Summary
F-BAR proteins function in diverse cellular processes by linking membranes to the actin cytoskeleton. Through oligomerization, multiple F-BAR domains can bend membranes into tubules, though the physiological importance of F-BAR-to-F-BAR assemblies is not yet known. Here we investigate the F-BAR domain of the essential cytokinetic scaffold, Schizosaccharomyces pombe Cdc15, during cytokinesis. Challenging a widely held view that membrane deformation is a fundamental property of F-BARs, we report that the Cdc15 F-BAR binds but does not deform membranes in vivo or in vitro, and six human F-BAR domains including those from Fer and RhoGAP4 share this property. Nevertheless, tip-to-tip interactions between F-BAR dimers are critical for Cdc15 oligomerization and high avidity membrane binding, stabilization of contractile ring components at the medial cortex, and the fidelity of cytokinesis. F-BAR oligomerization is also critical for Fer and RhoGAP4 physiological function, demonstrating its broad importance to F-BAR proteins that function without membrane bending.
Graphical Abstract
Introduction
Diverse membrane remodelers and scaffolds interact with and modulate the organization of biological membranes during many cellular processes including cell division, endocytosis, and motility (Bezanilla et al., 2015) . The Bin/Amphiphysin/Rvs (BAR) domain superfamily is a central player in many of these processes (Frost et al., 2009). Structural studies have defined the family as crescent-shaped dimers of anti-parallel alpha-helix bundles (Peter et al., 2004). Several families exist within the BAR domain superfamily: classical BAR domains, inverse BAR domains (I-BAR), and Fer/CIP4 homology BAR (FCH-BAR, F-BAR) domains, each of which exhibits structural variation on the canonical crescent theme (Qualmann et al., 2011). F-BAR domains in particular are flatter elongated versions of classical BAR domains (Henne et al., 2007; Shimada et al., 2007) involved in many actin-driven processes (Roberts-Galbraith and Gould, 2010).
Multiple BAR and F-BAR domains possess the ability to deform membranes at high concentration (Itoh et al., 2005; Peter et al., 2004; Tsujita et al., 2006). BAR domains do this by binding and imposing their intrinsic curvature on membranes, in some cases inserting amphipathic helices into the bilayer (Gallop et al., 2006; Masuda et al., 2006) or collaborating with adjacent membrane binding domains (Pang et al., 2014). In contrast, F-BAR domain tubulation is thought to arise from a cooperative mechanism wherein assemblies of shallowly curved F-BARs bound to a membrane collectively impose a curvature (Frost et al., 2008; Shimada et al., 2007; Yu and Schulten, 2013). A pioneering structural study of CIP4 F-BAR-coated membrane tubules revealed numerous interactions between F-BAR dimers; lateral interactions between dimers were strictly essential for tubule formation, while tip-to-tip interactions stabilized the tilt of F-BAR assemblies to result in a consistent tubule diameter (Frost et al., 2008).
Despite this detailed example and the tubulation model that has arisen from it (Frost et al., 2009; Mim and Unger, 2012; Qualmann et al., 2011; Suetsugu et al., 2009), it remains unknown if contacts between F-BAR dimers are important for CIP4’s in vivo functions. In fact, while the ability of multiple F-BAR domains to tubulate membranes has been noted since the earliest characterization of the domain (Itoh et al., 2005; Kamioka et al., 2004; Shimada et al., 2007; Tsujita et al., 2006), the connection between tubulation activity in vitro and protein function in vivo is not clearly established. A straightforward example is F-BAR domain function during endocytosis, where distinct membrane curvatures are generated and resolved as a vesicle is nucleated and budded from the plasma membrane (Doherty and McMahon, 2009). It was initially hypothesized that F-BAR domains would bind to or induce distinct membrane curvatures throughout this process (possibly predictable from their domain structure) (Qualmann et al., 2011). However, the membrane curvatures induced by endocytic F-BARs in vitro fail to strictly correlate with endocytic vesicle size at the time of F-BAR protein association (Taylor et al., 2011). In other processes requiring F-BAR proteins like cytokinesis, it is unclear how thin membrane tubules might contribute. Despite inconclusive experimental evidence linking in vitro tubulation activity to physiological function, the importance of F-BAR domain oligomerization and membrane tubulation has become dogma in the field (Daumke et al., 2014; Frost et al., 2009; Mim and Unger, 2012; Qualmann et al., 2011). In fact, it is unknown if all F-BAR domains oligomerize, if contacts between dimers are important for physiological F-BAR protein functions, and if membrane tubulation or deformation is a default consequence of these interactions.
To test the importance of inter-F-BAR domain interactions and F-BAR domain-induced tubulation in a cellular process, we studied Schizosaccharomyces pombe Cdc15, the founding member of the Pombe Cdc15 Homology (PCH) scaffolding family, a large subfamily of F-BAR domain containing proteins (Roberts-Galbraith and Gould, 2010). Cdc15 is essential for cytokinesis in S. pombe, directly binding the membrane and scaffolding multiple contractile ring (CR) proteins to ensure proper CR formation and stability (Carnahan and Gould, 2003; Fankhauser et al., 1995; Hachet and Simanis, 2008; Ren et al., 2015; Roberts-Galbraith et al., 2009; Wachtler et al., 2006; Willet et al., 2015). Indeed, Cdc15 is proposed to be a major membrane tether for the CR in both S. pombe and S. japonicas (Gu et al., 2015; Roberts-Galbraith et al., 2010). Here, we find that the Cdc15 F-BAR domain binds but does not bend or tubulate membranes in vitro or in vivo, a characteristic we extend to six human F-BAR domains. Nevertheless, Cdc15 utilizes contacts between F-BAR dimers to organize into linear oligomers. We reveal the structural basis for oligomerization and membrane binding of Cdc15, and determine the importance of these functions in vivo using endogenous oligomerization and membrane binding mutants. As further examples, we show F-BAR domain oligomerization interactions are critical for in vivo functions of the tyrosine kinase regulator of cell motility and adhesion, Fer (Greer, 2002; Itoh et al., 2009), and RhoGAP4, an inhibitor of cell motility (Vogt et al., 2007). Our results provide the first evidence for the importance of interactions between F-BAR dimers for in vivo function of F-BAR proteins in the absence of membrane tubulation.
Results
The Cdc15 F-BAR domain binds membranes and is essential for function
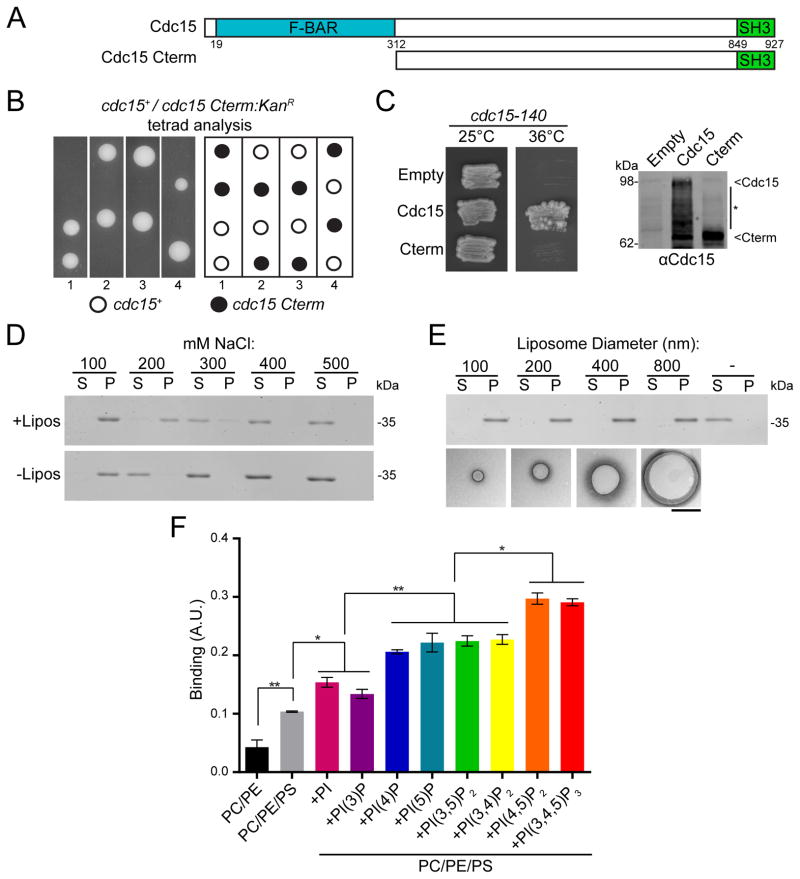
Although Cdc15 is the founding member of the PCH family and is essential for cytokinesis, the importance of its F-BAR domain has not been tested. Thus, we replaced one allele of cdc15 in a diploid with a version lacking sequences encoding the F-BAR domain (Figure 1A). Tetrad dissection of the resultant heterozygous diploid showed that the truncation allele did not give rise to viable progeny (Figure 1B). Furthermore, a construct lacking the F-BAR domain did not rescue a cdc15-140 strain at the restrictive temperature (Figure 1C). Together, these data indicate that the F-BAR domain is essential for Cdc15 function.
