Abstract
Paternal diet can impact metabolic phenotypes in offspring, but mechanisms underlying such intergenerational information transfer remain obscure. Here, we interrogate cytosine methylation patterns in sperm obtained from mice consuming one of three diets, generating whole genome methylation maps for 4 pools of sperm samples and for 12 individual sperm samples, as well as 61 genome-scale methylation maps. We find that “epivariation”, either stochastic or due to unknown demographic or environmental factors, was a far stronger contributor to the sperm methylome than was the diet consumed. Variation in cytosine methylation was particularly dramatic over tandem repeat families, including ribosomal DNA (rDNA) repeats, but rDNA methylation was strongly correlated with genetic variation in rDNA copy number, and was not influenced by paternal diet. These results identify loci of genetic and epigenetic lability in the mammalian genome, but argue against a direct role for sperm cytosine methylation in dietary reprogramming of offspring metabolism.
INTRODUCTION
The environmental conditions experienced by an organism can influence the phenotypes of future generations. In mammals, examples in which ancestral environmental conditions affect offspring phenotype include relatively well-studied effects of maternal dietary treatments on offspring metabolism (Harris and Seckl, 2010; Li et al., 2011), but also a number of cases in which paternal environmental conditions affect phenotypes in offspring (Rando, 2012). Paternal effect paradigms include toxin exposure, stress paradigms, and a wide variety of dietary manipulations. In rodents, males subjected to various dietary perturbations – including low protein diet, high fat diet, caloric restriction, and intermittent fasting – sire offspring with altered glucose and lipid metabolism, relative to males consuming a matched control diet (Anderson et al., 2006; Carone et al., 2010; Jimenez-Chillaron et al., 2009; Ng et al., 2010; Radford et al., 2012; Radford et al., 2014; Watkins and Sinclair, 2014; Wei et al., 2014). In humans, epidemiological studies have linked ancestral nutrition to children’s and grandchildren’s rates of diabetes, obesity, and cardiovascular disease (Lumey et al., 2007; Pembrey et al., 2006). Despite this wealth of phenomenology, the mechanistic basis by which fathers influence their children’s phenotype remains obscure.
Cytosine methylation is one of the best-characterized epigenetic marks in mammals (Cedar and Bergman, 2012; Deaton and Bird, 2011; Feng et al., 2010) and is often implicated in transgenerational inheritance paradigms (Daxinger and Whitelaw, 2012). Paternal diet can affect cytosine methylation in offspring tissues; for instance, we previously reported that paternal low protein diet causes a reproducible ~10% change in cytosine methylation at an enhancer of the lipid regulator Ppara in offspring livers (Carone et al., 2010). However, in this case and in many related cases, the cytosine methylation changes reported in offspring somatic tissues were not present in sperm of the treated males, indicating that these methylation changes are established at some point following fertilization, and thus cannot be the gametic information responsible for metabolic reprogramming of offspring. Such observations have motivated a number of investigators to focus on cytosine methylation in sperm of control and treated animals. Two recent studies reported reproducible cytosine methylation changes in sperm of males subject to in utero undernutrition (Radford et al., 2014), or subject to diet and drug-induced prediabetes (Wei et al., 2014), with a subset of these methylation changes persisting in offspring tissues.
However, these and other cytosine methylation changes reported in paternal effect paradigms are quantitatively modest, with 10–20% changes in methylation being typical. In principle, modest methylation differences at individual CpGs cannot account for penetrant offspring phenotypes because of the “digital” nature of sperm. As each sperm carries a single haploid genome, a CpG with 20% methylation thus means that 1 in 5 sperm are methylated at that CpG, and a change from 20% to 40% methylation at a single CpG merely alters the frequency of sperm with a methylated CpG from 1 in 5 sperm to 2 in 5 sperm. As fertilization involves the fusion of a single sperm with a single oocyte, modest methylation changes at individual CpGs at best should only alter penetrance of a phenotype across a set of siblings. That said, if small yet consistent methylation changes occur independently across multiple (presumably adjacent) CpGs in sperm, and methylation levels are integrated across a locus to alter phenotype in offspring, then digital sperm could in principle exert continuous control in a penetrant manner across a set of siblings. Given the moderate resolution of prior studies on dietary effect paradigms in mammals, it remains plausible that single-nucleotide resolution whole genome maps of dietary effects on cytosine methylation could uncover genomic loci with the potential to penetrantly transmit paternal information to offspring.
Beyond environmentally-directed cytosine methylation changes, variability in cytosine methylation among control animals, known as “epivariation” (Irizarry et al., 2009; Whitelaw and Whitelaw, 2008), also contributes to the sperm epigenome. For example, we previously reported that siblings on different diets exhibited more similar sperm methylation patterns than did control animals that were more distantly related (Carone et al., 2010), despite using inbred animals. Therefore, it is of great interest to uncover the relative contributions of epivariation versus environmental influences on the sperm epigenome.
We therefore sought to investigate the effect of post-weaning diet, and of epivariation, on cytosine methylation patterns in murine sperm. Genome-scale methylation assays of 61 sperm samples confirmed our prior findings that epivariation has a greater influence over the epigenome than any of the environmental conditions tested. Whole genome analyses highlighted tandem repeat regions as being particularly susceptible to variability in cytosine methylation, and follow-up studies on rDNA and other repeats revealed that tandem repeat methylation was highly variable between inbred mice, but did not exhibit consistent responses to paternal diet. Cytosine methylation at the rDNA loci was heritable from father to child, and was linked to rDNA copy number variation, emphasizing a key role for genetic variation in driving variation in the “epigenome”, even in inbred animals. Our results do not support a role for cytosine methylation changes in sperm as the central mechanism underlying post-weaning effects of paternal diet on offspring metabolism.
