in Figure 4, tumors assigned the same Gleason score may display different microvessel configurations. To capture such differences, we attempt to model the spatial distribution of microvessels using quantitative histomorphometry (QH), which uses computerized image based feature analysis to model the appearance of disease on digitized histopathology [21,22]. As previous work has shown that MVD is associated with Gleason grades [23], we individually assess each microvessel QH feature in terms of its ability to predict low vs. intermediate and high Gleason grades in order to retain the most informative microvessel QH features for subsequent correlative analysis with in vivo DCE MRI.
Figure 4.
(a, g) Microvessel (CD31) stained histology tumor patches from two tumors of same Gleason score. (b, h) Microvessels are segmented using HNCut [29]. (c, i) Ellipsoids enclosing the microvessels provide measurements of shape such as eccentricity, a measure of circularity, which is depicted via ellipsoid color (red=ellipse, blue=circle). Microvessel architectural features are extracted from graphical representations of microvessel distribution: (d, j) Voronoi diagram (e, k) Delaunay triangulation and (f, l) Minimum spanning tree
The purpose of this study is to identify in vivo DCE MRI markers of prostate cancer risk by examining the correlations between kinetic features and pathological prognostic markers, Gleason score and microvessel architecture. Figure 1 shows our radiology-pathology correlation framework which employs image registration, segmentation, and feature extraction methods to study associations between radiologic and pathologic attributes within spatially aligned regions of interest.
Figure 1.
Overview of the radiology-pathology correlation workflow for identifying in vivo DCE MRI markers of prostate cancer risk. Ground truth cancer annotations are mapped from pseudo-whole mount histology sections onto in vivo MRI using image registration methods. Kinetic features are extracted from tumor voxels and assessed in terms of their ability to distinguish low from intermediate/high grade tumors and for robustness to temporal resolution. Microvessels are segmented from cancerous regions on CD31 stained histology sections to extract quantitative histomorphometric features of microvessel arrangement. Select microvessel features that are able to distinguish between low and intermediate/high grade disease are then correlated with kinetic features found to be robust to temporal resolution.
MATERIALS AND METHODS
Dataset Description
This retrospective study was approved by the Institutional Review Board, which waived informed consent. Subjects imaged by 3 Tesla DCE MRI and T2w MRI between 2009 and 2011, who subsequently underwent radical prostatectomy at University of Pennsylvania were eligible for this study. A total of 54 subjects were identified. Two cases where DCE MRI did not follow routine protocol were excluded, leaving 52 potential cases. 23 of the 52 cases were selected and formed our study population (age range: 46–69; mean age: 59.3) in order to include a spectrum of cases with different Gleason scores.
All MRI studies were performed on a 3 Tesla MRI scanner (Verio©, Siemens, Erlangen, Germany) using a dedicated endorectal coil (Medrad©, Pittsburgh, PA). Axial T1 and T2w imaging was performed with 3 mm slice thickness and 1 mm gap. Imaging FOV was 14 cm, and acquisition matrix size was 256 by 128–179. DCE MRI was performed with T1w VIBE imaging at 3 mm slice thickness, with 24 cm FOV and matrix 256 by 192. Temporal resolution varied based on the number of prescribed slices (min: 8.13 sec; max: 30.83 sec). 5, 8, 2, 1, 3 and 4 cases had temporal resolutions between 8–12, 12–16, 16–20, 20–24, 24–28 and 28–32 seconds, respectively. The effect of varying temporal resolutions on the extracted feature values was studied on the 5 cases with the highest temporal resolution. Readers are referred to the section, Experimental Design for DCE MRI Kinetic Feature Evaluation, for details of the experiment. Twenty phases of imaging were performed for most cases, with IV gadolinium injection, 0.07 mmol/kg Multihance© (Bracco Diagnostics, Monroe Township, NJ) at a rate of 2 mL/sec beginning 30 seconds after scan initiation.
The surgically resected prostate gland, after fixation in formalin, was sectioned in a plane perpendicular to the urethral axis from apex to base into 3–4 mm slices. Each slice was then divided into 4 quadrants, stained with H&E and digitized by the Aperio® whole slide scanner at 20x magnification and 0.5um/pixel resolution. For each case, 2–3 consecutive slices (8 to 12 quadrants) containing representative regions of tumor were selected for CD31 staining. Immunohistochemistry (IHC) of formalin fixed paraffin embedded tissue was performed on a Leica Bond™ instrument using the Bond Polymer Refine Detection System. Epitope retrieval was done for 10 minutes with Enzyme 1 solution. Antibodies against CD31 were used at 1:25 dilution. Tumor regions were annotated by pathologist (NS, 8.5 years of experience) on the digitized H&E as well as CD31 stained slides using ImageScope (Aperio) software. The pathologist determined Gleason grades for each considered slice, which were then used to categorize each patient as having low (at least one slice with 3+3 and remaining slices with primary grade 3), intermediate (at least one slice with 4+3 or all slices with 3+4) or high grade (at least one slice with 4+4) prostate cancer. Based on this categorization, the data cohort consisted of 7 low, 11 intermediate and 5 high grade prostate cancer patients.