Figure 1. The Cdc15 F-BAR domain binds membranes and is essential for function.
A) Cdc15 domains and alleles. B) Cdc15 alleles from (A) integrated into cdc15/cdc15::ura4+ diploids were sporulated and tetrads were dissected. Relevant genotypes are indicated. C) Cdc15 constructs from (A) were expressed from the nmt81 promoter in a cdc15-140 strain. Right) Anti-Cdc15 immunoblot of construct-expressing cells. D) Co-pelleting between the Cdc15 F-BAR domain (19-312) and Folch fraction liposomes at different NaCl concentrations. S = supernatant, unbound; P = pellet, bound. E) Co-pelleting between the Cdc15 F-BAR domain and Folch fraction liposomes extruded to various diameters. Representative negative stain electron micrographs of liposomes are shown. Scale bar = 500 nm. F) Bio-Layer Interferometry binding assay between the Cdc15 F-BAR domain and synthetic liposomes composed of 69.9% DOPC, 0.1% Biotin-PE, 10% DOPS when indicated (PS), 5% of different phosphatidylinositols when indicated, and the remainder DOPE. *, p<0.05; **, p<0.01, one-way ANOVA. Error bars indicate standard error of the mean (SEM).
We next tested the membrane binding properties of the Cdc15 F-BAR domain (residues 19-312). We found that it pelleted with liposomes composed of Folch fraction lipids rich in phosphorylated phosphatidylinositols (PIPs) in a salt-dependent manner (Figure 1D), without any dependence on liposome size (Figure 1E). At low salt, the domain pelleted by itself, a behavior that will be explained below. Using a Bio-Layer Interferometry (BLI) binding assay (Abdiche et al., 2008), the lipid specificity of the Cdc15 F-BAR was tested (Figure 1F). Liposomes enriched in phosphatidylserine (PS) and PIP/PIP2 increased Cdc15 binding, and Cdc15 particularly favored liposomes containing PIPs phosphorylated at the 4- and 5- positions of the inositol ring.
Cdc15 represents a class of F-BARs that do not bend or tubulate membranes
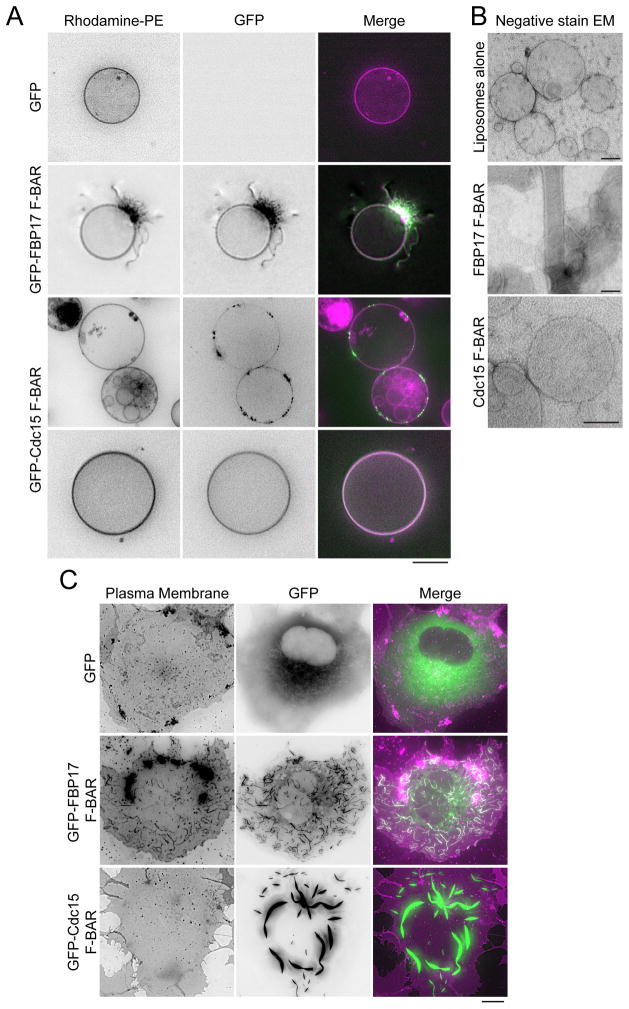
We used three assays to determine if the Cdc15 F-BAR domain is capable of remodeling or tubulating the membrane when present at high concentrations. First, GFP-Cdc15 F-BAR was mixed with giant unilamellar vesicles (GUVs) composed of a generic blend of lipids including negatively charged PS and PI(4)P (Figure 2A). The Cdc15 F-BAR bound and coated the GUVs, often clustering on the surface. However, unlike the previously characterized human FBP17 F-BAR domain (Frost et al., 2008; Tsujita et al., 2006), which formed tubules emanating from the GUVs, the Cdc15 F-BAR did not deform GUVs even when coating the surface at high concentration (Figure 2A, bottom panel; Figure S1). A similar binding experiment was performed with small (800 nm) liposomes and membrane deformation was examined by negative stain electron microscopy (EM). As with GUVs, tubules were observed when the FBP17 F-BAR domain was added, but not when Cdc15 was (Figure 2B). Third, whereas overexpression of the GFP-FBP17 F-BAR domain in mammalian COS-7 cells induced many short plasma membrane tubules as previously described (Kamioka et al., 2004) (Figure 2C), overexpression of the GFP-Cdc15 F-BAR domain did not tubulate the membrane and instead concentrated in ribbon-like clusters. Taken together, these data indicate that the Cdc15 F-BAR domain binds membranes but does not deform or tubulate them.
Figure 2. The Cdc15 F-BAR domain binds but does not bend or tubulate membranes.
A) 10–30 μm GUVs (composed of 69% DOPC / 15% DOPE / 10% DOPS / 5% PI(4)P / 1% Rhodamine-PE) mixed with 10 μM GFP, GFP-FBP17 (1-319), or GFP-Cdc15 (19-312) F-BAR constructs. Scale bar = 10 μm. B) 800nm extruded liposomes (composed of 70% DOPC / 15% DOPE / 10% DOPS / 5% PI(4)P) mixed with 10 μM of the indicated F-BAR domain and examined with negative stain EM. Scale bars = 100nm. C) GFP, GFP-FBP17 (1-319), or GFP-Cdc15(19-312) expressed in COS-7 cells and co-stained with CellMask Orange plasma membrane dye. Scale bar = 10 μm.