RESULTS
Genome-wide methylation profiles of four sperm pools
Previous studies from our lab and several others (reviewed in (Rando and Simmons, 2015)) document an effect of paternal diet on offspring phenotype, and in vitro fertilization (IVF) studies in our lab reveal that dietary information is carried in sperm in both Low Protein and High Fat diet paradigms (Sharma et al, submitted). We have therefore initiated an extensive series of genome-wide studies in mouse sperm to attempt to identify dietary effects on the sperm epigenome. Here, we focus on cytosine methylation patterns in sperm isolated from males maintained from weaning until sexual maturity – from 3 weeks of age until 10–14 weeks of age – on various diets. We focused on animals maintained on Control, Low Protein (10% instead of 19% protein, with remaining calories contributed by sucrose), or High Fat (60% instead of 20% fat calories) diets. For all experiments described below, mature sperm were isolated from cauda epididymis and vas deferens, and stringently washed with somatic cell lysis buffer, with all preparations being >99% pure as assessed by microscopy.
In order to characterize the cytosine methylation landscape genome-wide, we first carried out whole genome shotgun bisulfite sequencing (WGBS) of genomic DNA isolated from sperm. We generated four pools of sperm samples from paired sets of sibling animals weaned to alternative diets, and included a separate matched set of Control siblings for the Low Protein pool (n=8 animals in each pool) and for the High Fat (n=7) pool (Supplemental Table S1). This pooling strategy should be relatively robust (but see below) to epivariation between non-littermates (Carone et al., 2010), as paired sets of siblings underlie the Control/Low Protein comparison, and the Control/High Fat comparison. On average, 1.4 billion reads were generated for each pool, yielding an average of 47-fold mean genomic coverage. This sequencing depth and experimental design exceeds the “gold standard” 30X depth commonly achieved in WGBS datasets, sufficient to enable the robust identification of differentially-methylated regions associated with small methylation differences (Ziller et al., 2015).
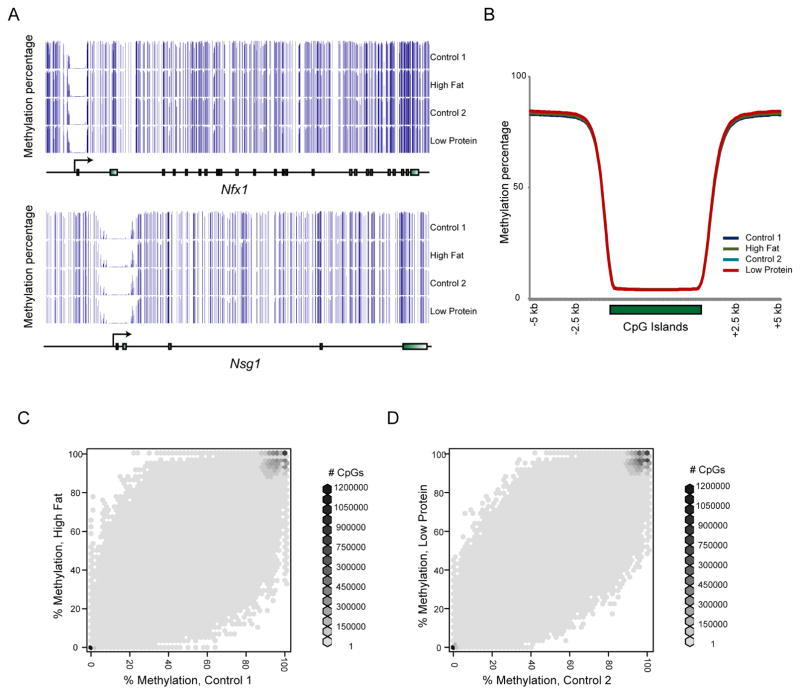
As previously observed (Molaro et al., 2011), sperm are overall highly methylated, with regions of focal hypomethylation occurring at CpG-rich regulatory elements such as promoters and enhancers (Figures 1A–B). Consistent with our prior analysis using MeDIP-Seq (Carone et al., 2010), the methylome was generally insensitive to diet (Figures 1C–D and Supplemental Figure S1) – the majority of CpGs were either completely unmethylated or methylated, and these CpGs were unaffected by paternal diet, while individual CpGs with intermediate methylation levels (20–80% methylated) exhibited modest changes (up to ~20–30% methylation differences) between samples. Consistent with studies of somatic cell methylation patterns (Irizarry et al., 2009), CpGs that were differentially-methylated between samples were commonly found ~1–2 kB from CpG islands, in so-called CpG island “shores” (see below).
Figure 1. Whole genome cytosine methylation in murine sperm.
(A) Examples of typical methylation profiles. For the two genes shown, cytosine methylation data for each of the four libraries – Control 1, High Fat, Control 2, and Low Protein – are shown. Each vertical bar represents the methylation percentage for a single CpG. Typical here is a general background of complete methylation, with hypomethylation occurring at CpG islands such as promoters. (B) Average cytosine methylation for each of the four libraries plotted over CpG islands and surrounding DNA. CpG islands were length-normalized for this visualization. (C–D) Scatterplots for individual CpG methylation levels between the matched Control and High Fat pools (C) or between matched Control and Low Protein pools (D). Data are shown for the 80% CpGs with the greatest read depth (n=16.1 and 16.6 million CpGs for C and D). See also Supplemental Figure S1.
