SUMMARY
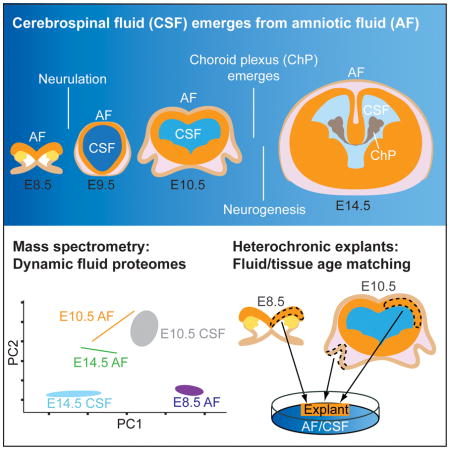
After neural tube closure, amniotic fluid (AF) captured inside the neural tube forms the nascent cerebrospinal fluid (CSF). Neuroepithelial stem cells contact CSF-filled ventricles, proliferate and differentiate to form the mammalian brain, while neurogenic placodes, which generate cranial sensory neurons, remain in contact with the AF. Using in vivo ultrasound imaging, we quantified the expansion of the embryonic ventricular-CSF space from its inception. We developed tools to obtain pure AF and nascent CSF, before and after neural tube closure, and define how the AF and CSF proteomes diverge during mouse development. Using embryonic neural explants, we demonstrate that age-matched fluids promote Sox2-positive neurogenic identity in developing forebrain and olfactory epithelia. Nascent CSF also stimulates Sox2-positive self-renewal of forebrain progenitor cells, some of which is attributable to LIFR signaling. Our resource should facilitate the investigation of fluid-tissue interactions during this highly vulnerable stage of early brain development.
Graphical abstract
INTRODUCTION
Morphogenesis of the cerebrospinal fluid (CSF)-filled ventricular system requires a series of highly stereotyped, conserved events (Bjornsson et al., 2015; Lowery and Sive, 2009; Massarwa and Niswander, 2013; Wilde et al., 2014). Tremendous changes in ambient fluid composition and brain morphology occur at embryonic day (E) 8.5-E10.5 in the mouse embryo, yet the relationship between these changes is poorly understood. At E8.5, the neuroectoderm destined to become the mammalian brain is exposed to amniotic fluid (AF). As the anterior neural tube closes, the AF captured inside the neural tube becomes the nascent CSF, and the entire mammalian brain subsequently develops around CSF-filled ventricular spaces (Ek et al., 2005; Zappaterra and Lehtinen, 2012). Subsequently, the neurogenic placodes that give rise to cranial sensory neurons (Moody and LaMantia, 2015) remain exposed to AF, while the developing brain is exposed to CSF. As brain development progresses, the forebrain neuroectoderm consists of neuroepithelial cells that rapidly divide to expand the neural progenitor cell pool (Bjornsson et al., 2015). Over subsequent days, the neuroepithelium becomes patterned, with well-defined gene expression domains along the anterior-posterior and dorsal-ventral axes, ultimately giving rise to the wealth of cells in distinct regions of the mammalian brain (Grove and Monuki, 2013; Rallu et al., 2002a). The progressive lineage restriction of neural progenitors and transition to radial glial progenitors of the cerebral cortex occurs during this period. While pressurization of the ventricles is essential for normal brain development to proceed (Desmond and Jacobson, 1977; Lehtinen and Walsh, 2011), little is known about the signals delivered by the AF versus nascent CSF and whether they instruct the development of adjacent epithelia during these early stages of brain development.
Cell intrinsic genetic programs regulate progenitor cell lineage restriction (Shen et al., 2006). However, extrinsic signals play important roles in specifying progenitor identity as well (Hunter and Hatten, 1995; LaMantia et al., 1993; McConnell and Kaznowski, 1991). The interplay between genetic programs (Manzini and Walsh, 2011; Shen et al., 2006), CSF (Lehtinen et al., 2011; Lun et al., 2015b), meningeal (Chatzi et al., 2013; Siegenthaler et al., 2009), and vascular (Kokovay et al., 2010; Shen et al., 2004; Tavazoie et al., 2008) signals help coordinate and synchronize brain development at later ages. At these later stages, secreted signals in the embryonic CSF (Lehtinen et al., 2011), many of which originate from the choroid plexus (Huang et al., 2009b; Lun et al., 2015a; Yamamoto et al., 1996), instruct the proliferation and survival of polarized radial glial progenitor cells as well as adult neural stem cells, in part via signal transduction at the cilium (Higginbotham et al., 2013; Tong et al., 2014; Yeh et al., 2013). Proteomic approaches have begun to characterize the composition of the CSF during later stages of development in different species (e.g. Chang et al., 2015; Dziegielewska et al., 1981; Lehtinen et al., 2011; Lun et al., 2015a; Parada et al., 2005; Zappaterra et al., 2007), as well as in adult (e.g. Dislich et al., 2015). However, around the time of neural tube closure (E8.5-E10.5), the choroid plexus does not exist (Lun et al., 2015b), and the meninges as well as the brain’s vasculature are only beginning to form (Bjornsson et al., 2015). The only source of soluble extrinsic cues for neural stem cells at this time is the AF and nascent CSF, which contact the apical surface of all neural stem cells. Nevertheless, it is not known whether distinct AF and nascent CSF instruct early neuroepithelial cells and very few clues exist regarding the identity of such instructive signals. Indeed, even the basic proteomic composition of early embryonic AF and nascent CSF remains undefined.