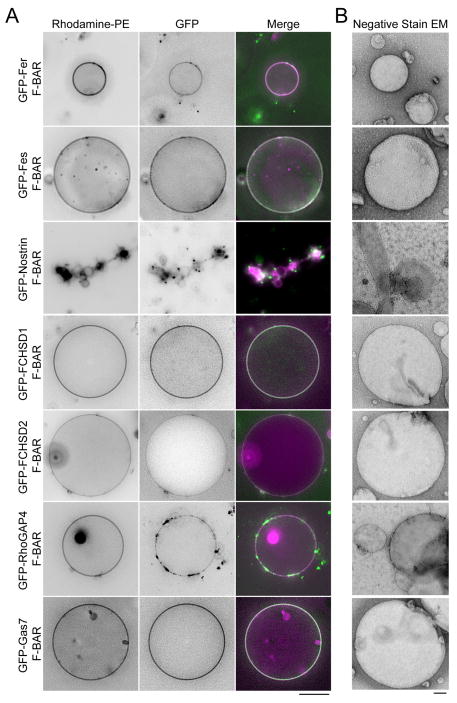
Lack of membrane deformation in cell culture overexpression assays has been reported previously only for human Fer (Tsujita et al., 2006) and S. cerevisiae Hof1 F-BAR domains (Moravcevic et al., 2015). Out of 22 total F-BAR proteins in humans, 15 have been shown to tubulate membranes with either positive (CIP4, TOCA1, FBP17, PSTPIP1/2, FCHO1/2, PACSIN1/2/3) or negative (SRGAP1/2A/2B/2C/3) curvature in vitro and/or upon overexpression in vivo (Table S1). To determine the total number of human F-BAR domains that do not deform membranes, we tested the remaining seven (Fer, Fes, Nostrin, FCHSD1/2, Gas7, RhoGAP4) in vitro with GUVs and smaller liposomes, and in vivo by expression in COS-7 cells (Figures 3 and S1–S2; Table S1). The Nostrin F-BAR tubulated membranes in all assays, but the six other F-BAR domains did not tubulate membranes in any assay (Figure 3A–B, S2) although Fer and RhoGAP4, like Cdc15, clustered on the GUVs’ surface (Figure 3A). The assemblies formed by the 6 non-tubulating F-BARs in COS-7 cells were dynamic as determined by Fluorescence Recovery after Photobleaching (FRAP) (Figure S3A). Additional evidence that they fold correctly in vitro and in vivo was obtained by circular dichroism, wherein each GFP-F-BAR displayed a strong near-UV signal above that of GFP alone, indicative of folded molecules (Figure S3B). We conclude that a significant number of F-BAR domains bind but do not bend membranes.
Figure 3. A significant fraction of human F-BAR domains do not bend membranes.
A) 10–30 μm GUVs (composed of 69% DOPC / 15% DOPE / 10% DOPS / 5% PI(4)P / 1% Rhodamine-PE) mixed with 10 μM of the indicated GFP-F-BAR domain constructs. Scale bar = 10 μm. B) 800nm extruded liposomes (composed of 70% DOPC / 15% DOPE / 10% DOPS / 5% PI(4)P) mixed with 10 μM of the indicated F-BAR domain and examined with negative stain EM. Scale bar = 100nm. See also Figures S1–3.
The Cdc15 F-BAR oligomerizes in a tip-to-tip manner
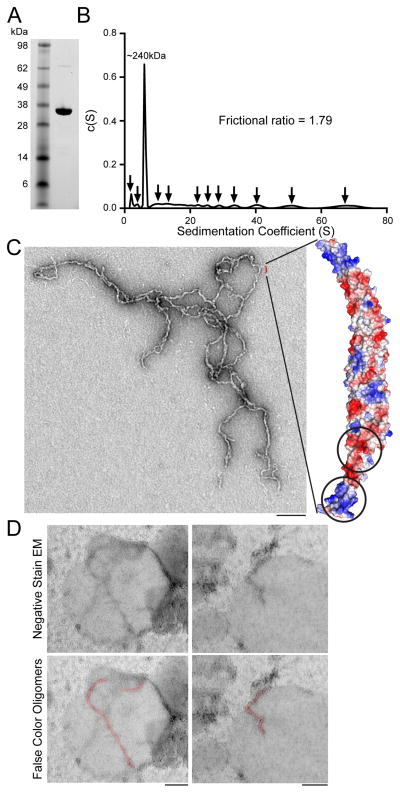
In membrane-tubulating F-BAR domains, interactions between adjacent F-BAR dimers, as identified in CIP4, are important for building the assemblies necessary to collectively bend membranes (Frost et al., 2008). The Cdc15 F-BAR domain forms oligomers (Roberts-Galbraith et al., 2010), suggesting similar interactions may exist despite a lack of tubulation activity. Indeed, as salt concentration was lowered, the Cdc15 F-BAR (Figure 4A) formed multiple oligomeric species when assayed by sedimentation velocity analytical ultracentrifugation (AU) (Figure 4B). A prominent peak comprising ~40% of the sample was present at ~240 kDa, corresponding to an assembly of four F-BAR dimers, while the remaining protein formed a heterogeneous mix of smaller and larger species (Figure 4B, arrows). The frictional ratio of the sample was 1.79, indicating a non-spherical shape of complexes in solution and negative stain EM of the sample revealed thin, linear oligomers (Figure 4C, left). These oligomers appeared to exist primarily via tip-to-tip interactions between F-BAR domain wings, as minimal branching and no sheet-like structures were observed (beyond the expected overlap from settling onto a 2D EM grid). The formation of these oligomers explains the F-BAR domain’s sedimentation in the absence of liposomes at low salt concentrations (Figure 1D, 100 mM NaCl), and suggests a mechanism for Cdc15 F-BAR domain clustering on GUVs (Figure 2A). Cdc15 F-BAR oligomers were also visualized bound to liposomes by EM (Figure 4D). We also investigated the six human non-tubulating F-BAR domains for evidence of oligomerization in solution using AU (Figure S4). Each of these F-BAR domains exhibited oligomeric species of two (Fer) or more (Fes, FCHSD1, FCHSD2, RhoGAP4, Gas7) dimers in 50 mM NaCl.
Figure 4. The Cdc15 F-BAR domain organizes into tip-to-tip oligomers.
A) Coomassie stained SDS-PAGE of purified recombinant Cdc15 F-BAR domain (19-312). B) Analytical ultracentrifugation trace of the Cdc15 F-BAR domain in 50 mM NaCl. Multiple oligomer species are indicated by arrows. C) Negative stain electron micrograph of the Cdc15 F-BAR domain at 50 mM NaCl with electrostatic potential map of a Phyre2 structural homology model for scale. Putative patches interacting during oligomerization are circled. D) Cdc15 F-BAR domain mixed with liposomes and examined by negative stain EM. Bottom panels include false color highlighting of observable oligomers. All scale bars = 100 nm. See also Figure S4.