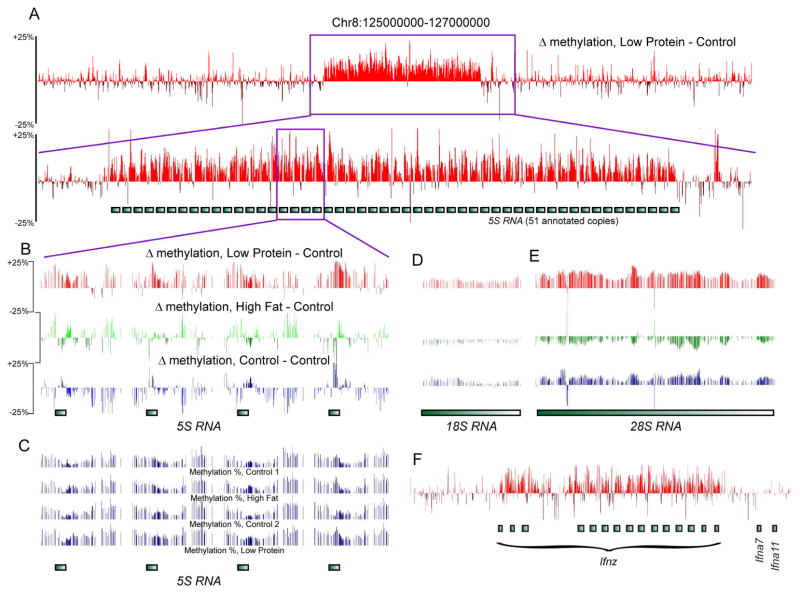
Differentially-methylated CpGs between sample pairs were identified using a sequencing depth-sensitive statistic. As modest (~10%) methylation changes at any individual CpG in sperm cannot account for penetrant phenotypic effects on offspring (see Introduction), we therefore searched specifically for short (300 bp) clusters of CpGs showing consistent dietary effects on methylation (Supplemental Table S2 – similar results were obtained with 100 and 1000 bp windows). The majority of loci identified occurred in tandem repeat regions, notably the rDNA clusters (Aldrich and Maggert, 2015), but also including gene families such as the interferon zeta (Ifnz) gene family, defensins, cytochrome P450 genes, Mrgpra/b genes, Skint genes, and many others (Figure 2 and Supplemental Figure S2). These loci were generally hypermethylated in the Low Protein sample, and hypomethylated or unaffected in the High Fat sample, relative to the matched Controls. A smaller set of CpGs, such as those located in the pseudoautosomal region of the X chromosome, exhibited the opposite methylation behavior (Supplemental Figures S2D–E). Beyond these relatively long differentially-methylated regions, we found few dietary effects on cytosine methylation, with most methylation differences being modest (~10%) changes occurring at a handful of CpGs located in CpG island shores (see example in Supplemental Figure S2E). Although repetitive elements have been implicated in transgenerational effects of some dietary perturbations (Waterland and Jirtle, 2003), averaged methylation over repeat elements was also generally unaffected by diet (Supplemental Table S3).
Figure 2. Methylation differences primarily occur over repeated gene families.
(A) Methylation changes over the 5S rDNA locus on chromosome 8, shown as Low Protein minus its matched Control pool – positive values indicate hypermethylated loci in Low Protein diet. Top panel shows 2 MB of chromosome 8 surrounding the 5S rDNA repeats, bottom panel is a zoom-in as indicated. (B) Zoom-in on 4 repeats of the 5S rDNA locus, showing Low Protein minus Control, High Fat minus Control, and Control 1 minus Control 2, as indicated. For all three images, the scale runs from −25% to +25%. (C) Absolute methylation levels (from 0 to 100%) for the loci shown in (B). (D–F) Additional loci hypermethylated in the Low Protein pool, as in (B). See also Supplemental Figures S2–3.
As 10% methylation changes occurring over small numbers of cytosines are unlikely to account for penetrant dietary effects on offspring, we focused on methylation changes over gene family clusters such as the rDNA and Ifnz clusters.
Genome-scale analysis of epivariation
Methylation differences identified in pooled animal samples could result from modest but penetrant changes across many animals, or could be driven by larger changes occurring in a subset of animals, as might be observed with highly “epivariable” loci. Indeed, cytosine methylation at rDNA repeats has previously been shown to vary significantly between individual inbred mice (Shiao et al., 2005; Shiao et al., 2012). Consistent with this latter possibility, most of the apparent dietary effects on cytosine methylation described above, not only over tandem repeat gene clusters but more generally, also exhibited substantial methylation differences between our two Control pools (Figure 2, Supplemental Figures S1D and S2).
We took two approaches to further explore dietary effects on methylation and to characterize epivariation across multiple individual animals. First, we generated relatively low-depth (~7X coverage) WGBS datasets for 12 individual sperm samples obtained from trios of littermates split to Control, Low Protein, or High Fat diet (Supplemental Table S1). These data did not recapitulate the dietary effects on tandem repeat loci observed in the WGBS pools (Supplemental Figures S3A–B), strongly arguing that the apparent dietary effects described above were driven by epivariation. To generally identify epivariable genomic loci across individual sperm samples, we calculated average methylation level for all 300 bp tiles (filtered for minimum coverage and methylation levels), and identified the 2000 tiles exhibiting maximal variance across the individual sperm samples. Clustering these data did not group sperm samples by diet (Supplemental Figure S3C), further supporting the observation that epivariation between animals is a far greater contributor to the sperm methylome than is the diet consumed by a given animal.
To investigate epivariation across a greater number of individual sperm samples, we next turned to reduced-representation bisulfite sequencing (RRBS) to characterize the methylome at ~4% of CpGs across the mouse genome, with a preference for CpGs located in CG-rich regulatory elements (Bock et al., 2010). This dataset included 61 sperm samples (Supplemental Table S1) isolated from males on one of several treatment regimens, primarily Control, Low Protein, or High Fat diets. Our methylation data recapitulated known features of the mammalian sperm methylome, with a bimodal distribution of methylation values, a strong anti-correlation between CpG density and methylation levels, and maintenance of cytosine methylation over repeat elements even at relatively high CpG density (Supplemental Figures S4A–E).