To provide a resource to formulate and answer critical questions of how early neural stem cells in the central as well as peripheral nervous system are regulated by the adjacent fluid proteomes, we developed techniques to collect AF and nascent CSF from developing embryonic mouse before and after neural tube closure. We demonstrate the dynamic capacity of these fluids to signal via cardinal morphogenetic signaling pathways including Sonic hedgehog (Shh), Bone morphogenetic proteins (Bmps), and retinoic acid (RA). Using quantitative mass spectrometry (MS) of these fluids, we define their progressive proteomic differentiation from their common AF origin at E8.5, through E10.5 and E14.5 when distinct AF and CSF compartments exist. We developed a neural explant culture system to establish that age-matched fluid-tissue interactions promote Sox2-positive neurogenic identity in developing forebrain neuroepithelium as well as peripheral olfactory epithelium. Nascent CSF promotes Sox2-positive self-renewal of forebrain neural stem cells, and we demonstrate a role for signaling via the LIFR in this process. Thus, our comprehensive resource defines divergence between the AF and CSF proteomes and validates the functional significance of these differences using in vitro and ex vivo approaches, facilitating the investigation of fluid-tissue interactions during the initial formation of the nervous system.
RESULTS
Rapid morphological changes in brain-ventricular system during early development
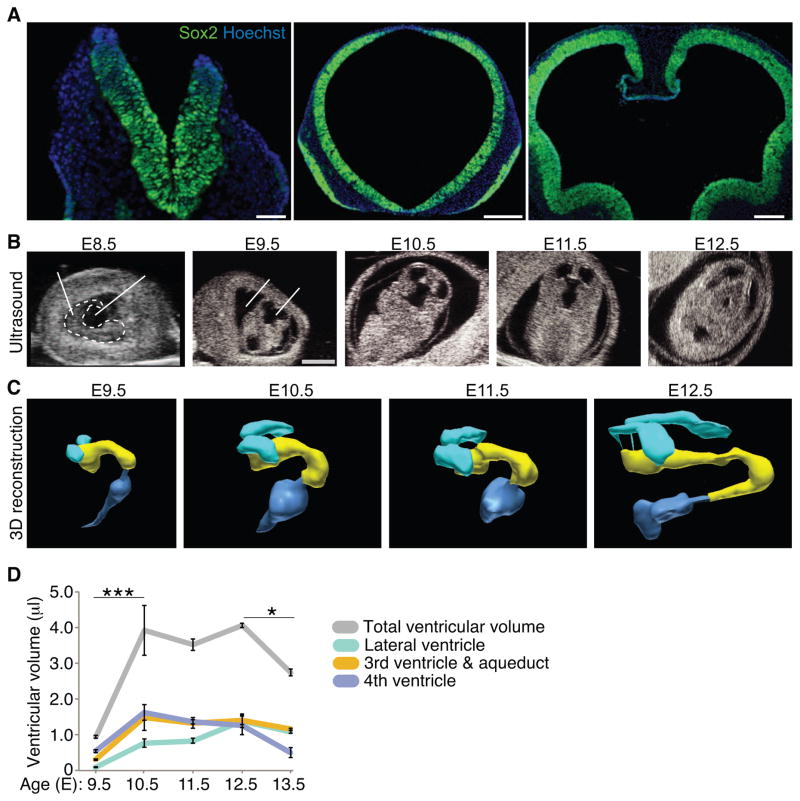
In the vertebrate embryo, the transition from an open anterior neural plate contacted by AF to a closed forebrain adjacent to CSF-filled ventricles is a key morphogenetic event in brain development. At E8.5 in mouse, the neuroectoderm, which is home to Sox2-positive neuroepithelial cells, directly contacts the AF (Fig. 1A). As the anterior neural tube closes, the AF captured inside the neural tube becomes the nascent CSF (Ek et al., 2005; Zappaterra and Lehtinen, 2012). By E9.5, Sox2-positive neuroepithelial progenitor cells in the developing forebrain directly contact the CSF (Fig. 1A), and the forebrain continues to expand around this CSF-filled ventricle (Fig. 1A). To evaluate the influence of these morphogenetic changes for defining the CSF as a distinct entity, we performed in utero ultrasound imaging of live embryos (Fig. 1B) and quantified regional differences in the establishment and expansion of the ventricular system during this early phase of brain development (Fig. 1C, D). Following neural tube closure at E9, the lateral ventricles continuously expanded until E12.5 (E9.5: 0.092 ± 0.015 μl; E12.5: 1.392 ± 0.193 μl; t-test, p < 0.005, n=3; Fig. 1C, D), while third and fourth ventricle expansion was most prominent between E9.5-E10.5 (Fig. 1C, D). Ventricular volume in live embryos was quite distinct from that measured in fixed tissue in a separate set of samples. Paraformaldehyde fixation caused shrinkage of the ventricles by approximately 60% (data not shown). Taken together, following its definition with the closure of the anterior neural tube, the mouse ventricular system undergoes dynamic morphological changes in a ventricle-specific manner.
Figure 1. Expansion of the mouse ventricular system following neural tube closure.