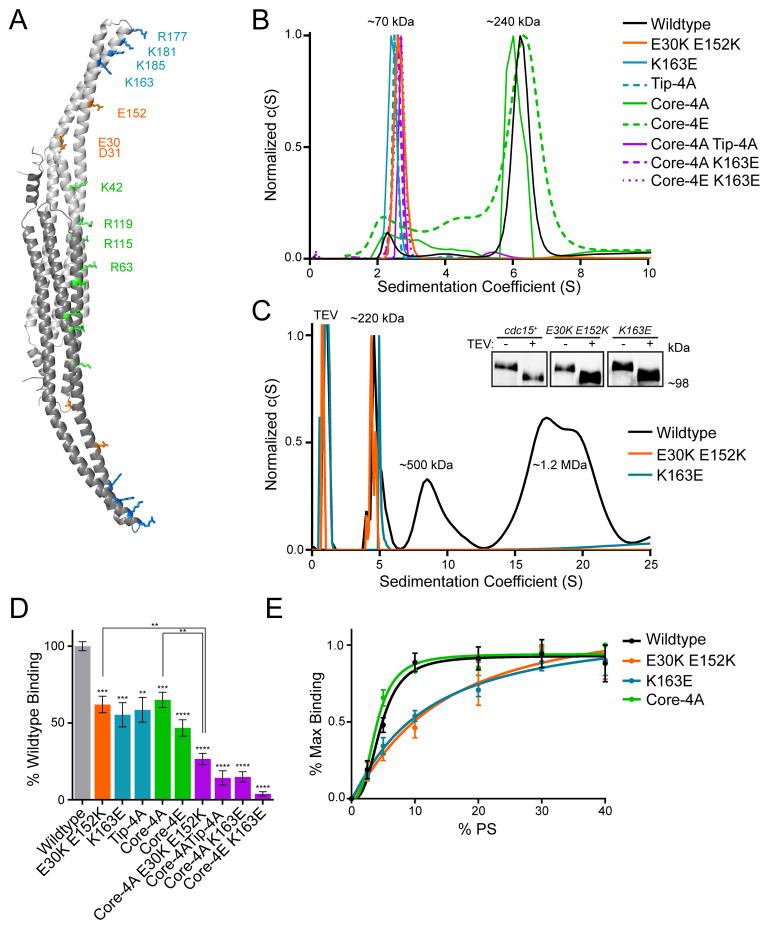
To understand the structural basis for Cdc15’s F-BAR domain oligomerization, we generated a model of the domain with the Protein Homology/analogY Recognition Engine v2.0 (Phyre2) (Kelley and Sternberg, 2009) using the homologous S. cerevisiae Hof1 F-BAR domain as a template structure, as Hof1 is the only non-tubulating F-BAR domain whose structure is available (Lippincott and Li, 1998; Moravcevic et al., 2015) (Figure 4C, right). Based on the observed dependence of oligomerization on ionic strength and the appearance of the linear oligomer by EM, we reasoned that charged residues located on the sides of the F-BAR domain’s wing tips might mediate F-BAR-to-F-BAR interaction. In an electrostatic potential map of the Cdc15 model, a prominent positively charged patch exists at the tip of the domain, wrapping around the concave face to one side, while a negatively charged cleft is located proximally on the wing (Figure 4C, circled on right). We hypothesized that interaction between these two patches could support oligomerization.
Structural basis for Cdc15’s F-BAR domain oligomerization and membrane binding
To test this oligomerization model, we identified residues located in the positive (K163, R177, K181, and K185) and negative (E30, D31, and E152) patches on each F-BAR domain tip and wing, respectively (Figure 5A). Charge reversal mutations in the identified wing (E30K E152K) and tip (K163E) residues or a Tip-4A mutation (K163A, R177A, K181A, and K185A) abolished oligomerization of the F-BAR domain in vitro (Figure 5B and S5A). To ensure these mutations eliminated oligomerization in vivo as well, we purified the double E30K,E152K and single K163E mutants from S. pombe, and confirmed loss of oligomerization by AU (Figure 5C) (Roberts-Galbraith et al., 2010). These two mutations also eliminated the clustering of the Cdc15 F-BAR domain when overexpressed in COS-7 cells (compare Figure 2C and S5C). That disruption of either of these two oppositely charged patches abolished oligomerization supports our reciprocal electrostatic interaction model of Cdc15 oligomerization.
Figure 5. Structural basis for Cdc15 F-BAR domain oligomerization and membrane binding.
A) Phyre2 homology model of the Cdc15 F-BAR (based on Hof1, PDB: 4WPE) with dimer subunits colored in dark and light grey. Putative charged residues involved in either oligomerization (blue and orange) or membrane binding (green and blue) are labeled. B) Analytical ultracentrifugation traces of Cdc15 F-BAR domain mutants. C) Analytical ultracentrifugation trace of purified full length Cdc15 and mutants purified from S. pombe. A TEV contaminant from the purification is indicated. Inset) Anti-Cdc15 Western blot of purified Cdc15-TEV-2xProtA and mutants before and after TEV cleavage. D) Bio-Layer Interferometry binding assay between 1 μM Cdc15 F-BAR domain mutants and 100 nm liposomes composed of 69.9% DOPC / 15% DOPE / 10% DOPS / 5% PI(4)P / 0.1% Biotin-PE. **, p<0.01; ***, p<0.001; ****, p<0.0001, one-way ANOVA. E) Normalized saturation binding curves of Cdc15 and mutant F-BAR domains to liposomes composed of 50% DOPC, increasing amounts of DOPS, and the remainder DOPE. Tip-4A consists of K163A, R177A, K181A, K185A while Core-4A/E consist of K42A/E, R63A/E, R115A/E, R119A/E. All error bars indicate SEM. See also Figure S5.
We next determined the mechanism of Cdc15 F-BAR domain membrane binding and the interplay between this function and oligomerization. By analogy to other F-BAR domains, we reasoned that positively charged residues in the concave central core region (K42, R63, R115, and R119) might mediate membrane binding (Figure 5A, green residues). As predicted, mutation of these residues to alanine or glutamate (Core-4A and Core-4E) reduced binding of the F-BAR to liposomes in vitro (Figure 5D). However, membrane binding was not abolished, indicating the involvement of additional residues. Interestingly, mutations of either the positive or negatively charged tip patches that mediate oligomerization also decreased binding to membranes (Figure 5D and S5B). Since mutation of either positively (K163E) or negatively charged (E30K E152K) residues resulted in a similar decrease in equilibrium binding, this impairment likely arises from loss of oligomerization and a consequent decrease in the avidity of the Cdc15 F-BAR domain for the membrane. Supporting this idea, combining a strictly membrane binding mutation (Core-4A) with the acidic oligomerization mutant (E30K E152K) decreased equilibrium binding below the level of a Core-4A mutation alone (Figure 5D). Furthermore, oligomerization mutants displayed a loss of cooperative binding to membranes, with Hill coefficients of 1.1 ± 0.3 (K163E) and 1.2 ± 0.4 (E30K E152K) compared to 2.9 ± 0.8 for wildtype (Figure 5E).
Since Core-4A and Core-4E mutants retained some membrane binding, we tested whether the basic residues at the F-BAR domain tips contributed both to oligomerization and also directly to membrane binding. Combination mutations (Core-4A Tip-4A, Core-4A K163E, and Core-4E K163E) decreased binding to very low levels, indicating that the residual membrane binding of core mutants is mediated by positive residues at the tips (Figure 5D). The positively charged patch, which extends from the concave face of wing tips around the sides of the wing, therefore, appears to have a dual function of binding membranes and mediating oligomerization.