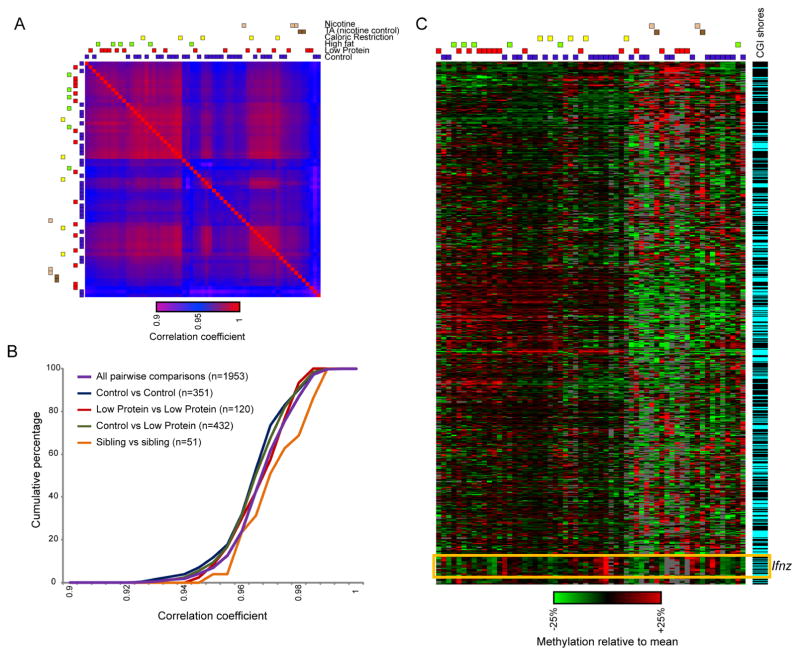
Our sperm methylation data were highly reproducible, with an average inter-sample correlation coefficient of 0.967 (Figure 3, Supplemental Figure S4F). Interestingly, clustering sperm samples by their methylome did not group animals based on diet (Figure 3A). Indeed, the distribution of correlations between pairs of Control animals, or between pairs of Low Protein animals, was indistinguishable from the distribution of correlations between Control and Low Protein animals (Figure 3B Supplemental Figure S4G). This is consistent with our prior observation (Carone et al., 2010), made using MeDIP-Seq, that sibling animals maintained on different diets had more similar sperm methylomes than did non-sibling pairs of Control animals. We confirm and extend this observation, finding that sperm from sibling animals were epigenetically more similar, whatever the dietary conditions experienced by the siblings, than were sperm samples obtained from non-siblings maintained on identical diets (Figure 3B). Together, these results emphasize a dominant role for epivariation, possibly stochastic (Schmitz et al., 2011) or potentially induced by unaccounted-for demographic factors (time of year, number of littermates, etc.), in the sperm epigenome. These results are generally consistent with observations from mouse somatic tissues (Feinberg and Irizarry, 2010), Arabidopsis (Schmitz et al., 2011), and other organisms identifying variability in methylation patterns between nominally genetically-identical organisms maintained under controlled conditions.
Figure 3. Epivariation among 61 sperm samples.
(A) Correlations between 61 individual RRBS libraries. The environmental conditions for each male are indicated as colored boxes. In addition to Control, Low Protein, and High Fat diets, data include animals subject to Caloric Restriction (60% of the diet mass consumed by ad libitum fed Controls), nicotine (administered 200 μg/mL free base in saccharine-sweetened drinking water), and the matched tartaric acid control. (B) Cumulative distribution plots for inter-sample correlations, for the indicated animal pairs. (C) Heatmap showing 748 regulatory elements (promoters, distal CpG islands, and CpG island shores) as rows, with all CpGs within each element averaged. Heatmap shows zero-centered data, grouped both by animal and by regulatory element. Right panel indicates CpG island shores as blue bars, showing that these represent the majority of epivariable loci in this dataset. See also Supplemental Figures S4–5.
To constrain hypotheses regarding the mechanistic basis for this epivariation, we sought to uncover the genomic loci subject to high levels of epivariation. For each individual CpG, we calculated the average methylation level for all 61 animals, then identified 3,396 CpGs exhibiting the greatest variation across our set of 61 animals. Consistent with prior observations in somatic cells (Irizarry et al., 2009), we found an overabundance of epivariable CpGs in CpG island “shores” (Supplemental Figures S5A and S5B). Similar results were also obtained using averaged data for various regulatory elements (promoters, non-promoter CpG islands, and CpG island shores). Figure 3C shows 748 epivariable regulatory elements, grouped both by sperm sample and by regulatory element, revealing an enrichment for CpG island shores. We have been unable to identify animal parameters (weight at sacrifice, etc.) that explain the clustering of sperm samples observed here, and it is of course possible that variation at these loci is stochastic and not environmentally-responsive. Together, these data reveal that epivariation, rather than diet, is the primary driver of variation in cytosine methylation between murine sperm samples, and identify a small number of genomic regions that exhibit unusually high levels of variation relative to the rest of the genome.