Pseudo Whole Mount Histology Reconstruction and Co-registration with MRI
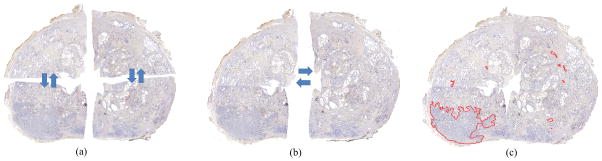
Prostate quadrants were stitched into pseudo whole mount sections using Histostitcher [24,25], an interactive program that assembles digitized histologic fragments into a contiguous slice as shown in Figure 2. At least three corresponding points between adjacent quadrants were manually selected. Transformations required to maximally align these points were then computed, applied and stored. The same transformations were then reapplied to binary images of quadrant annotations in order to obtain prostate cancer masks for pseudo whole mount sections.
Figure 2.
Reconstruction of whole-mount prostate sections using HistoStitcher [24,25] (a) Sectioned quadrants of surgical specimens are (b) stitched into hemispheres which are then combined to obtain (c) pseudo whole-mount section (pWMHs) that is annotated for cancer (red outline).
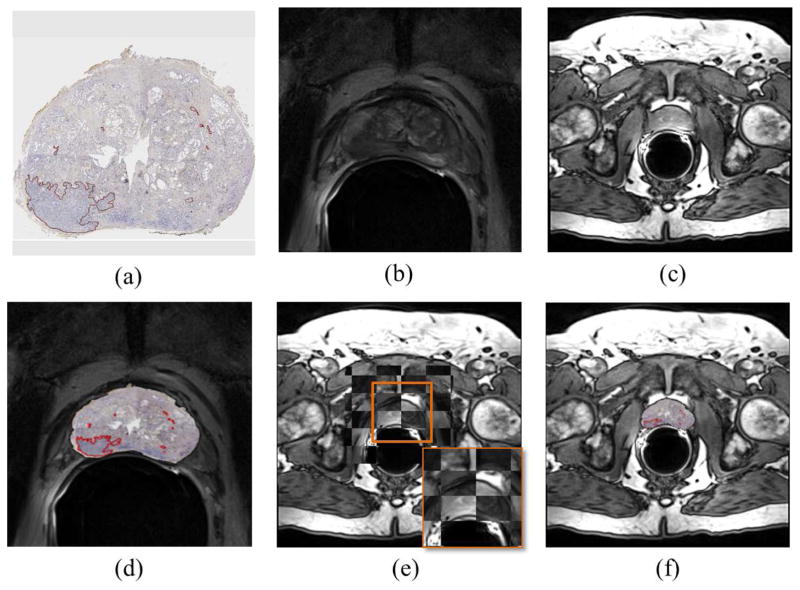
In order to map cancer annotations from reconstructed histologic sections onto the MRI (Figure 3), correspondences between T2w MRI, H&E and CD31 stained slices were identified by expert radiologist and pathologist based on distances between slices and major anatomical landmarks [26]. Subsequently, CD31 stained sections were registered with corresponding T2w MRI slices using thin plate splines (TPS), a landmark based registration method that interpolates the transformation at each voxel such that it (i) maximizes overlap between target and template landmarks, establishing accurate spatial correspondences while (ii) minimizing the bending energy to generate smooth transformations [27]. Manually selected landmarks were used to align prostate boundaries and visible internal structures (such as urethra) between the moving histology image and the target T2w MRI image. T2w MRI volume was resampled and aligned with DCE MRI volume using BRAINS resampling module in 3D Slicer [28] which allowed for mapping of cancer extent from T2w MRI to DCE MRI.
Figure 3.
Histology-MRI co-registration scheme used to map cancer annotations from (a) CD31 stained pseudo whole-mount radical prostatectomy section onto its corresponding (b) T2w and (c) DCE MRI slices. (d) Histology is first registered to T2w MRI, following which (e) the transformation needed to align T2 and DCE is learned and (f) re-applied to the T2-transformed histology section to obtain histology-DCE MRI registration.