(A) Sox2-positive neuroepithelial cells (green) contact the AF before neural tube closure at E8.5 (left panel), and the nascent CSF following neural tube closure at E9.5 (middle panel) and E10.5 (right panel). Nuclei counterstained with Hoechst. Scale bars: 50μm (left panel); 200μm (middle, right panels). (B) Representative ultrasound images of developing mouse (E8.5-E12.5). Dashed white line (left panel) outlines embryo. Scale bar: 2mm. (C) 3D reconstruction of ventricular system from ultrasound images sequences captured in (B) from E9.5-E12.5 with the following color representation: turquoise: lateral ventricles; yellow: third ventricle and aqueduct; dark blue: fourth ventricle. (D) Quantification of ventricle size based on 3D reconstructions of ultrasound image sequences (B, C, data not shown; ventricular volume (μl) represented as mean ± SEM: Lateral ventricles: E9.5 = 0.09 ± 0.02; E10.5 = 0.77 ± 0.12; E11.5 = 0.83 ± 0.07; E12.5 = 1.39 ± 0.19; E13.5 = 1.08 ± 0.05; Third ventricle and aqueduct: E9.5 = 0.31 ± 0.03; E10.5 = 1.48 ± 0.37; E11.5 = 1.33 ± 0.15; E12.5 = 1.40 ± 0.11; E13.5 = 1.16 ± 0.03; Fourth ventricle: E9.5 = 0.54 ± 0.04; E10.5 = 1.63 ± 0.22; E11.5 = 1.36 ± 0.06; E12.5 = 1.27 ± 0.27; E13.5 = 0.50 ± 0.14; Total ventricular volume: E9.5 = 0.94 ± 0.04; E10.5 = 3.92 ± 0.70; E11.5 = 3.51 ± 0.16; E12.5 = 4.06 ± 0.06; E13.5 = 2.74 ± 0.09; n = 3; ANOVA statistical analyses of total ventricular volume represented as (*) p < 0.05 and (***) p < 0.0001; n=3).
Age-matched AF and CSF support neurogenic cell fate
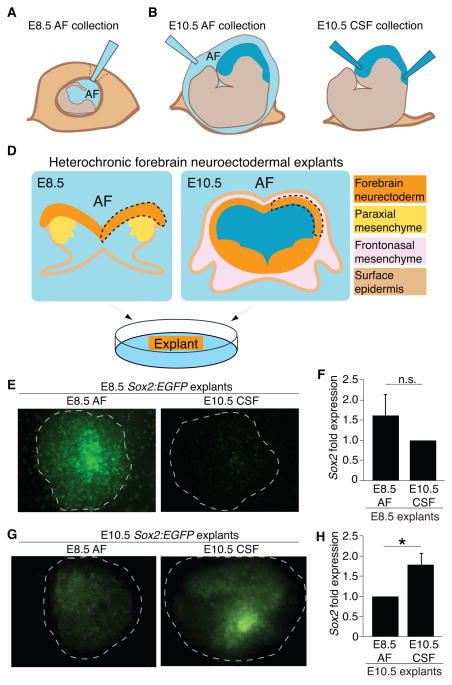
Given the striking change in fluid environment that neural tube closure imposes on the developing neuroectoderm, we tested if E8.5 AF and E10.5 CSF have the capacity to differentially instruct neuroepithelial cell fate before and after neural tube closure. We first developed methods to collect pure samples of AF and CSF at these early ages. To collect E8.5 AF, the decidua and parietal yolk sac were removed to expose the visceral yolk sac, and AF was withdrawn from the amniotic cavity (Fig. 2A). The E8.5 embryo did not appear to incur damage due to AF collection (Fig. S1A, S1B). AF was similarly collected from the amniotic cavity at E10.5 (Fig. 2B). To collect E10.5 CSF, the extra-embryonic membranes were removed and CSF was withdrawn from the telencephalic vesicles, aqueduct, and fourth ventricle (Fig. 2C). Following fluid collection, samples were verified for purity (based upon absence of red blood cells or other particulate matter) and processed (Zappaterra et al., 2013). Standard litters (10–12 embryos) yielded the following volumes: E8.5 AF = 10–15μl; E10.5 AF = 180–225μl; E10.5 CSF = 30–40μl. CSF yield was comparable to the ventricular size calculated by ultrasound (e.g. E10.5 ventricular volume = 3.9 ± 0.7μl/embryo; Fig. 1D).
Figure 2. Heterochronic explants show tissue-fluid age matching promotes Sox2-positive progenitors.
Schematics of (A) E8.5 AF collection; (B) E10.5 AF collection; (C) E10.5 CSF collection; (D) Heterochronic forebrain neuroectodermal explant dissections: isolated neuroepithelium (dashed line) placed on membrane with ventricular surface down contacting AF or CSF. (E) E8.5 explants grown on E8.5 AF or E10.5 CSF. Sox2:EGFP suggests age-matched fluid best supports neural stem cell identity (E8.5 explant with E8.5 AF [n=8] or with E10.5 CSF [n=8]). (F) qPCR of explants cultured as in (E) suggest that E8.5 AF more favorably supports E8.5 explants compared to E10.5 explants. (E8.5 AF = 1.63 ± 0.49; E10.5 CSF = 1.00; n=3 of 4 pooled explants each, p = n.s., Mann-Whitney). Two experiments (n=2) of 4 pooled explants each were excluded from statistical analyses. While E8.5 AF supported these E8.5 explants, the experiments had undetectable Sox2 expression in the E10.5 CSF condition, suggesting a role for age-matched AF and CSF in cell survival and demonstrating experimental variability at this age (see also Discussion). (G) E10.5 explants grown on E8.5 AF [n=9] or with E10.5 CSF [n=11]. Note increased GFP expression for age-matched CSF vs. non-age-matched CSF experiments. (H) qPCR of explants cultured as in (G) show that E10.5 CSF more favorably promotes Sox2 expression in E10.5 explants compared to E8.5 AF (E8.5 AF = 1.00; E10.5 CSF = 1.80 ± 0.29; n=3 of 4 pooled explants each, p < 0.05, Mann-Whitney). See also Figure S1.