Cdc15 oligomerization defects impair cytokinesis
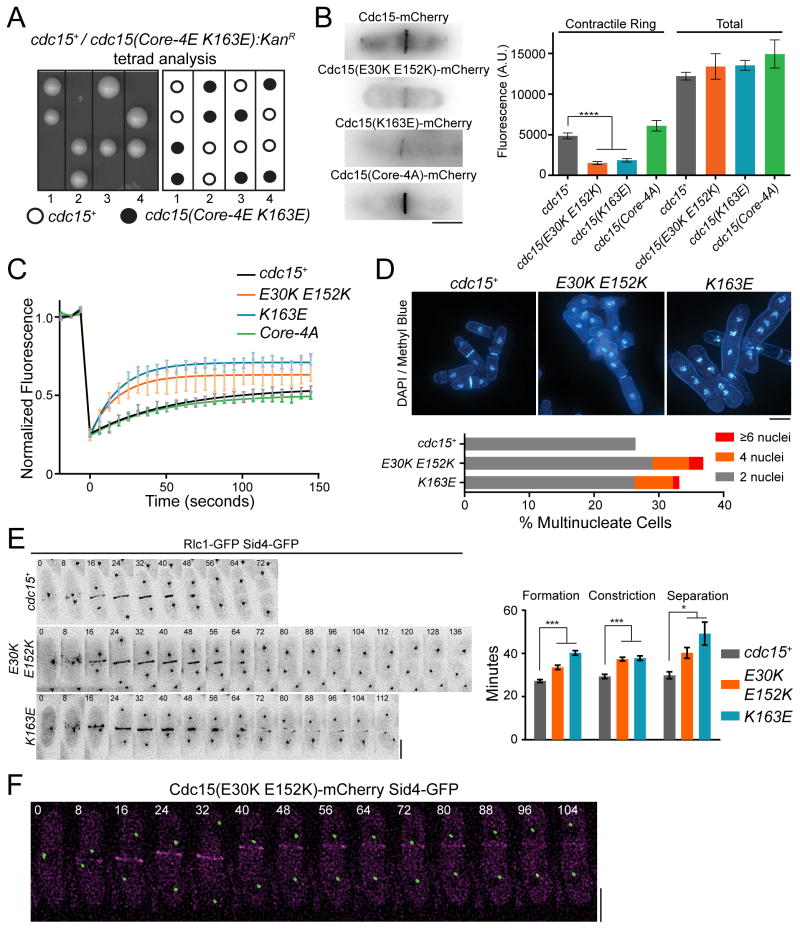
Next, we investigated the function of the Cdc15 F-BAR domain’s membrane binding and oligomerization activities in vivo by integrating mutants compromising one or both functions into Cdc15’s endogenous locus. The most defective membrane-binding mutant (Core-4E K163E) was inviable (Figure 6A), indicating that membrane binding is strictly required for Cdc15’s function. Although we were able to recover other strains producing Cdc15 mutants strongly deficient in membrane binding (e.g. Core-4A Tip-4A), they failed cytokinesis regularly and became multinucleate (Figures S6A–B), supporting the conclusion that membrane binding is an essential Cdc15 function.
Figure 6. Oligomerization is critical for a stable Cdc15 scaffold at the division site and robust cytokinesis.
A) A complete membrane binding Cdc15 mutant is inviable. The Cdc15(Core-4E K163E) mutation (Figure 5) was integrated into a cdc15/cdc15::ura4+ diploid, sporulated, and tetrads were dissected. Relevant genotypes are indicated. B) Comparison of cdc15(E30K E152K)-mCherry, cdc15(K163E)-mCherry, and cdc15(Core-4A)-mCherry to cdc15+-mCherry fluorescence at the division site. Right) Quantification of Cdc15-mCherry contractile ring and whole cell fluorescence intensities of the indicated strains. n>50 for each genotype. C) Recovery after photobleaching of Cdc15-GFP and mutants. Lines represent least squares fit of recovery, n > 30 for each genotype. D) DAPI (DNA) and Methyl Blue (cell wall / septum) staining of cdc15+ and cdc15 mutants. Bottom) Number of nuclei per cell in cdc15+ and cdc15 mutants. n>400 for each genotype. E) Representative time-lapses (left) and quantification (right) of cytokinesis in the indicated strains. n>30 for each genotype. F) Time-lapse imaging of a cdc15(E30K E152K)-mCherry sid4-GFP cell as cytokinesis fails. All scale bars = 4 μm. All error bars indicate SEM. *, p<0.05; ***, p<.001; ****, p<.0001, one way ANOVA. See also Figures S6–7.
To distinguish between the contribution of oligomerization as opposed to both oligomerization and direct membrane binding, we focused on the mutant of the negatively charged patch (E30K E152K) in further experiments but also assayed the basic charge reversal mutant K163E. mCherry-tagged E30K E152K and K163E were present at only ~31% or ~38%, respectively, of wildtype levels at the division site, while the Core-4A mutant defective in membrane binding but still capable of oligomerization was present at wildtype levels (Figure 6B). Wildtype Cdc15 is highly static in CRs with a mobile fraction (Fm) of 29.5% and a slow recovery half time (t1/2) of 38.5 seconds, as determined by FRAP (Figure 6C) (Roberts-Galbraith et al., 2009). This indicates that Cdc15 is stably “glued” on the membrane at the division site, which we hypothesized was due to extensive oligomerization and membrane interactions. To test this, we compared the dynamics of the oligomerization mutants to wildtype Cdc15 (Figure 6C) and found that oligomerization-defective Cdc15 mutants were significantly more dynamic with t1/2 = 12.7 seconds (E30K E152K) and 12.9 seconds (K163E), and increasingly mobile with Fm = 37.6 % (E30K E152K) and 47.5% (K163E). This increased dynamicity is due to loss of oligomerization, as the Core-4A mutant deficient only in membrane binding (Figure 5B, D–E) resembled wildtype with Fm = 26.1% and t1/2 = 30.8 seconds. The Core-4A mutant therefore appears to retain enough membrane binding capability via electrostatic interactions of tip residues with the membrane and oligomerization to function normally in the CR.
Cdc15 partners with several other proteins at the CR; therefore, we tested whether reduced levels and increased dynamicity of the Cdc15 oligomerization mutants affected partner recruitment (Figure S6C–D). We found that the SH3 domain interactors Rgf3, Fic1, Pxl1, and Spa2 (Ren et al., 2015; Roberts-Galbraith et al., 2009), in addition to F-BAR domain interactor Cdc12 (Carnahan and Gould, 2003; Willet et al., 2015), were all significantly diminished at pre-constriction CRs in both cdc15(E30K E152K) and cdc15(K163E) mutants. However, CR proteins not known to directly bind Cdc15 including F-actin (measured by LifeAct-mCherry), Myo2, and β-glucan synthases Bgs1 and Bgs4 were unchanged in abundance at pre-constriction CRs. Decreased recruitment of binding partners was a function of oligomerization defects alone, as the cdc15(Core-4A) mutant had no effect on downstream recruitment levels (Figure S6C–D), as expected with normal levels and stability at the division site.
To further understand the importance of linear F-BAR oligomerization to Cdc15’s function in cytokinesis, we studied oligomerization mutant cells using live cell microscopy. Oligomerization-defective mutants displayed phenotypes indicative of compromised cytokinesis (Figure 6D). Positive and negative residue charge reversal mutations exhibited identical phenotypes, supporting our previous conclusion that both are involved in mediating F-BAR domain oligomerization. Combining the E30K E152K mutation with truncation of the C-terminal SH3 domain exacerbated the cytokinesis defects as might be expected if SH3-domain interactions help to further stabilize Cdc15 at the CR (Figure S6A–B) (Roberts-Galbraith et al., 2009). That cytokinesis defects result from loss of Cdc15 F-BAR domain oligomerization is further supported by synthetic lethal genetic interactions between oligomerization mutants and other mutations in cytokinesis genes including myo2-E1, rng2-D5, imp2Δ, cdc4-8, in addition to synthetic sickness in combination with cps-191 and cdc12-112 (Figures S6E and S7). Interestingly, oligomerization mutants were also synthetically lethal in combination with pxl1Δ and sick with fic1Δ, two Cdc15 SH3 binding partners.