Microvessel Segmentation and Feature Extraction
Vascular structures expressing CD31 within tumor regions were segmented using Hierarchical Normalized Cuts (HNCut) as shown in Figure 4 [29]. HNCut combines frequency weighted mean shift (FWMS) and normalized cuts (NCut) in a hierarchical manner at multiple resolutions to identify differently stained biomarker regions corresponding to user selected color swatch.
To capture the overall MVD within the tumor, we compute features using the following two definitions of densities: (1) ratio of microvessel area to tumor area (MVDarea), (2) number of microvessels per unit area of tumor (MVDnum). In addition, graph based and nearest neighbor based QH features were computed. Graph based features capture the overall architecture of the structures of interest using graphical constructs while the nearest neighbor features seek to capture density in local neighborhoods. To extract graph based features, centroids of segmented CD31 stained microvessels were used as nodes which were connected using Delaunay, Voronoi and Minimum spanning tree graphs. A total of 24 statistical features, listed in Table 1, were then extracted from these graphical representations of microvessel distribution [30]. In addition, 24 nearest neighbor (NN) features were extracted by computing statistics of distance to NN around each microvessel and the number of microvessels within a given pixel radius. Another 24 statistical shape features of microvessel area, perimeter, orientation, length of major and minor axes and eccentricity were included in the analysis, resulting in a total of 77 features.
Table 1.
List of extracted prostate cancer microvessel and DCE MRI features
| Modality | Type | Features | # of Features |
|---|---|---|---|
| CD31 Stained Histology | Density | Ratio of total microvessel and tumor area across all tumor foci (total MVDarea), maximum (max MVDarea) and standard deviation of (std MVDarea) of the ratio of microvessel and tumor area; Maximum (max MVDnum) and standard deviation (std MVDnum) of the number of microvessels per unit area | 5 |
| Shape | Mean, median, standard deviation and disorder of area, eccentricity, major axis length, minor axis length, orientation and perimeter | 24 | |
| Architectural: Voronoi Diagram | Mean, standard deviation, minimum-maximum ratio and disorder of polygon area, perimeter and chord length | 12 | |
| Architectural: Delaunay Triangulation | Mean, standard deviation, minimum-maximum ratio and disorder of triangle area and side length | 8 | |
| Architectural: Minimum Spanning Tree (MST) | Mean, standard deviation, minimum-maximum ratio and disorder of edge length | 4 | |
| Architectural: Nearest Neighbors | Mean, standard deviation and disorder of (a) distance to 3, 5 and 7 nearest neighbors (NN) and (b) NN in 10, 20, 30, 40, 50 pixel radius (PR) | 24 | |
| DCE MRI | Per voxel SI Kinetic Curve | Mean and variance of maximum uptake (MU), time to peak (TTP), initial gradient (IG), washout gradient (WG), enhancement (En), enhancement ratio (EnR), initial area under the kinetic curve at 30 and 60 seconds (IAUC 30, IAUC 60), total area under the curve (IAUC) | 18 |
Experimental Design for Microvessel Feature Selection
Quadratic discriminant analysis (QDA) was applied using leave-one-out cross validation strategy on the entire data cohort to independently evaluate each feature in terms of its ability to distinguish between patients with low grade and intermediate/high grade tumors. For each feature, a QDA classifier was constructed, which enabled the identification of a quadratic surface that optimally separates inputs from different classes. Area under the receiver operating characteristic curve (AUC), accuracy, balanced accuracy, sensitivity and specificity, were computed to assess classification performance of each feature. Finally, features with an AUC > 0.7 were retained for subsequent correlation analysis with DCE MRI.
DCE MRI Feature Extraction
Mapping of cancer extent from histology to DCE MRI presents an opportunity for interrogation of tumor contrast kinetics at per-voxel level. Kinetic curves were computed at each voxel by averaging over a 3×3 neighborhood and the following kinetic features were computed: maximum uptake (MU), time to peak (TTP), initial gradient (IG), washout gradient (WG), enhancement (En), enhancement ratio (EnR), initial area under the curve (IAUC) at 30, 60 seconds and total area under the curve. Maximum uptake was defined as the maximum signal intensity and time to peak was defined as the time at maximum signal intensity. Initial enhancement was computed as the initial slope of the signal intensity vs. time curve while enhancement ratio was the ratio of initial enhancement and maximum uptake. To obtain initial gradient and washout gradient as defined by Chen et al.[10], relative enhancement (RE) curves were obtained where RE(t) = (SI(t)post − SIpre)/SI(t)post × 100. Initial gradient was computed as the slope between 10% and 70% of maximum enhancement and the washout gradient was computed as the slope at the last 60 seconds of the RE curve. Initial gradient, washout gradient and initial area under the curve features were calculated by linearly interpolating RE curves.