We then developed a protocol for forebrain neuroectodermal explants at E8.5 and E10.5 and tested for the instructive capacity of AF and nascent CSF. Once the neuroepithelium was separated from mesenchyme and surface ectoderm (both are sources of local signals that might confound assessment of AF or CSF signaling capacity: LaMantia et al., 2000; Tucker et al., 2008), explants were exposed to AF or CSF from different ages (Fig. 2D). Heterochronic forebrain neuroectodermal explants using Sox2:EGFP knock-in mice (Ellis et al., 2004) suggested that progenitor cell identity is differentially regulated by secreted signals received from E8.5 AF vs. E10.5 CSF. Survival as well as the neurogenic phenotype of immature neuroectodermal cells at E8.5, as assessed by Sox2:EGFP in explants, appeared optimally supported by age-matched E8.5 AF (Fig. 2E, F). Conversely, E10.5 CSF optimally supported E10.5 neuroectodermal cells (Fig. 2G, H). These data demonstrate that compartment/fluid/tissue age-matching—either via changes in osmolality or through distinct signaling capacities—contributes to the maintenance of neurogenic progenitor cell fate.
Dynamism of AF and CSF, and age-dependent instructive capacities of AF
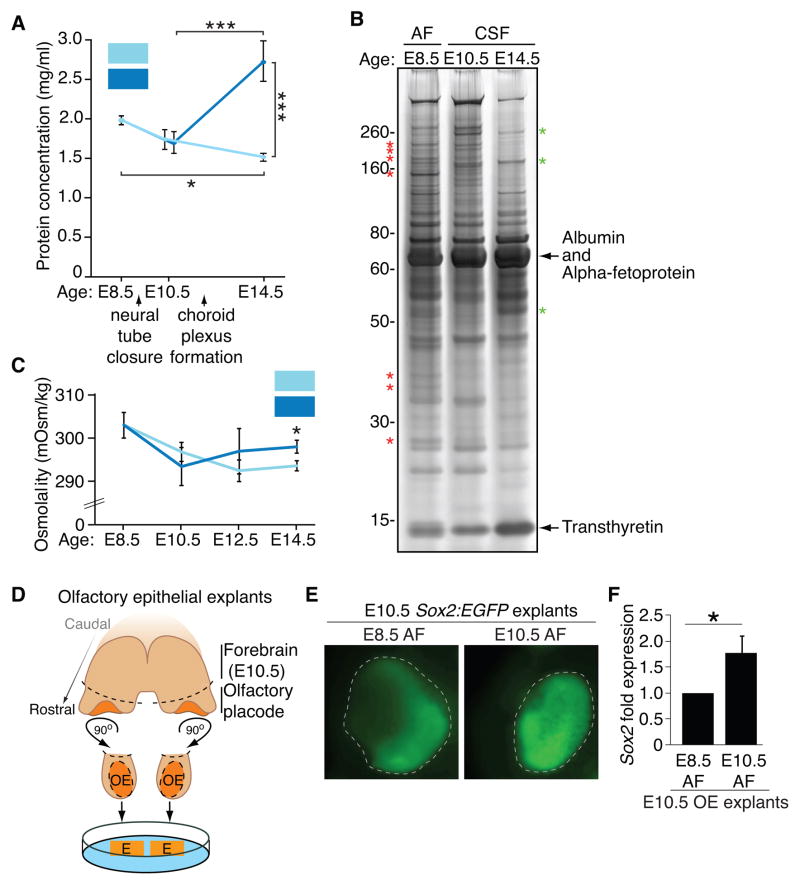
There is very little insight into the quantitative changes in soluble factors that occur during the time that the CSF becomes a distinct fluid compartment from the AF. Measurement of the total protein concentration in AF and nascent CSF suggested that the proteomes of these fluids are tightly regulated. The total AF and CSF protein concentrations started to decrease from E8.5-E10.5 (Fig. 3A), and the total AF protein concentration continued to decrease to E14.5 (Fig. 3A). In contrast, the total CSF protein concentration sharply increased from E10.5-E14.5 (Fig. 3A), consistent with previous studies (Ek et al., 2005; Lun et al., 2015a). Unexpectedly the E14.5 CSF proteome appeared less complex in comparison with E8.5 AF and E10.5 CSF when analyzed by silver staining (Fig. 3B). The increase in E14.5 CSF protein concentration was apparently due to robust increases in certain proteins, such as albumin, which appeared saturated by silver staining (Fig. 3B) and which we verified by quantitative MS to increase approximately two-fold between E10.5 CSF to E14.5 CSF (albumin mean spectral counts ± SEM: E10.5 CSF = 97.1 ± 16.6; E14.5 CSF = 201.6 ± 23.6; Mann-Whitney, p<0.05, n=3; see also Table S1).
Figure 3. Early AF and CSF undergo dynamic changes, and age-matched AF supports olfactory placode development.