Extensive methylation variability at the rDNA repeats
Although most of the genomic loci that exhibit changes in methylation between sperm pools in our WGBS dataset are not covered by RRBS reads, a subset of the CpGs in the rDNA and the Ifnz clusters are represented in our RRBS dataset, and are among the most highly epivariable loci in this dataset (Figure 3C). These data, along with the failure of our low-coverage WGBS data to recapitulate dietary effects on these tandem repeats (Supplemental Figure S3), indicate that the apparent diet-related changes in methylation in our WGBS dataset are likely to result from high levels of epivariation at these loci that was not eliminated by our sample pooling strategy. To further explore dietary effects on cytosine methylation at epivariable loci, we carried out pyrosequencing to quantitatively assay cytosine methylation at the 45S rDNA loci across more than 200 sperm samples. We also measured cytosine methylation levels at additional loci that show apparent dietary effects in our WGBS dataset, and at a number of loci previously reported to exhibit nutrient-related changes in cytosine methylation (Wei et al., 2014), in smaller numbers of sperm samples. Pyrosequencing measurements were robust between individual bisulfite conversions of the same DNA (Supplemental Figure S6A), were correlated between sperm and testis samples from the same male (Supplemental Figure S6B), and were independently validated for a subset of animals using a methylation-sensitive restriction enzyme-based assay (not shown).
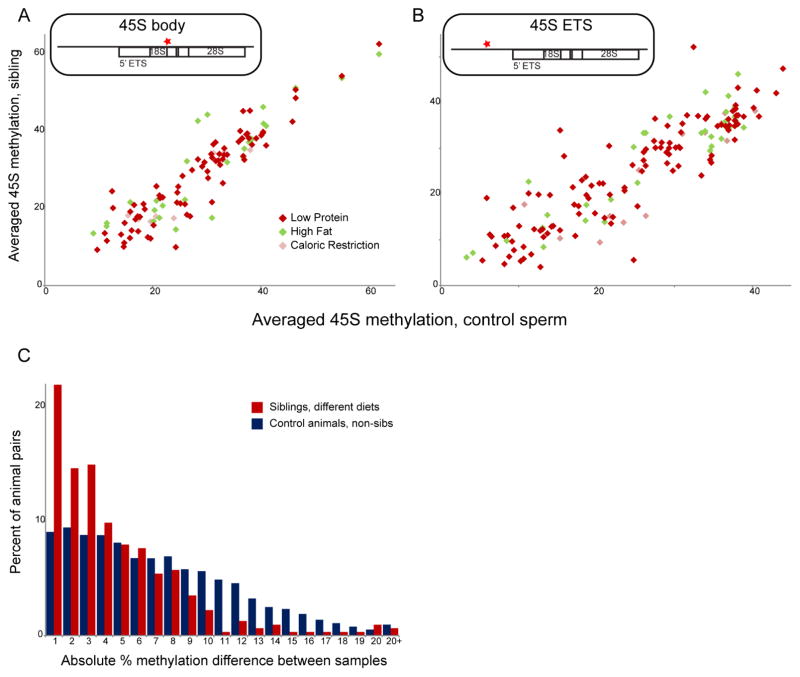
Bisulfite pyrosequencing of over 200 sperm samples (223 and 289 samples analyzed for 45S body and ETS regions, respectively) confirmed extensive epivariation at two locations within the 45S rDNA locus, with methylation levels at individual cytosines in the 45S spacer promoter varying from ~10% to ~60% between sperm samples (Figures 4A–B). Although methylation differed by as much as 20% between animals for many pairs of littermates on different diets, neither Low Protein nor High Fat diet showed consistent effects on methylation at this locus. As with other aspects of the sperm epigenome, variation between sibling animals, whatever their diet, was significantly lower than variation between non-sibling pairs of animals (Figure 4C). These data confirm that the apparent effect of diet on rDNA methylation in sperm observed in our WGBS dataset results from extensive animal-to-animal variation at this locus, which was not averaged out in our 8 animal pools. Moreover, we find that several other regions apparently subject to dietary influence on cytosine methylation from our WGBS dataset, or from prior studies on paternal prediabetes (Wei et al., 2014), similarly do not exhibit changes in cytosine methylation in response to Low Protein diet (Supplemental Figure S6C and not shown). As our mouse strain background and our dietary regimen differ from this prior report, this last finding does not necessarily dispute reported findings, but overall we conclude that few, if any, reproducible changes in cytosine methylation occur in sperm as a result of consumption of Low Protein or High Fat diets.
Figure 4. Lack of consistent dietary effects on rDNA methylation.
(A–B) Scatterplot comparing 45S methylation levels for Control animals (x axis) along with matched siblings raised on various diets (y axis). Schematic above each panel shows a red star indicating the location of pyrosequencing primer pair (primer pairs encompass 10 CpGs and 3 CpGs for (A) and (B), respectively). Although siblings on different diets exhibit up to ~20% changes in methylation at the 45S rDNA locus, such dietary effects are not observed consistently across paired sperm samples. (C) Absolute rDNA methylation differences plotted for pairs of sibling males (red bars), or for Control animals from different litters (blue bars). Note that Control-Control comparisons only include comparisons between C57 animals, or between FVB animals, given that C57 and FVB strains backgrounds exhibit consistent differences in rDNA copy number and methylation. See also Supplemental Figure S6.