Experimental Design for DCE MRI Kinetic Feature Evaluation
The effect of temporal resolution on kinetic features was studied by evaluating the concordance between feature values extracted from the original and temporally downsampled kinetic curves. For this experiment, we considered a subset of the data comprising 5 cases with high temporal resolutions ranging between 8–12 seconds. At each tumor voxel, three temporally downsampled kinetic curves were generated by considering every second, third and fourth time frame which resulted in curves with temporal sampling of roughly 16, 24 and 32 seconds. These simulated curves spanned the range of resolutions found in our data cohort. Intraclass correlation coefficient (ICC) was used to estimate the concordance of feature values extracted from the original and downsampled kinetic curves.
In addition, each kinetic feature was evaluated in terms of its ability to distinguish low from intermediate and high grade prostate cancer using the entire data cohort. A QDA classifier was constructed independently for each feature statistic (Table 2) capturing the distribution of feature values over all tumor voxels from each patient. Classification was performed using leave-one-out cross validation strategy in order to account for the small sample size considered in this work. The resulting AUC was then plotted against ICC to identify kinetic features that are robust to temporal resolution and are predictive of low grade prostate cancer.
Table 2.
Microvessel features predictive of low vs. intermediate/high grade prostate cancer with an AUC > 0.7 as evaluated by leave-one-out cross validation using QDA classifier. Microvessel features with the highest balanced accuracies (BACC) are shown in bold. The table also lists maximally correlated DCE MRI feature with their corresponding Spearman’s correlation coefficients and false discovery rates (FDR). Mean washout gradient (WG) is maximally correlated with most microvessel features and is significantly (FDR<0.05) correlated with mean Delaunay triangle area of microvessel arrangement, one of the most predictive features with high sensitivity and specificity. Significant correlations are denoted by (*).
| # | Type | Feature | AUC | ACC | BACC | SEN | SPE | MAX CORR (FDR) | MAX CORR FEATURE |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Architectural | Disorder in 20 PR | 0.84 | 0.87 | 0.79 | 0.57 | 1.00 | 0.63 (0.06) | Mean WG |
| 2 | Architectural | Disorder in 30 PR | 0.78 | 0.83 | 0.71 | 0.43 | 1.00 | −0.55 (0.02*) | Mean IAUC 30 |
| 3 | Architectural | Mean Side Length (Delaunay) | 0.76 | 0.83 | 0.71 | 0.43 | 1.00 | 0.58 (0.03*) | Mean WG |
| 4 | Architectural | Mean Triangle Area (Delaunay) | 0.74 | 0.83 | 0.79 | 0.71 | 0.88 | 0.52 (0.02*) | Mean WG |
| 5 | Shape | Mean microvessel area | 0.74 | 0.78 | 0.64 | 0.29 | 1.00 | 0.36 (0.04*) | Mean WG |
| 6 | Density | Total MVDarea | 0.73 | 0.78 | 0.64 | 0.43 | 0.94 | 0.38 (0.03*) | Var IG |
| 7 | Architectural | Mean MST Edge Length | 0.72 | 0.83 | 0.71 | 0.43 | 1.00 | 0.59 (0.03*) | Mean WG |
| 8 | Architectural | Mean distance to 5 NN | 0.72 | 0.83 | 0.71 | 0.43 | 1.00 | 0.58 (0.02*) | Mean WG |
| 9 | Architectural | Mean distance to 7NN | 0.72 | 0.83 | 0.71 | 0.43 | 1.00 | 0.58 (0.02*) | Mean WG |
| 10 | Architectural | Mean distance to 15 NN | 0.72 | 0.83 | 0.71 | 0.43 | 1.00 | 0.57 (0.02*) | Mean WG |
| 11 | Architectural | Average NN in 30 PR | 0.72 | 0.70 | 0.50 | 0.00 | 1.00 | −0.61 (0.03*) | Mean WG |
| 12 | Architectural | Average NN in 40 PR | 0.71 | 0.74 | 0.73 | 0.71 | 0.75 | −0.62 (0.04*) | Mean WG |
| 13 | Architectural | Average NN in 50 PR | 0.71 | 0.74 | 0.73 | 0.71 | 0.75 | −0.60 (0.03*) | Mean WG |
| 14 | Architectural | Std Side Length (Delaunay) | 0.71 | 0.83 | 0.71 | 0.43 | 1.00 | 0.58 (0.02*) | Mean WG |
| 15 | Shape | Mean microvessel major axis length | 0.71 | 0.78 | 0.68 | 0.43 | 0.94 | 0.40 (0.03*) | Mean WG |
Statistical Analysis
Spearman’s rank correlation test was used to assess strengths of associations between select kinetic features (ICC > 0.5) and select microvessel features (AUC > 0.7). The test seeks a general monotonic trend without assuming linearity and is robust to outliers. The resulting matrix of pairwise correlation coefficients was visualized as a heatmap, which we term radiohistomorphometric map. Statistical significance of each pairwise correlation was assessed using false discovery rate (FDR) to account for multiple hypothesis testing [31]. All statistical analyses were performed in Matlab (Mathworks, USA).