(A) Total protein concentration of AF decreases from E8.5–E14.5; total CSF protein concentration increases from E10.5-E14.5 (mean protein concentration [mg/ml] ± SEM: E8.5 AF = 1.99 ± 0.06; E10.5 AF = 1.73 ± 0.13; E10.5 CSF = 1.70 ± 0.14; E14.5 AF = 1.51 ± 0.05; E14.5 CSF = 2.73 ± 0.26; n = 4 for all except E10.5 CSF, n=5; ANOVA; (*) p < 0.05 (***) p < 0.0001). (B) Silver stain of 1 μl AF or CSF suggests highest protein complexity in E8.5 AF. Asterisks denote representative proteins enriched in E8.5 AF (red) or E14.5 CSF (green). (C) Osmolality of CSF decreases from E8.5-E10.5, and then increases. Osmolality of AF decreases from E8.5-E14.5 (mean osmolality [mOsm/Kg] ± SEM: E8.5 AF = 303.0 ± 3.0 (n=3); E10.5 AF = 296.8 ± 2.2 (n=5); E10.5 CSF = 293.4 ± 4.4 (n=5); E12.5 AF = 292.5 ± 2.5 (n=4); E12.5 CSF = 297.0 ± 5.3 (n=3); E14.5 AF = 293.7 ± 1.2; E14.5 CSF = 298.0 ± 1.5; n=3; ANOVA, p < 0.05). Osmolality of E14.5 CSF is higher compared to E14.5 AF (t-test, (*) p < 0.05). (D) Schematic of E10.5 olfactory epithelium dissections: pairs of bilateral olfactory epithelial explants (denoted by dashed lines) placed on porous membrane contacting the AF. (E) E10.5 olfactory epithelial explants grown on E8.5 or E10.5 AF. Sox2:EGFP shows age-matched fluid most favorably supports neural stem cell identity (n=4). (F) qPCR of E10.5 OE explants cultured as in (E) show E10.5 AF more favorably promotes Sox2 expression in E10.5 OE explants compared to E8.5 AF (E8.5 AF=1.00; E10.5 AF = 1.78 ± 0.32; n=3 of 4 pooled explants each, p < 0.05, Mann-Whitney).
As the ventricular system expands during the course of early development, the CSF draws fluid from the AF, a process suggested to be driven in part by an osmolarity gradient (Alonso et al., 1998; Gato and Desmond, 2009). Despite the changes observed in total protein concentration between the fluids (Fig. 3A), we did not detect differences in global osmolality between AF and CSF during this time frame (Fig. 3C). These data agree with the model in which high colloid osmotic pressure, which typically constitutes a small fraction of the total osmotic pressure and is generated locally by individual or groups of proteins, drives ventricular expansion (Ek et al., 2005).
We next tested if the AF, like the CSF, harbors instructive signaling activities for adjacent tissues. We asked whether a cranial neurogenic placodal epithelium, where neural stem cells for peripheral neurons are established and maintained (Moody and LaMantia, 2015), depends selectively on the signaling capacity of the AF, which contacts these structures. We developed an E10.5 olfactory placode explant culture system (Fig. 3D), which gives rise to the olfactory sensory neurons and which under normal conditions directly contacts the E10.5 AF. The neurogenic phenotype of developing E10.5 olfactory epithelial cells was optimally supported by E10.5 AF vs. E8.5 AF, as assessed by Sox2:EGFP (Fig. 3E, F), and the lateral to medial patterning of Sox2 (Rawson et al., 2010; Tucker et al., 2010) was also optimally maintained in the presence of E10.5 AF. These data demonstrate that the AF can regulate cranial placode neuroepithelial cells in an age-and tissue-specific manner.
Quantitative proteomic analyses reveal dynamic transition from AF to CSF
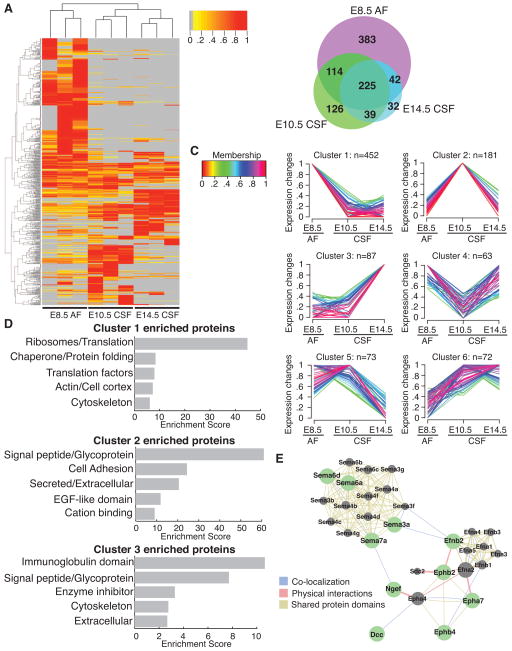
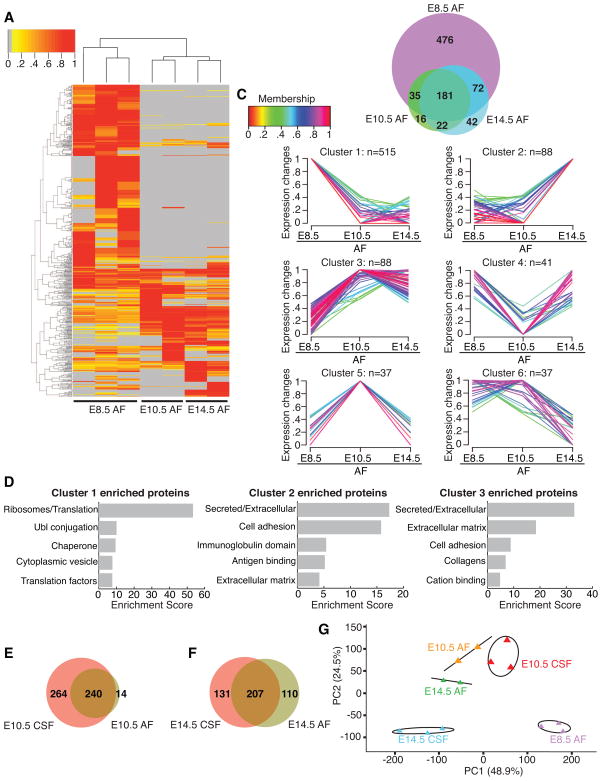
To characterize the differentiating CSF proteome as it emerges from E8.5 AF, we performed quantitative MS using label-free spectral counts on E8.5 AF, E10.5 CSF, and E14.5 CSF. Consistent with the silver staining results (Fig. 3B), E8.5 AF had the most complex proteome (764 proteins identified), followed by E10.5 CSF (504 proteins), while E14.5 CSF had the least complex proteome (410 proteins) (Fig. 4A, B, Table S1). All three fluid compartments shared 225 proteins (e.g. Apoa4, Fstl1) (Fig. 4A, B, Table S1). Beyond these shared constituents, we found 383 proteins exclusive to E8.5 AF (e.g. Eef1g, Rpl6), whereas 126 and 32 were exclusive to E10.5 CSF (e.g. Actg1, Efnb2) and E14.5 CSF (e.g. Blbp/Fabp7, Cdh13), respectively (Fig. 4A, B, Table S1).