To understand how impaired Cdc15 oligomerization compromises cytokinesis, we imaged cdc15+, cdc15(K163E), and cdc15(E30K E152K) strains with Rlc1-GFP and Sid4-GFP as markers of the CR and spindle pole body (SPB), respectively, to analyze the kinetics of cytokinesis (Figure 6E). We defined CR formation as the time from SPB separation to onset of CR constriction, constriction as the time from Rlc1-GFP ingression to disappearance at the division site, and separation as time from Rlc1-GFP disappearance to cell separation. In both E30K E152K and K163E mutants, each step of cytokinesis was extended (Figure 6E, right). We additionally observed cells that failed to divide, consistent with the 5–10% multi-nucleation rate. By imaging cdc15(K163E)-mCherry cells, we observed that in cells failing cytokinesis, Cdc15 rings were formed, but slid from the cell middle and disassembled during anaphase (Figure 6F and Movies S1–S2). These data indicate that Cdc15 oligomerization facilitates its stable membrane binding and scaffolding function at the cell division site.
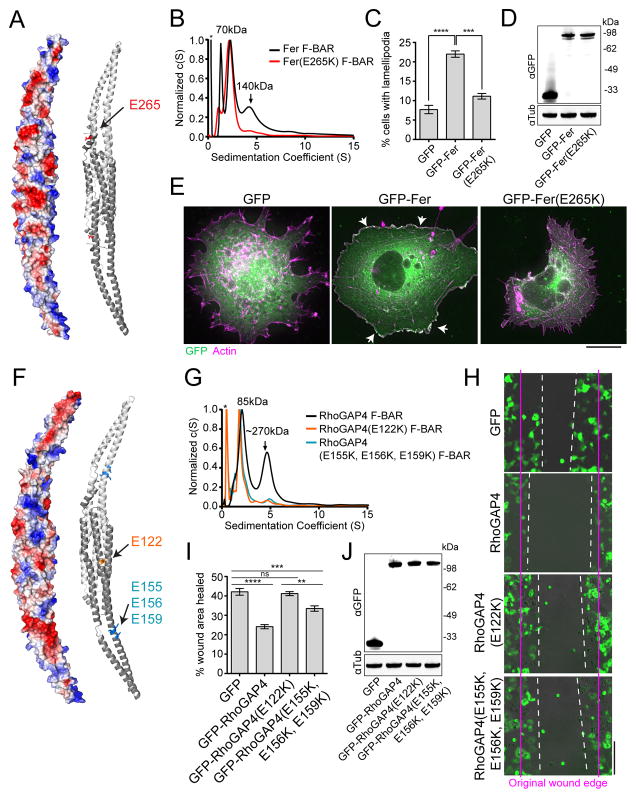
To determine if the importance of F-BAR oligomerization extends to any of the human F-BARs studied above that also do not tubulate membranes, we focused on Fer and RhoGAP4 because they have easily assayable functions in lamellipodia formation and cell migration, respectively (Itoh et al., 2009; Vogt et al., 2007). We generated structural models of the Fer and RhoGAP4 F-BAR domains with Phyre2 (Figure 7A, F) and identified acidic patches on the side of each F-BAR. By testing each patch, we found specific ones that, when mutated, eliminated Fer and RhoGAP4 F-BAR oligomerization in vitro (Figure 7A–B, F–G). We then tested the functional consequence of these mutations in the context of their respective full-length proteins. Whereas exogenously expressed wildtype Fer drove lamellipodia formation as shown previously (Itoh et al., 2009), the Fer E265K oligomerization mutation prevented this activity (Figure 7C–E). Furthermore, the E122K or E155K, E156K, E159K mutations in RhoGAP4 blocked its ability to inhibit cell migration in a wound healing assay (Vogt et al., 2007) (Figure 7H–J). Together, these data demonstrate that F-BAR oligomerization but not membrane bending is functionally important in several biological contexts.
Figure 7. Oligomerization is critical for Fer and RhoGAP4 function.
A) Phyre2 homology model (based on Fes, PDB:4DYL) of the Fer F-BAR domain with dimer subunits colored in dark and light grey. An electrostatic potential map (left) and a cartoon model (right) with oligomerization residues highlighted are shown. B) Analytical ultracentrifugation trace of purified Fer and Fer(E265K) F-BAR domains. Small MW contaminant indicated by *. C–E) GFP, GFP-Fer, or GFP-Fer(E265K) were expressed in COS-7 cells (D), stained with Rhodamine-phalloidin (E), and quantified for the presence of lamellipodia (C). Scale bar = 10 μm. F) Phyre2 homology model (based on CIP4, PDB:2EFK) of the RhoGAP4 F-BAR domain with dimer subunits colored in dark and light grey. An electrostatic potential map (left) and a cartoon model (right) with oligomerization residues highlighted are shown. G) Analytical ultracentrifugation trace of purified Fer and Fer(E265K) F-BAR domains. Small MW contaminant indicated by *. H–J) GFP, GFP-RhoGAP4, GFP-RhoGAP4(E122K), and GFP-RhoGAP4(E155K, E156K, E159K) were expressed in COS-7 cells (J). A wound was formed with a P200 pipet tip (H) and migration into the wounded area was quantified after 8 hours (I). Scale bar = 100 μm. All error bars indicate SEM from 3 experiments. ***, p<0.001; ****, p<0.0001, one-way ANOVA.
Discussion
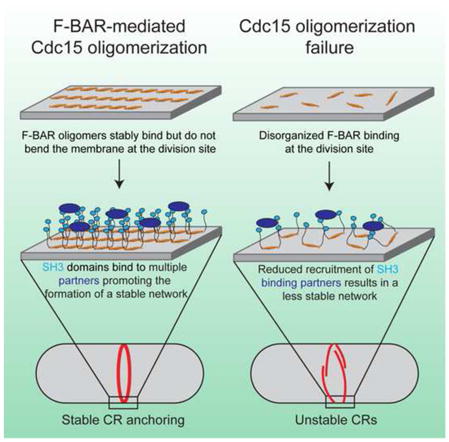
In this study, we found that the Cdc15 F-BAR domain, unlike many previously described F-BAR domains, binds but does not bend membranes in vivo or in vitro, challenging the widely held view that tubulation is a fundamental function of F-BAR domains. Six human F-BAR domains exhibit similar behavior, suggesting that a significant fraction of F-BAR proteins function primarily as molecular tethers. Tuned by post-translational modification, these F-BAR proteins transiently link different machinery to the membrane in a variety of biological contexts without changing membrane contour. Oligomerization is critical for these F-BAR domains’ functions; Fer and RhoGAP4 rely upon F-BAR oligomerization for their functions in cell migration, while the Cdc15 F-BAR domain forms linear oligomers that are critical for efficient membrane binding and robust localization to the division site. Indeed, Cdc15 oligomers appear to serve as a major anchor for the CR at the membrane.
Diverse modes of F-BAR oligomerization
Lateral and tip-to-tip interactions have been observed between dimers in CIP4 F-BAR domain in vitro tubule reconstructions (Frost et al., 2008). Lateral interactions were essential in CIP4 for bending membranes into tubules, while tip-to-tip contacts performed an organizational role of aligning dimers and resisting helical tilting of F-BAR domain assemblies (Frost et al., 2008; Shimada et al., 2007). In the Cdc15 F-BAR, tip-to-tip contacts between adjacent dimers organized them into linear strands. In Fer and RhoGAP4, residues on the wings and core of F-BAR dimers were important for oligomerization, suggesting lateral F-BAR to F-BAR contacts. Interestingly, the residues identified in Cdc15, Fer, and RhoGAP4 are distinct from those identified in CIP4 tubule reconstructions. This suggests that the F-BAR family possesses multiple distinct modes of oligomerization which we propose result in different organizations on the membrane, not all of which produce membrane deformation. Crystal structures of non-tubulating F-BAR domains may also reveal any primary structural differences that preclude their abilities to bend membranes.