RESULTS
Microvessel Features Discriminative of Low from Intermediate and High Grade Prostate Cancer
As shown in Table 2, QDA classification results revealed 15 microvessel features which were independently able to distinguish low from intermediate/high grade prostate cancer on a per-patient basis with an AUC > 0.7. This feature subset comprised of 12 architectural features, 1 density measure based on total microvessel area (MVDarea) and 2 shape features. Of the 12 architectural features, 4 were graph-based features obtained from delaunay and minimum spanning tree graphs and 8 were measures of local microvessel density based on distance to nearest neighbors and number of nearest neighbors within a given pixel radius around each microvessel. According to the balanced accuracy measure, disorder in the number of nearest neighbors in a 20 pixel radius and mean delaunay triangle area were the best performing features with balanced accuracies of 0.79. By contrast, the density feature had a lower balanced accuracy of 0.64 whereas the two morphologic features, mean microvessel area and mean microvessel major axis length, had balanced accuracies of 0.64 and 0.68, respectively. Taking into consideration all the evaluation metrics, mean delaunay triangle area of microvessel architecture appears to be the best performing feature with high AUC, accuracy, balanced accuracy, sensitivity and specificity.
Stability and Discriminability of DCE MRI Features for Prostate Cancer Risk Assessment
Figure 5 shows the trade-off between feature stability with respect to temporal resolution and discriminability with respect to Gleason scores for all kinetic features. Although variance in initial area under the curve at 60 seconds (var IAUC 60) appears to be highly discriminative of Gleason scores with an AUC > 0.9, it has low ICC indicating that the feature is highly sensitive to temporal resolution. Mean enhancement ratio, variance washout gradient and variance initial gradient on the other hand have ICC > 0.6 and are predictive of low grade prostate cancer with an AUC > 0.75.
Figure 5.
Scatter plot showing DCE MRI kinetic feature performance (AUC) in distinguishing low from intermediate/high grade tumors and feature robustness (ICC) to temporal resolution. An ideal marker is expected to be both robust to temporal resolution and discriminable with an ICC and AUC of 1. The closest feature to the ideal (1,1) coordinate is mean enhancement ratio (EnR) which has an ICC of 0.96 and AUC of 0.78.
Radiohistomorphometric Maps Associate in vivo DCE MRI Features with Microvessel Characteristics on CD31 Stained ex vivo Pathology
Pairwise correlation between histology and MRI features were examined on per-patient basis, wherein feature statistics were computed within the tumor across all annotated slices for each patient. Figure 6 shows the correlation heatmap of pairwise associations between the 15 microvessel features which were found to have an AUC > 0.7 in predicting low grade tumor and 12 DCE MRI features which were found to be robust to temporal resolution with an ICC > 0.5. As can be seen in Table 2 and Figure 6, most of the architectural features were maximally and significantly (FDR < 0.05) correlated with mean washout gradient. In particular, mean washout gradient was most highly and significantly correlated with features capturing the average number of nearest neighbors within specified pixel radius (PR) (max rho = −0.62, FDR = 0.04), followed by minimum spanning tree feature (max rho = 0.59, FDR = 0.03), mean distance to specified NN features (max rho = 0.58, FDR = 0.02) and Delaunay graph features (max rho = 0.58, FDR = 0.02). However, kinetic features were poorly correlated with area based measure of microvessel density (max rho = 0.38, FDR = 0.03) and shape features (max rho = 0.40, FDR = 0.03). Among the kinetic features, variance in enhancement ratio, variance in washout gradient and mean initial gradient appear to have the weakest correlations with all microvessel features.
Figure 6.