Figure 4. Mass spectrometry reveals dynamic changes in CSF proteome as it differentiates from E8.5 AF to E14.5 CSF.
(A) Heatmap of normalized spectral counts reveals differential protein availability between E8.5 AF, E10.5 CSF and E14.5 CSF. Unsupervised hierarchical clustering grouped the three biological replicates of each fluid together. Each replicate contains 30μg total CSF protein pooled from multiple embryos. Spectral counts were scaled such that for each protein, the sample with the highest spectral count is set as one. Grey indicates undetected protein (i.e. 0 spectral counts). Proteins with peptides detected in at least two samples were included in the heatmap. (B) Total of 961 proteins were identified across all samples. Venn diagram depicts numbers of proteins exclusive to or shared between distinct fluid compartments. (C) Unsupervised clustering using GProX partitioned proteins into 6 clusters, revealing different temporal expression patterns for each cluster. Number of proteins in each cluster (n) is indicated above each graph. 33 proteins showed less than two-fold difference in availability between the fluids, and were not included in clustering. Membership indicates how well proteins fit into the general profile of cluster. (D) Functional annotation clustering of proteins in GProX clusters 1, 2 and 3 using DAVID. The top five enriched functional clusters are shown. (E) Proteins in GProX cluster 2 that are members of the axon guidance pathway were subjected to network analysis using GeneMania (Warde-Farley et al., 2010). Green nodes represent proteins members of cluster 2, whereas grey nodes represent related proteins. Lines connecting the nodes denote the relationship between the proteins. See also Table S1 and Figure S1.
Unsupervised clustering using GProX (Rigbolt et al., 2011) partitioned the proteomes into six individual clusters with distinct temporal expression patterns as the fluids diverged from E8.5 AF (Fig. 4C, Table S1). Cluster 1 consisted of 452 proteins that were more abundant at E8.5 AF than at later stages. Enrichment analysis using DAVID webtool (Huang et al., 2008; Huang et al., 2009a) showed that ribosomal proteins and other proteins involved in translation constituted the most enriched class of proteins in cluster 1 (Fig. 4D, Table S1). The early neuroepithelium releases membrane particles (Marzesco et al., 2005). Consistent with these findings, immunostaining analyses of some ribosomal protein subunits suggested to be enriched in E8.5 AF based on our MS analyses (e.g. Rps12 and Rpl11) were expressed in the E8.5 neuroepithelium at higher levels than in the E10.5 neuroepithelium (Fig. S1C, D), strongly suggesting that ribosomal protein content in E8.5 AF arises from normal membrane shedding that occurs during this age. Cluster 2 (181 members) represented proteins that were more abundant in E10.5 CSF than in E8.5 AF and E14.5 CSF. The most enriched class within cluster 2 was glycoproteins (Fig. 4D, Table S1), many of which were membrane proteins (e.g. cadherins, prominin1) and extracellular matrix (ECM) proteins (e.g. collagens, laminins, reelin), likely reflecting the active proliferation and membrane shedding of the neuroepithelium at E10.5. Supporting these data, KEGG pathway analysis of cluster 2 showed that ECM-receptor interaction (p-value = 5.89 × 10−9) and focal adhesion (p-value = 6.84 × 10−6) were the two most enriched pathways within this cluster (Table S1). The third most enriched pathway was axon guidance (p-value = 4.03 × 10−5, Table S1), with members such as semaphorins, ephrin receptors, ephrin-B2, and Dcc (netrin-1 receptor) (Fig. 4E, Table S1), reflecting the active cellular movement in the embryo at this age. Whether these activities have active roles in instructing migration, as has been suggested for CSF-Slit in adult (Sawamoto et al., 2006), remains to be determined. A number of proteins involved in canonical signaling pathways, including Bmp1, Mstn, Dkk3, and Tgfbr3, were also found in cluster 2 (Table S1), demonstrating that these activities are already present in E10.5 CSF, before the development of the choroid plexus. The availability of these proteins was downregulated by E14.5, suggestive of roles in early brain development. Cluster 3 (87 members) represented proteins enriched in E14.5 CSF, and the most enriched class within this cluster was proteins with immunoglobulin domains. Taken together, these data demonstrate that the AF-to-CSF transition during neural tube closure is accompanied by dynamic changes in protein availability and putative functions.