Heritability of rDNA methylation
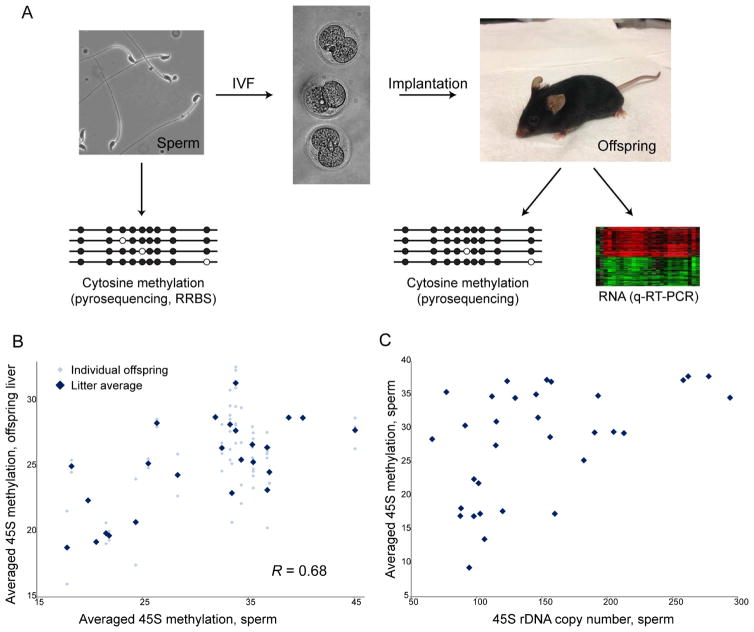
Although we do not find consistent dietary effects on sperm rDNA methylation, the differences in cytosine methylation at the rDNA repeats in sperm could nonetheless be epigenetically inherited from fathers to offspring, and could potentially influence offspring phenotype. We established a pipeline, based on in vitro fertilization (IVF), in which we could jointly characterize the epigenome in a sperm sample, and characterize the phenotype of offspring generated using a small aliquot of the very same sperm sample (Figure 5A). Because many paternal effect paradigms, including our system using paternal Low Protein diet, alter mRNA abundance for lipid metabolism genes in offspring livers (Carone et al., 2010; Radford et al., 2012), we characterized not just rDNA methylation but also Sqle expression in offspring livers. Across 25 sperm samples and 75 paired offspring, rDNA methylation levels in the sperm were significantly maintained in offspring livers (Figure 5B).
Figure 5. Linking the sperm epigenome to offspring phenotype.
(A) Schematic of system used to link the paternal sperm epigenome to offspring phenotype. For a given sperm sample, 5% was used to generate offspring via IVF and surgical implantation. Ninety-five percent of the sperm sample was used for analysis of cytosine methylation, and methylation and mRNA abundance data from matched offspring were obtained. Importantly, the very same sperm sample used to generate offspring was also used for molecular analysis. (B) Heritability of rDNA methylation patterns. 45S promoter methylation was analyzed by pyrosequencing for 25 sperm samples and 75 matched offspring livers. Data are shown for individual CpGs (circles) as well as averaged for the 3 CpGs. (C) Cytosine methylation at the rDNA repeats is correlated with rDNA copy number. X axis shows rDNA copy number in sperm samples as assayed by digital droplet PCR, with y axis showing average 45S methylation for the same sperm sample. See also Supplemental Figure S6.
The high heritability of cytosine methylation at the rDNA loci was surprising given the near-global erasure of paternal methylation patterns upon fertilization in mammals (Feng et al., 2010; Smith et al., 2012). As tandem repeat loci such as the rDNA loci are subject to relatively rapid copy number changes (Aldrich and Maggert, 2015; Paredes et al., 2011; Shiao et al., 2012), we hypothesized that the variation in cytosine methylation at the 45S locus described above might reflect a homeostatic response to rapid genetic variation in rDNA copy number. We used digital PCR to quantitate rDNA copy number in 33 sperm samples, finding a nearly 5-fold range in rDNA copy number between individual sperm samples (Figure 5C). Consistent with our hypothesis, rDNA methylation was positively correlated with rDNA copy number (Figure 5C), strongly suggesting that the apparent epigenetic inheritance of rDNA methylation levels is instead secondary to genetic inheritance of rDNA copy number.
DISCUSSION
In this study, we investigate dietary effects on the landscape of cytosine methylation in mature sperm in detail. We find no consistent effects of Low Protein or High Fat diet on the sperm methylome. Instead, a variety of repeat loci are subject to dramatic epivariation between siblings, and more detailed study of the rDNA loci reveals that this “epivariation” is linked to genetic variation in copy number at this locus. Interestingly, a recent report in flies linked male diet to rDNA copy number changes in offspring (Aldrich and Maggert, 2015), but here we find no consistent dietary effect on methylation or copy number of these loci in mice in response to two dietary challenges. Taken together, our results argue strongly that paternal dietary effects on offspring metabolism, at least in our system where diet is consumed from weaning onwards, are not mediated by cytosine methylation in sperm.
A wide variety of paternal exposures have been reported to influence offspring phenotype in mammals (Rando, 2012), but at present the mechanistic basis by which paternal information is passed to progeny remains unknown for all such paradigms. Although the paternal environment has been reported to cause changes in cytosine methylation in sperm in several paternal effect paradigms, in all reported cases the changes in cytosine methylation are relatively modest, with 10–20% changes in methylation at a handful of loci being typical. As each individual sperm carries a single haploid copy of the genome, such methylation changes reflect a change in the fraction of sperm which carry a methylated CpG at a given location – such population changes in “digital” sperm are unlikely to cause penetrant effects on offspring phenotype. This motivates a focus in this study on clusters of CpGs, which could potentially act in concert to exert penetrant “analog” effects on offspring phenotype.
However, we fail to identify any significant changes in the sperm methylome that could be confirmed in extensive follow-up studies, arguing against the hypothesis that the sperm methylome is responsible for paternal dietary effects on offspring metabolism, at least for Low Protein and High Fat diets. Instead, we confirm and extend prior findings (Carone et al., 2010) that “epivariation” between control animals that are not littermates is a far stronger influence over the sperm epigenome than is an animal’s diet. Epivariation is most notable over CpG island shores, and across tandem repeat regions, including rDNA. However, detailed follow-up at the rDNA loci in our system reveals that variability in cytosine methylation at these clusters is correlated with changes in rDNA copy number, strongly suggesting that rDNA methylation differences between animals reflect homeostasis of rRNA production. This last result almost certainly explains the strong inheritance of methylation levels at the 45S locus (Figure 5), with genetic inheritance of a relatively labile tandem repeat resulting in apparent inheritance of methylation levels.