Correlation heatmap showing Spearman’s correlation coefficient (red=negative correlation, blue=positive correlation) between each pair of microvessel (horizontal axis) and DCE MRI features (vertical axis). The sizes and colors of the squares are proportional to the correlation coefficient. Correlations that are not statistically significant with false discovery rate (FDR) ≥ 0.05 are marked in gray with an x.
DISCUSSION
Overdiagnosis and overtreatment of prostate cancer has contributed to the emerging need for in vivo imaging markers of prostate cancer risk. Identification of such markers may enable selection of suitable candidates for active surveillance, where patients are actively observed and only treated upon signs of disease progression. In this paper, we introduced a quantitative image analysis framework for radiology-pathology convergence that allowed us to identify promising DCE MRI markers that were (i) correlated with a subset of quantitative histomorphometric features of microvessels, (ii) discriminative of low vs. intermediate/high Gleason scores and appeared to be (iii) robust to temporal resolution. Challenges in extracting and relating quantitative descriptors of tumor appearance on radiology and pathology were addressed by employing automated segmentation, quantitative histomorphometric feature extraction and image co-registration methods.
This study identified promising potential markers, initial enhancement ratio and washout gradient, the latter of which was previously shown to be discriminative of Gleason scores [10,11]. While previous work additionally showed associations between Gleason scores and pharmacokinetic features, temporal resolutions of the studies in our data cohort limited us from considering these features as they are known to be sensitive to temporal resolution [32]. The datasets considered in previous work had temporal resolutions of 2 seconds [10] and 3 seconds [11] whereas the maximum resolution of the data considered in this study was 8 seconds. Thus, in addition to constraining our analysis to kinetic features, we also attempted to study feature robustness using temporally downsampled kinetic curves to simulate those extracted at lower temporal resolutions. However, the simulated curves do not take into account changes in spatial resolution that accompany changes in temporal sampling on imaging. As such, we assumed that none of the other image characteristics are significantly affected by changes in temporal resolution.
The most commonly used clinical criteria for selection of patients for active surveillance include only patients with GS ≤ 6 [33]. However, a long term follow up study [34] showed that the 10 and 15 year prostate cancer specific survival rate was 98% and 94%, respectively, for a data cohort that included patients with both low risk (GS≤ 6) and favorable intermediate risk (GS 3+4) cancer. Such findings suggest that merely relying on GS may result in exclusion of patients with intermediate GS who may benefit from active surveillance. Thus, by considering correlates of additional hallmarks associated with cancer such as tissue vascularity, this work attempts to identify imaging markers that are potentially better suited for active surveillance. One of the major findings of this work was that DCE MRI kinetic features strongly correlate with graph based architectural features and measures of microvessel density within local neighborhoods. Previous work studying correlations between microvessel and DCE MRI parameters in the context of prostate cancer have only considered pharmacokinetic features [20,35] using images acquired at temporal resolutions that are not reflective of those acquired in clinical settings. Findings of this study suggest that it may be possible to infer characteristics of tissue vascularity with kinetic features computed from DCE MRI acquired using standard clinical protocol.
This study, however, also had limitations. Firstly, we considered a small data cohort obtained from a larger set of patients who were eligible for this study. Time and budget constraints associated with sectioning, staining, and annotation of radical prostatectomy specimens limited the study to only 23 patients. Secondly, there is no practical method for assessing registration accuracy at per-voxel level. While attempts were made to minimize errors in the x-y plane by selecting anatomical landmarks visible on both histology and MRI, identifying slice correspondences to ensure accurate mapping in the z plane is a highly challenging task. Furthermore, microvessel features for correlative analysis were selected based on their ability to separate tumors of different Gleason scores due to absence of outcome information. Numerous studies have reported the limitations of Gleason grading system, primarily due to its associated inter-, and intra-observer variability [36]. These limitations are particularly pertinent to tumors with intermediate grades, which form the largest group in this study’s data cohort.
In conclusion, this work presented a radiology-pathology correlation framework that enabled identification of promising in vivo DCE MRI markers of prostate cancer risk. Future directions of this work include interrogating the imaging markers identified in this study on a larger data cohort to evaluate their ability to predict longer term disease outcome. In addition, we would like to explore features from other MP-MRI protocols such as T2w MRI and diffusion weighted imaging (DWI) that may be informative of tumor aggressiveness by the virtue of their correlation with other histologic and immunohistochemical biomarkers of prostate cancer risk.