Canonical signaling activities in AF and CSF
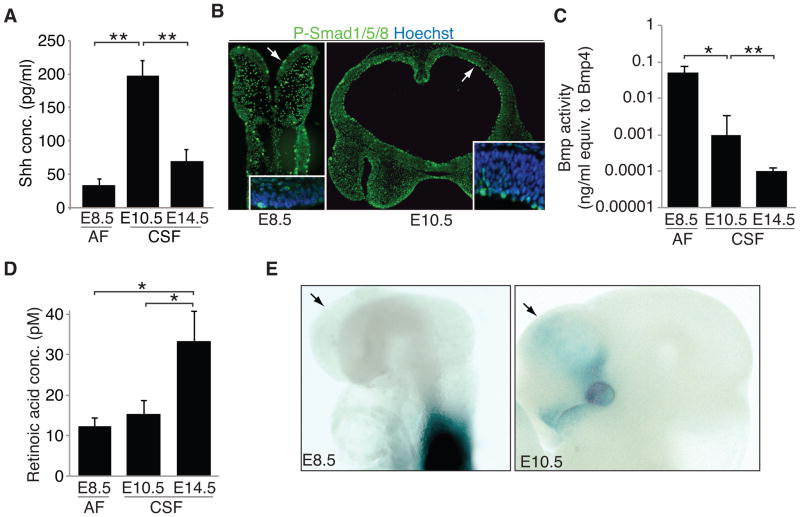
A number of canonical signals (e.g. Shh, Bmps, and RA) act on neural stem cells in the developing brain during the time when the CSF becomes a distinct fluid compartment (Wilde et al., 2014), and we found that these activities are tightly regulated in the nascent CSF during this time frame. First, from our MS analysis, the non-canonical Shh receptors, Boc and Gas1, were detected in E10.5 CSF (Table S1). Consistent with these data, CSF-Shh, quantified by ELISA, peaked in E10.5 CSF (Fig. 5A). The fourth ventricle choroid plexus secretes Shh into the CSF at later ages (Huang et al., 2010; Lun et al., 2015a); however, the choroid plexi have not yet developed at E10.5. Shh enrichment in the nascent CSF supports a model in which secretion of signals from cells adjacent to the ventricles, and/or diffusion of signals from surrounding tissues (e.g. floor plate), contribute to the CSF proteome, akin to secretion of Sema3B into the CSF in the spinal cord (Arbeille et al., 2014).
Figure 5. Canonical signaling activities present in AF and nascent CSF.
(A) Shh levels measured in E8.5 AF, E10.5 CSF, and E14.5 CSF by ELISA peaked in E10.5 CSF (mean Shh concentration [pg/ml] ± SEM: E8.5 AF = 31.13 ± 4.34; E10.5 CSF = 197.58 ± 12.71; E14.5 CSF = 70.68 ± 14.25). (B) P-SMAD 1/5/8/-positive staining progenitor cells along ventricular surface of E8.5 (left panel) and E10.5 (right panel) forebrain (arrows). Inset: high magnification image. Nuclei counterstained with Hoechst. (C) Bmp activity measured in E8.5 AF, E10.5 CSF, and E14.5 CSF as luciferase signal in clonally derived Bmp-sensitive cell line. Responses were compared to linear responses generated by pure Bmp4 (ng/ml) in the same cell line (data not shown). Overall Bmp activity decreases in nascent CSF (mean Bmp activity + SEM: E8.5 AF = 0.05 + 0.02; E10.5 CSF = 0.001 + 0.002; E14.5 CSF = 9.9×10−5 + 3.4×10−5; E8.5 AF vs. E10.5 CSF, t-test, p < 0.05; E10.5 CSF vs. E14.5 CSF, t-test, p < 0.01; data represent duplicate runs of biological replicates, n=3, from E8.5 AF and E14.5 CSF and n=2 for E14.5 CSF). (D) Retinoic acid (RA) activity measured in E8.5 AF, E10.5 CSF, and E14.5 CSF as luciferase signal in clonally derived RA-sensitive cell line. Responses were compared to linear responses generated by pure RA in the same cell line (data not shown). Overall RA activity increases in E14.5 CSF (mean RA activity + SEM: E8.5 AF = 12.3 + 2.0; E10.5 CSF = 15.3 + 3.4; E14.5 CSF = 33.4 + 7.4; t-test, p < 0.01; n=3; data represent duplicate runs of biological replicates, n=3, at each age. (E) RA indicator mice at E8.5 (left panel) with open neural tube and E10.5 (right panel) with closed neural tube. Arrows: presumptive, developing forebrain. RA signaling is not detected in the forebrain neuroectoderm of the open neural tube at E8.5. Signaling in the somites and spinal cord reflects local RA production by the somites (Stavridis et al., 2010; Vermot et al., 2005). At E10.5, RA signaling activity in the forebrain and head is modest, confined to the ventro-lateral forebrain, eye and nose, all reflecting local neural crest mesenchyme sources of RA (Haskell and LaMantia, 2005; LaMantia et al., 1993).
Previous studies have demonstrated activation of Bmp signaling in apical progenitor cells along the length of the dorsal telencephalic ventricular surface (Doan et al., 2012; Lehtinen et al., 2011), and we found Bmp signaling activity to change dynamically in the nascent CSF. Bmp4 availability measured by ELISA showed a trend to increase from E8.5 AF to E14.5 CSF, but was not significant (mean Bmp4 [nmol/ml] ± SEM: E8.5 AF = 745.1 ± 111.7; E10.5 CSF = 941.6 ± 201.4; E14.5 CSF = 1018.3 ± 231.4; n=3, not significant). We observed progressively increased phospho-SMAD1/5/8-positive staining cells along the ventricular surface during neurulation (mean forebrain phospho-SMAD1/5/8 frequency/mm ± SEM: E8.5 = 28.0 ± 1.6 [n=8]; E9.5 = 75.3 ± 3.2 [n=5]; E10.5 = 83.0 ± 2.9 [n=6]; E8.5 vs. E9.5, t-test, p < 0.0001; E9.5 vs. E10.5, t-test, p < 0.05 and Figure 5B), suggesting that these cells have the ability to transduce Bmp signals from the AF and CSF. In contrast, overall Bmp activity progressively decreased from E8.5 AF to E14.5 CSF, as revealed by a luciferase-based assay in a Bmp-sensitive cell line (Fig. 5C). Finally, retinoic acid (RA) activity was similarly low between E8.5 AF and E10.5 CSF, and increased by E14.5 (Fig. 5D), after choroid plexus formation, consistent with a requirement during initial neural tube formation to “protect” the anterior neural plate from the posteriorizing influence of RA (Durston et al., 1989), followed by increased RA signaling in more mature forebrain neural stem cells (Fig. 5E) (Haskell and LaMantia, 2005; LaMantia et al., 1993; Lehtinen et al., 2011; Siegenthaler et al., 2009). Together, these data show that signaling activities with established roles in brain development are dynamically available in the AF and nascent CSF and parallel changes in signaling in the neural progenitors.