Interestingly, rDNA copy number and cytosine methylation in paternal sperm were anti-correlated with Sqle expression in offspring (Supplemental Figures S6D–E). This is consistent with prior observations of widespread gene regulatory consequences of rDNA copy number in organisms such as flies (Paredes et al., 2011) and humans (Gibbons et al., 2014). However, as detailed above, rDNA copy number and cytosine methylation are not consistently altered by diet. Thus, we conclude that random or unexplained variation in the rDNA locus explains a subset of the variation in cholesterol metabolism that is observed among inbred nearly-isogenic animals, but is not responsible for the effects of paternal diet on metabolism. Given that offspring metabolism is altered in response to paternal diets, paternal stress, maternal diets and stress, and even brief embryo culture (Rando and Simmons, 2015), it will be interesting to determine where and how these ancestral inputs converge during development. Ongoing studies on paternal dietary effects are focused on chromatin packaging, RNAs, and DNA-binding proteins as potential carriers of paternal dietary information.
EXPERIMENTAL METHODS
Animal husbandry
Mice used in this study included C57Bl6/J and FVB/NJ strains from Jackson Laboratories. Animals were maintained on-site in accordance with approved IACUC protocols.
Dietary regimens
The 61 animals in the epivariation dataset included animals consuming standard laboratory chow, a defined Control diet (Bioserv AIN-93g), a Low Protein diet based on AIN-93g (10% of protein rather than 19%, remaining mass made up with sucrose), a High Fat diet 60% Fat based on Ain-93g (Bioserv S3282), as well as animals provided with nicotine hydrogen tartrate (200 μg/mL nicotine, reported as free base) in drinking water sweetened with 2% saccharine to increase palatability, or animals provided with tartaric acid and saccharine water alone. Animals were placed on diet at weaning (21 days) until mating or sacrifice (10–12 weeks).
Isolation of epididymal sperm DNA
Animals were sacrificed by isoflurane administration followed by cervical dislocation. For sperm isolation, cauda epididymis and vas deferens was rapidly dissected and punctured, and was incubated in 500 μL of human tubule fluid (HTF – Millipore MR-070-D) at 37 C for 30 minutes. Supernatant was removed, sperm were pelleted at 2000 g for 5 minutes, washed once with 1 mL water and then pelleted. Sperm were washed again with 1 mL PBS, and pelleted again. Sperm were resuspended in 400 μL DNA Lysis Buffer (10 mM Tris, 5 mM EDTA, 0.5% SDS, and 200 mM NaCl) with 10mM DTT, and incubated at 37ºC for 30 minutes. Sperm were subjected to needle homogenization. 20 μg/mL Proteinase K was added to the homogenate and incubated at 55ºC for 16 hours. DNA was extracted with Phenol:Chloroform:Isoamyl Alcohol and precipitated with 100% EtOH.
In vitro fertilization
In vitro fertilization was performed according to “Manipulating the Mouse Embryo” Second Edition (Nagy, 2003). FVB/NJ mice were used as oocyte donors, and sperm was isolated from males fed dietary regimes as above. Fertilization took place in 250 μL HTF media covered in mineral oil, pre-gassed in 5% CO2 at 37ºC. Swiss Webster Females between 25 and 35 grams were used as 2-cell stage embryo recipients via unilateral oviduct transfer.
Reduced Representation Bisulfite Sequencing (RRBS)
RRBS was carried out as previously described (Boyle et al., 2012). Briefly, genomic DNA was digested with MspI, ends were filled-in, and fragments were A-tailed. DNA fragments were ligated to methylated barcoded adaptors. DNA was subjected to bisulfite conversion and PCR amplified. Cleanup and size selection were performed with SPRI AMPure XP beads.
Whole Genome Bisulfite Sequencing (WGBS)
Control and Low Protein WGBS pools were generated from 8 paired animal samples in which one sibling was weaned to Control diet and the other sibling was weaned to Low Protein diet (Table S1). Control/High Fat pools were generated using a similar approach using 7 animal pairs. For each pool, 1 μg of genomic DNA was contributed by each animal. DNA was sheared to an average length of ~100–500 bp with a Covaris sonicator, fragment ends were cleaned up and A-tailed. Fragments were ligated to pre-methylated Illumina paired-end adaptors, bisulfite converted, and PCR-amplified. Libraries were subjected to paired-end 50 bp sequencing on Illumina HiSeq sequencers, yielding an average of 1.4 billion reads and 140 billion base pairs of sequence.