Cdc15 F-BAR domain’s linear mode of oligomerization is important for efficient membrane binding. Our data suggest two interconnected factors contribute to weakened membrane binding when oligomerization is impaired: 1) Loss of cooperativity effectively lowers the amount of protein bound to membrane at a given concentration and 2) loss of Cdc15 oligomers’ avidity (as each oligomer subunit contains membrane binding contacts) destabilizes membrane association. Cooperative binding has been observed for human FBP17 and Drosophila Nwk F-BAR domains, which may also arise from oligomerization of F-BAR dimers (Becalska et al., 2013; Itoh et al., 2005). Cooperative membrane binding through oligomerization may serve as a mechanism to concentrate and stabilize F-BAR domain localization at sites of action, similar to Cdc15 concentration at the division site.
Diversity among F-BAR domains is also seen with the choice of residues used for membrane binding. Of F-BARs studied in mechanistic detail, S. pombe Cdc15 and human FBP17, CIP4, and FCHo2, utilize unique charged residues on their concave surfaces, located both in the core and wings, to associate with membranes (Frost et al., 2008; Henne et al., 2007; Shimada et al., 2007). Human PACSIN F-BAR domains additionally utilize a small loop inserted into the membrane bilayer (Wang et al., 2009), and S. cerevisiae Rgd1p contains a specific PIP binding patch at the base of each wing (Moravcevic et al., 2015). This patch may be loosely conserved in additional F-BAR domains, though none have been shown to have the strong PIP specificity observed in Rgd1p.
Cdc15 oligomerizes to robustly scaffold a network of cytokinesis proteins
There are approximately ~16,100 copies, or 8050 dimers, of Cdc15 at the division site (Wu and Pollard, 2005), and F-BAR dimers are approximately 220 Å in length (Moravcevic et al., 2015; Shimada et al., 2007). Assuming a 3.5 μm inner cell diameter, linear oligomers of Cdc15 aligned perpendicular to the cell’s long axis could, in a simple model, circle the division site ~16 times. Considering a ~25 Å width of one F-BAR dimer, oligomers lying next to each other would yield a scaffolding platform a minimum of ~40 nm wide, easily within estimates of CR width of 100–200 nm (Kamasaki et al., 2007). The filamentous linear oligomerization of Cdc15, with each repeating F-BAR domain unit binding the membrane, generates a strong avidity for the membrane. Consequently, these oligomers serve as a stable scaffolding platform for the CR, reinforced by the many stabilizing interactions mediated by Cdc15’s C-terminal SH3 domain (Ren et al., 2015; Roberts-Galbraith et al., 2009). Accordingly, when Cdc15 oligomerization is prevented, this platform weakens and becomes increasingly dynamic, leading to impairment in cytokinetic fidelity. The state of Cdc15 oligomerization is controlled during a normal cell cycle via cell cycle-regulated phosphorylation (Roberts-Galbraith et al., 2010); dephosphorylation allows oligomerization while re-phosphorylation antagonizes it. Inhibition of oligomerization via phosphorylation during CR constriction could aid significantly in CR disassembly. In the future, it will be interesting to determine exactly how phosphorylation modulates F-BAR-to-F-BAR interactions within the context of the full-length protein.
CR sliding and/or instability have been observed in other situations with compromised Cdc15 levels or function (Arasada and Pollard, 2014; Hachet and Simanis, 2008; Roberts-Galbraith et al., 2009; Wachtler et al., 2006), as well as when glucan chain synthesis is compromised (Arasada and Pollard, 2014; Liu et al., 2000; Muñoz et al., 2013; Pardo and Nurse, 2003; Stachowiak et al., 2014), suggesting an intimate link between the function of glucan synthase enzymes, Cdc15, and CR integrity. In S. pombe, it is essential that a primary and secondary septum forms behind the constricting CR (Liu et al., 1999; Muñoz et al., 2013; Proctor et al., 2012). Considering these data and our results, we propose two factors contribute to CR instability when Cdc15 function is defective. First, the physical connection between the plasma membrane and the CR is weakened. Second, Cdc15’s direct binding partners that influence septum formation (Bohnert and Gould, 2012; Morrell-Falvey et al., 2005; Ren et al., 2015; Roberts-Galbraith et al., 2009; Tajadura et al., 2004) including the Rho-GEF Rgf3, essential for activating Rho1 and the Bgs1 glucan synthase, are reduced in abundance at the CR. A weakened membrane anchor and insufficient septum deposition likely contribute to CR instability, allowing CR sliding from the cell middle.
A significant fraction of F-BARs do not deform membranes
With the addition of our data, all human F-BAR domains have been tested for their ability to bend membranes into tubules at high concentration (Table S1). Eleven domains positively bend membranes into tubules, five induce negative curvature tubules, and here we find an additional six (Fer, Fes, FCHSD1/2, RhoGAP4, and Gas7) that bind but are unable to bend or tubulate membranes, similar to S. pombe Cdc15. Fer and Fes are non-receptor protein-tyrosine kinases with broad roles in actin cytoskeletal arrangement during cell motility and in cell-cell adhesions (Greer, 2002; Itoh et al., 2009). FCHSD1/2 are the apparent human orthologs of Drosophila nervous wreck 1/2 proteins; interestingly, Drosophila Nwk1 is able to bend membranes into “ridges and scallops” (Becalska et al., 2013), which we did not observe for human homologs FCHSD1/2. RhoGAP4 contains a GTPase activating domain and is involved in cell motility and axon growth (Vogt et al., 2007). Gas7 is primarily expressed in quiescent cells and is involved in forming actin-based protrusions (Ju et al., 1998; You and Lin-Chao, 2010). The lack of a functional connection between these proteins suggests the membrane binding and oligomerization properties of each F-BAR domain are tuned to its specific cellular function. It will be interesting to clarify the diverse structural mechanisms and roles of F-BAR oligomerization in other F-BAR proteins in their respective physiological functions.
Experimental Procedures
Detailed methods are available in Supplemental Experimental Procedures.
Yeast Strains, Media, and Genetics
S. pombe strains (see Supplemental Experimental Procedures, “S. pombe strains used in this study”) were grown in yeast extract (YE) media or 4X YE for large scale purifications. cdc15 mutants were integrated at the endogenous locus by transforming cdc15+/cdc15::ura4+ diploids with pIRT2-cdc15 mutant constructs containing 750bp flanks. Haploid integrants resistant to 5-FOA were isolated and verified by sequencing.
Cell Culture
COS-7 cells were grown in DMEM media + 10% FBS (Life Technologies). Cells were plated on glass slides or MatTek glass bottom dishes (MatTek Corporation, Ashland, MA) coated with fibronectin (Sigma-Aldrich) before transfection with pEGFP F-BAR constructs using Lipofectamine 3000 reagents (ThermoFisher).
Protein Purification
F-BARs and GFP-F-BARs were produced in Escherichia coli Rosetta2(DE3)pLysS cells and purified on cOmplete His-Tag resin (Roche) according to manufacturer’s protocol. His tags were removed by thrombin digestion and F-BARs were further purified on either a HiTrap Q SP anion exchange column (GE Healthcare) or a HiPrep Sephacryl S-100HR gel filtration column (GE Healthcare) and concentrated with Amicon Ultra Centrifugal Filters (Millipore). Full length Cdc15-TEV-2xProtA was purified from S. pombe pellets as previously described ((Roberts-Galbraith et al., 2010), see Supplemental Experimental Procedures).
Liposomes
All lipids were obtained from Avanti Polar Lipids. Liposomes were formed and liposome co-pelleting assays were performed as previously described (Itoh et al., 2005). Giant Unilamellar Vesicles (GUVs) were electro-formed on Indium-Tin-Oxide-coated (ITO) glass coverslips (Sigma-Aldrich) using a 10 Hz, 2.5 V sinusoidal current. GUVs were adjusted to 150 mM NaCl before F-BAR addition unless otherwise indicated. Recombinant GFP-F-BAR domains were mixed with GUVs at a final concentration of 10 μM before imaging in a 0.5 mm chamber. Liposome binding experiments were performed through Bio-Layer Interferometry on an Octet RED96 instrument (ForteBio, Menlo Park, CA) with streptavidin sensor tips (Abdiche et al., 2008). One-way ANOVA tests were used to determine significance values between binding conditions.