Acknowledgments
Grant Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01CA136535-01, R01CA140772-01, R21CA167811-01, R21CA179327-01; The DOD Prostate Cancer Synergistic Idea Development Award (PC120857); Department of Defense (W81XWH-13-1-0487), the Ohio Third Frontier Technology development Grant, the CTSC Coulter Annual Pilot Grant, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. New England Journal of Medicine. 2012;367(3):203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia J, Trock BJ, Cooperberg MR, et al. Prostate cancer mortality following active surveillance versus immediate radical prostatectomy. Clinical Cancer Research. 2012;18(19):5471–5478. doi: 10.1158/1078-0432.CCR-12-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klotz L. Cancer overdiagnosis and overtreatment. Current Opinion in Urology. 2012;22(3):203–209. doi: 10.1097/MOU.0b013e32835259aa. [DOI] [PubMed] [Google Scholar]
- 4.CP Active surveillance may be preferred option in some men with prostate cancer. 2011 [Google Scholar]
- 5.Bono A, Celato N, Cova V, et al. Microvessel density in prostate carcinoma. Prostate cancer and prostatic diseases. 2002;5(2):123–127. doi: 10.1038/sj.pcan.4500572. [DOI] [PubMed] [Google Scholar]
- 6.Buhmeida A, Pyrhonen S, Laato M, Collan Y. Prognostic factors in prostate cancer. Diagn Pathol. 2006;1(4):124. doi: 10.1186/1746-1596-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah RB. Current perspectives on the Gleason grading of prostate cancer. Archives of Pathology & Laboratory Medicine. 2009;133(11):1810–1816. doi: 10.5858/133.11.1810. [DOI] [PubMed] [Google Scholar]
- 8.Palisaar JR, Noldus J, Löppenberg B, et al. Comprehensive report on prostate cancer misclassification by 16 currently used low-risk and active surveillance criteria. BJU International. 2012;110(6b):E172–E181. doi: 10.1111/j.1464-410X.2012.10935.x. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro RH, Johnstone PA. Risk of Gleason grade inaccuracies in prostate cancer patients eligible for active surveillance. Urology. 2012;80(3):661–666. doi: 10.1016/j.urology.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y-J, Chu W-C, Pu Y-S, et al. Washout gradient in dynamic contrast-enhanced MRI is associated with tumor aggressiveness of prostate cancer. Journal of Magnetic Resonance Imaging. 2012;36(4):912–919. doi: 10.1002/jmri.23723. [DOI] [PubMed] [Google Scholar]
- 11.Vos EK, Litjens GJ, Kobus T, et al. Assessment of prostate cancer aggressiveness using dynamic contrast-enhanced magnetic resonance imaging at 3 T. European Urology. 2013;64(3):448–455. doi: 10.1016/j.eururo.2013.05.045. [DOI] [PubMed] [Google Scholar]
- 12.Thompson I, Thrasher JB, Aus G, et al. Guideline for the management of clinically localized prostate cancer: 2007 update. The Journal of Urology. 2007;177(6):2106–2131. doi: 10.1016/j.juro.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Amin MB, Lin DW, Gore JL, et al. The critical role of the pathologist in determining eligibility for active surveillance as a management option in patients With prostate cancer: Consensus statement with recommendations supported by the College of American Pathologists, International Society of Urological Pathology, Association of Directors of Anatomic and Surgical Pathology, the New Zealand Society of Pathologists, and the Prostate Cancer Foundation. Archives of Pathology and Laboratory Medicine. 2014;138(10):1387–1405. doi: 10.5858/arpa.2014-0219-SA. [DOI] [PubMed] [Google Scholar]
- 14.Kryvenko ON, Carter HB, Trock BJ, Epstein JI. Biopsy criteria for determining appropriateness for active surveillance in the Modern Era. Urology. 2014;83(4):869–874. doi: 10.1016/j.urology.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Djenaba JA, Soman A, Rim SH, Master VA. Recent trends in prostate cancer incidence by age, cancer stage, and grade, the United States, 2001–2007. Prostate cancer. 2012;2012 doi: 10.1155/2012/691380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark JR, Perner S, Stampfer MJ, et al. Gleason score and lethal prostate cancer: does 3+ 4= 4+ 3? Journal of Clinical Oncology. 2009;27(21):3459–3464. doi: 10.1200/JCO.2008.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bostwick DG, Wheeler TM, Blute M, et al. Optimized microvessel density analysis improves prediction of cancer stage from prostate needle biopsies. Urology. 1996;48(1):47–57. doi: 10.1016/s0090-4295(96)00149-5. [DOI] [PubMed] [Google Scholar]