In addition, 12 sperm samples from individual males were subject to WGBS using the TruSeq DNA Methylation kit (Illumina) according to the manufacturer’s instructions. To allow for multiplexed sequencing of libraries, individual libraries were barcoded using the TruSeq DNA Methylation Index PCR Primers (IIlumina). Size selection and DNA purification was performed using Ampure XP beads (Agencourt) according to the TruSeq DNA Methylation kit protocol. Library quantity was determined using the Library Quantification kit (Kapa Biosystems). Paired-end sequencing was performed on an Illumina NextSeq500 system in a 2 x 75 bp run with 30% PhiX Control (Illumina). Individual samples were sequenced to an average depth of ~140M reads (21 billion bp, ~7X coverage) per sample. Methylation data for most samples were moderately well-correlated (r~0.75 or greater), except for outlier sample “43HF” (r<0.5 with most samples), which was therefore not included in downstream analyses
Data Processing and Analysis
Technical replicas were merged together for the pooled WGBS dataset. Data were mapped against mm9 mouse genome with bsmap software v2.73 (Xi and Li, 2009). Default parameters were used for error rate and maximum number of equal best hits was selected as default as well. For the majority of analyses, if more than one read pair had the same sequence as other read pairs in the same sample dataset, only one of the identical read pairs was used for mapping and further analysis. Bsmap was also used to perform methylation calls. To get a methylation level for a given CpG, information from C from both strands was combined together. Differentially-methylated CpGs were discovered with the methylKit R package (Akalin et al., 2012). Discovered CpGs were merged using tiling arrays with 300nt windows to calculate p-values, q-values, and fold enrichment factors for High Fat and its Control, Low Protein and its Control. The data can be found in Table S2.
Data for the 12 individual sperm samples were processed similarly. methylKit was used to compute the fraction of methylated CpGs in each 300 base window across the genome. For the heatmap in Supplemental Figure S3C, tiles were filtered by requiring a mean CpG coverage of 5 reads (as recommended by Ziller et al).
Pyrosequencing
Cytosine methylation data for individual loci were generated using a Qiagen Pyromark Q24 pyrosequencer. Genomic DNA was bisulfite converted, and loci to be analyzed were amplified by PCR – primers are listed in supplementary materials. Amplified DNA was cleaned up and analyzed using the manufacturer’s protocol for the Pyromark Q24.
rDNA copy number analysis
Digital droplet PCR was used to measure rDNA copy number, performed according to the manufacturer’s protocol (Bio-Rad). Briefly, purified sperm DNA was digested with the restriction endonuclease DpnI. Reaction mixtures were made with target copy number variable sequence (for example, 45S rDNA) and internal control (Gapdh). Droplet generation was performed, followed by endpoint PCR. Droplets were read by QX200 droplet reader, and quantitation was performed using Quantasoft software. DdPCR data were well-correlated with lower precision rDNA copy numbers as measured by q-PCR (not shown).
Supplementary Material
Research Highlights.
Whole genome nucleotide resolution analyses of dietary effects on sperm methylation
There is no consistent effects of paternal diet on sperm cytosine methylation pattern
Epivariation between animals influence methylation pattern differences over diet
Identification of genetic and epigenetic variability over tandem repeats such as rDNAs
Acknowledgments
We would like to thank S. Jones and J. Gallant for training BC and JMS in in vitro fertilization. AM is supported by NIH grants R01HD078679 and R01DA036898 and the New York Stem Cell Foundation. AM Is a New York Stem Cell Foundation Robertson Investigator. OJR is supported by NIH grants R01HD080224, DP1ES025458, R01DA033664, and March of Dimes grant FY13-1268. ART and PDG are supported by NIH grant R01DA033664.
Footnotes
OJR, JMS, RWS, and BRC designed the experiments. JMS and BRC carried out animal husbandry and in vitro fertilization experiments. JMS and RS carried out q-PCR, q-RT-PCR, and pyrosequencing. MV, PDG, and ART carried out nicotine-related animal husbandry. RWS, MJZ, HG, and AM carried out RRBS and WGBS library construction. HPS, AK, MG, and OJR analyzed the data. OJR and JMS wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich JC, Maggert KA. Transgenerational inheritance of diet-induced genome rearrangements in Drosophila. PLoS genetics. 2015;11:e1005148. doi: 10.1371/journal.pgen.1005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, Alvord WG. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition. 2006;22:327–331. doi: 10.1016/j.nut.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature biotechnology. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, Holmes L, Meldrim J, Kelley F, Gnirke A, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome biology. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annual review of biochemistry. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature reviews Genetics. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science (New York, N Y. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Yu S, Lemos B. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nature communications. 2014;5:4850. doi: 10.1038/ncomms5850. [DOI] [PubMed] [Google Scholar]
- Harris A, Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Horm Behav. 2010;59:279–289. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature genetics. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, et al. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sloboda DM, Vickers MH. Maternal obesity and developmental programming of metabolic disorders in offspring: evidence from animal models. Exp Diabetes Res. 2011;2011:592408. doi: 10.1155/2011/592408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey L, Stein AD, Kahn HS, van der Pal-de Bruin KM, Blauw G, Zybert PA, Susser ES. Cohort Profile: the Dutch Hunger Winter Families Study. Int J Epidemiol. 2007 doi: 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo : a laboratory manual. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS genetics. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, et al. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS genetics. 2012;8:e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, et al. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science (New York, NY. 2014 doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Simmons RA. I’m Eating for Two: Parental Dietary Effects on Offspring Metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science (New York, N Y. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao YH, Crawford EB, Anderson LM, Patel P, Ko K. Allele-specific germ cell epimutation in the spacer promoter of the 45S ribosomal RNA gene after Cr(III) exposure. Toxicol Appl Pharmacol. 2005;205:290–296. doi: 10.1016/j.taap.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Shiao YH, Leighty RM, Wang C, Ge X, Crawford EB, Spurrier JM, McCann SD, Fields JR, Fornwald L, Riffle L, et al. Molecular and organismal changes in offspring of male mice treated with chemical stressors. Environ Mol Mutagen. 2012;53:392–407. doi: 10.1002/em.21701. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and cellular biology. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AJ, Sinclair KD. Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. American journal of physiology Heart and circulatory physiology. 2014;306:H1444–1452. doi: 10.1152/ajpheart.00981.2013. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nature methods. 2015;12:230–232. 231. doi: 10.1038/nmeth.3152. following 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.