Microscopy
S. pombe cells were imaged live at 25°C or fixed with ice-cold 70% ethanol and stained with Methyl Blue and DAPI. Time-lapse imaging was performed on log-phase cells using a CellASIC ONIX microfluidics perfusion system (Millipore), flowing YE media at 25°C through the chamber at 5 psi throughout imaging. Image stacks were deconvolved using Deltavision softWoRx imaging software. Image projections, intensity measurements, and FRAP analyses were performed with ImageJ software (http://rsweb.nih.gov/ij/). Lamellipodia were considered arc-shaped membrane edges with decreasing actin staining intensity with increasing distance toward the cell middle (Itoh et al., 2009). Quantification of COS-7 wound healing was performed by measuring the advancement of the leading cell edge into the wound after 8 hours as a percentage of the original wound area.
Electron Microscopy
Cdc15 F-BAR or F-BAR bound liposome samples were adsorbed to a glow discharged 200-mesh copper grid covered with carbon-coated collodion film (EMS, Hatfield, PA), stained with uranyl formate (Ohi et al., 2004), and imaged on a FEI Morgagni electron microscope at 100 kV with a 1K x 1K CCD camera (ATM, Woburn, MA).
Analytical Ultracentrifugation
Cdc15 or Cdc15 mutant F-BAR domains were diluted to a final concentration of 0.5 mg/mL and 50 mM NaCl to induce oligomerization before ultracentrifugation. Sedimentation experiments were run at 42,000 rpm at 4°C on an Optima XLI ultracentrifuge (Beckman-Coulter) and analyzed with SedFit (version 14.4d) using 250 scans collected ~2 min apart (Schuck, 2000). Size distributions were determined for a confidence level of p = 0.95 and resolution of n = 200.
Structural Modelling
F-BAR domain structural models were generated using the Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) (Kelley and Sternberg, 2009) using Hof1 (PDB: 4WPE), Fes (PDB: 4DYL), and CIP4 (PDB: 2EFK) crystal structures as templates for Cdc15, Fer, and RhoGAP4, respectively. Graphical representations of F-BAR models were generated using PyMOL (Schrodinger, LLC).
Supplementary Material
Acknowledgments
The authors thank Anna Feoktistova for outstanding technical assistance with biochemical experiments and Dr. Rachel Roberts-Galbraith and members of the Gould lab for critical review of the manuscript. FRAP experiments were performed through the VUMC Cell Imaging Shared Resource. BLI experiments were performed through the VUMC Antibody and Protein Resource. N.A.M. was supported by AHA fellowship 15PRE21780003. This work was supported by NIH grant GM101035 to K.L.G.
Footnotes
Author Contributions
Conceptualization, N.A.M., C.V.K., M.D.O., K.L.G.; Investigation, N.A.M.; Supervision of EM, M.D.O.; Supervision of structural modelling, C.V.K.; Writing, N.A.M., K.L.G.; Editing, N.A.M., C.V.K., M.D.O., K.L.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem. 2008;377:209–17. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Contractile Ring Stability in S. pombe Depends on F-BAR Protein Cdc15p and Bgs1p Transport from the Golgi Complex. Cell Rep. 2014;8:1–12. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Kelley CF, Berciu C, Stanishneva-Konovalova TB, Fu X, Wang S, Sokolova OS, Nicastro D, Rodal AA. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol Biol Cell. 2013;24:2406–18. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Gladfelter AS, Kovar DR, Lee WL. Cytoskeletal dynamics: A view from the membrane. J Cell Biol. 2015;209:329–337. doi: 10.1083/jcb.201502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast. PLoS Genet. 2012;8:e1003004. doi: 10.1371/journal.pgen.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–62. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O, Roux A, Haucke V. BAR Domain Scaffolds in Dynamin-Mediated Membrane Fission. Cell. 2014;156:882–892. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–44. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–17. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–6. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO. 2006;25:2898–910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat Rev Mol Cell Biol. 2002;3:278–89. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- Gu Y, Yam C, Oliferenko S. Rewiring of Cellular Division Site Selection in Evolution of Fission Yeasts. Curr Biol. 2015;25:1187–1194. doi: 10.1016/j.cub.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–16. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Kent HM, Ford MGJ, Hegde BG, Daumke O, Butler PJG, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–52. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Itoh T, Hasegawa J, Tsujita K, Kanaho Y, Takenawa T. The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci Signal. 2009;2:ra52. doi: 10.1126/scisignal.2000393. [DOI] [PubMed] [Google Scholar]
- Ju YT, Chang ACY, She BR, Tsaur ML, Hwang HM, Chao CCK, Cohen SD, Lin-Chao S. gas 7 : A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc Natl Acad Sci. 1998;95:11423–11428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–71. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem. 2004;279:40091–9. doi: 10.1074/jbc.M404899200. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–60. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Balasubramanian MK. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci. 2000;113:1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, McCollum D, Balasubramanian MK. Drc1p/Cps1p, a 1,3-β-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153:1193–1203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO. 2006;25:2889–97. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Alvarado D, Ferguson KM, Lemmon MA, Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM. Comparison of Saccharomyces cerevisiae F-BAR Domain Structures Reveals a Conserved Inositol Phosphate Binding Site. Structure. 2015;23:1–12. doi: 10.1016/j.str.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Den Haese G, Gould KL. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Muñoz J, Cortés JCG, Sipiczki M, Ramos M, Clemente-Ramos JA, Moreno MB, Martins IM, Perez P, Ribas JC. Extracellular cell wall β(1,3)glucan is required to couple septation to actomyosin ring contraction. J Cell Biol. 2013;203:265–82. doi: 10.1083/jcb.201304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, Fan J, Zhang Y, Zhang K, Gao B, Ma J, Li J, Deng Y, Zhou Q, Egelman EH, Hsu VW, Sun F. A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature. Dev Cell. 2014;31:73–86. doi: 10.1016/j.devcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–1574. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Proctor SA, Minc N, Boudaoud A, Chang F. Contributions of Turgor Pressure, the Contractile Ring, and Septum Assembly to Forces in Cytokinesis in Fission Yeast. Curr Biol. 2012;22:1–8. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let’s go bananas: revisiting the endocytic BAR code. EMBO. 2011;30:3501–15. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, Beckley JR, Gould KL. The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell. 2015;26:256–269. doi: 10.1091/mbc.E14-10-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: Functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T, Yokoyama S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–72. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Stachowiak MR, Laplante C, Chin HF, Guirao B, Karatekin E, Pollard TD, O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 2014;29:547–61. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2009;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Tajadura V, García B, García I, García P, Sánchez Y. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J Cell Sci. 2004;117:6163–6174. doi: 10.1242/jcs.01530. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–79. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt DL, Gray CD, Young WS, Orellana SA, Malouf AT. ARHGAP4 is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol Cell Neurosci. 2007;36:332–42. doi: 10.1016/j.mcn.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. Cell Cycle-dependent Roles for the FCH-Domain Protein Cdc15p in Formation of the Actomyosin Ring in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:3254–3266. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MVAS, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci. 2009;106:12700–5. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, Mcdonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol. 2015;208:391–399. doi: 10.1083/jcb.201411097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–4. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- You JJ, Lin-Chao S. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem. 2010;285:11652–66. doi: 10.1074/jbc.M109.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput Biol. 2013;9:e1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.