- 18.Mucci LA, Powolny A, Giovannucci E, et al. Prospective study of prostate tumor angiogenesis and cancer-specific mortality in the health professionals follow-up study. Journal of Clinical Oncology. 2009;27(33):5627–5633. doi: 10.1200/JCO.2008.20.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tretiakova M, Antic T, Binder D, et al. Microvessel density is not increased in prostate cancer: digital imaging of routine sections and tissue microarrays. Human Pathology. 2012 doi: 10.1016/j.humpath.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 20.van Niekerk CG, van der Laak JA, Hambrock T, et al. Correlation between dynamic contrast-enhanced MRI and quantitative histopathologic microvascular parameters in organ-confined prostate cancer. European Radiology. 2014;24(10):2597–2605. doi: 10.1007/s00330-014-3301-z. [DOI] [PubMed] [Google Scholar]
- 21.Doyle S, Feldman M, Shih N, Tomaszewski J, Madabhushi A. Cascaded discrimination of normal, abnormal, and confounder classes in histopathology: Gleason grading of prostate cancer. BMC Bioinformatics. 2012;13(1):282. doi: 10.1186/1471-2105-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeffler M, Greulich L, Scheibe P, et al. Classifying prostate cancer malignancy by quantitative histomorphometry. The Journal of Urology. 2012;187(5):1867–1875. doi: 10.1016/j.juro.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 23.Erbersdobler A, Isbarn H, Dix K, et al. Prognostic value of microvessel density in prostate cancer: a tissue microarray study. World Journal of Urology. 2010;28(6):687–692. doi: 10.1007/s00345-009-0471-4. [DOI] [PubMed] [Google Scholar]
- 24.Chappelow J, Tomaszewski JE, Feldman M, Shih N, Madabhushi A. HistoStitcher©: An interactive program for accurate and rapid reconstruction of digitized whole histological sections from tissue fragments. Comput Med Imaging and Graph. 35:557–567. doi: 10.1016/j.compmedimag.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toth R, Shih N, Tomaszewski J, et al. Histostitcher™: An informatics software platform for reconstructing whole-mount prostate histology using the extensible imaging platform framework. Journal of Pathology Informatics. 2014;5(1):8. doi: 10.4103/2153-3539.129441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao G, Bloch BN, Chappelow J, et al. Determining histology-MRI slice correspondences for defining MRI-based disease signatures of prostate cancer. Comput Med Imaging and Graph. 35:578–658. doi: 10.1016/j.compmedimag.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Bookstein FL. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Pattern Analysis and Machine Intelligence. 1989;11(6):567–585. [Google Scholar]
- 28.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magnetic resonance imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janowczyk A, Chandran S, Singh R, et al. High-Throughput Biomarker Segmentation on Ovarian Cancer Tissue Microarrays via Hierarchical Normalized Cuts. IEEE Transactions on Bio-Medical Engineering. 2011;59:1240–1252. doi: 10.1109/TBME.2011.2179546. [DOI] [PubMed] [Google Scholar]
- 30.Basavanhally AN, Ganesan S, Agner S, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. Biomedical Engineering, IEEE Transactions on. 2010;57(3):642–653. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 32.Fluckiger JU, Schabel MC, DiBella EV. The effect of temporal sampling on quantitative pharmacokinetic and three-time-point analysis of breast DCE-MRI. Magnetic resonance imaging. 2012;30(7):934–943. doi: 10.1016/j.mri.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Salomon L, Bastide C, Beuzeboc P, et al. CCAFU Recommendations 2013: Prostate cancer. Progres en urologie: journal de l’Association francaise d’urologie et de la Societe francaise d’urologie. 2013;23:69–101. doi: 10.1016/S1166-7087(13)70048-4. [DOI] [PubMed] [Google Scholar]
- 34.Klotz L, Vesprini D, Sethukavalan P, et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. Journal of Clinical Oncology. 2014:272–277. doi: 10.1200/JCO.2014.55.1192. [DOI] [PubMed] [Google Scholar]
- 35.Franiel T, Ludemann L, Rudolph B, et al. Prostate MR Imaging: Tissue Characterization with Pharmacokinetic Volume and Blood Flow Parameters and Correlation with Histologic Parameters 1. Radiology. 2009;252(1):101–108. doi: 10.1148/radiol.2521081400. [DOI] [PubMed] [Google Scholar]
- 36.Allsbrook WC, Jr, Mangold KA, Johnson MH, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: general pathologist. Human pathology. 2001;32(1):81. doi: 10.1053/hupa.2001.21135. [DOI] [PubMed] [Google Scholar]