Nascent CSF supports progenitor self-renewal
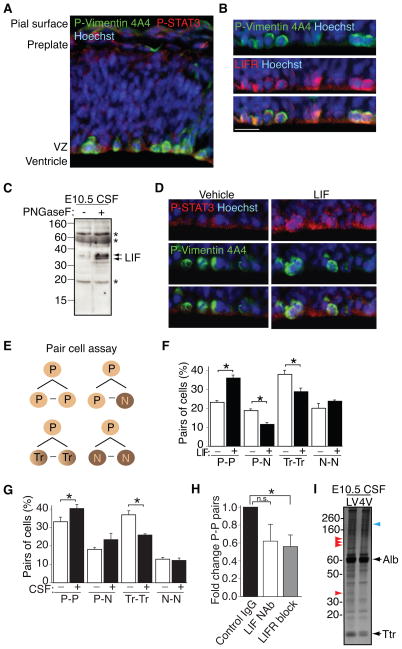
We next asked if these distinct, dynamic AF and CSF proteomes could be mined as a resource for signaling activities that might influence neural stem and progenitor progression. Proteomic analyses revealed the dynamic availability of many receptors from E8.5 AF-E14.5 CSF (e.g. TGFβ, EGFR, LIFR, representative of 41 proteins annotated as receptors by DAVID; Figs. S2A, B, C, Table S1). For example, availability of the Leukemia inhibitory factor (LIF) receptor (LIFR) declined during the course of AF to CSF fluid maturation (LIFR mean spectral counts ± SEM: E8.5 AF = 14.8 ± 1.2; E10.5 CSF = 7.8 ± 2.8; E14.5 CSF = not detected, n=3; Fig. S2C, Table S1). While LIFR was not detected by MS in E14.5 CSF, immunoblotting revealed bands consistent with some LIFR availability in E14.5 CSF (Fig. S2D). Soluble LIFR binds LIF and can inhibit its biological activity (Layton et al., 1992; Tomida, 2000). These observations suggest that neural tube closure could decrease access of an inhibitory factor into the CSF.
The LIF signaling pathway represents one compelling example of signaling activity in AF and nascent CSF. LIF regulates Sox2 expression in stem cells (Foshay and Gallicano, 2008; Niwa et al., 2009; Onishi and Zandstra, 2015; Pitman et al., 2004; Shimazaki et al., 2001). LIF also stimulates neural progenitor proliferation (Hatta et al., 2002) and influences cell fate at later developmental stages (Onishi and Zandstra, 2015). Thus, we tested if CSF-LIF mediated signaling regulates neural stem cells following neural tube closure. We observed membrane-associated LIFR and phospho-STAT3 in P-Vimentin-positive apical progenitor cells at E10.5 (Fig. 6A, B). In agreement with a previous report showing LIF availability in the CSF at E11 (Hatta et al., 2006), we detected LIF-reactive bands in E10.5 CSF treated with the deglycosylating enzyme PNGase F (Fig. 6C). Direct injection of recombinant LIF into E10.5 telencephalic ventricles ex vivo stimulated P-STAT3 activity in neuroepithelial progenitor cells (Fig. 6D). We then used a pair-cell assay to test for the functional effects of LIF on E10.5 neural progenitor self-renewal (Fig. 6E). Using Sox2 to identify neural progenitors and Tuj1 to identify neurons, we found that LIF stimulated self-renewal (progenitor-progenitor division; P-P) of Sox2-positive neural progenitors at the expense of differentiation (progenitor-neuron division; P-N) or transient cells that co-express Sox2 and Tuj1 (Transient-Transient division; Tr-Tr; Hutton and Pevny, 2011; Fig. 6F). Pair-cell assays using 20% E10.5 CSF revealed a similar ability to promote progenitor self-renewal (Fig. 6G). The frequency of Sox2-positive P-P divisions declined in cells cultured with CSF containing LIFR blocking or LIF neutralization (NAb) antibodies (Fig. 6H), showing that CSF-based signaling via the LIFR can promote neuroepithelial cell self-renewal, though other CSF-borne signals likely work in concert in vivo to regulate this process.
Figure 6. Nascent CSF promotes signaling and self-renewal in progenitor cells.
(A) E10.5 forebrain shows P-STAT3 (red) activity in P-Vimentin-positive (green) progenitor cells along ventricular surface. Nuclei counterstained with Hoechst. (B) Higher magnification images of E10.5 forebrain show LIFR (red) expression in P-Vimentin-positive (green) progenitor cells along ventricular surface. Nuclei counterstained with Hoechst. Scale bar, 20 μm. (C) Removal of N-linked glycans from E10.5 CSF with PNGase F reveals robust bands immunoreactive for LIF. (D) Intraventricular LIF injection (15 minutes; 200ng/ml) at E10.5 stimulates P-STAT3 (red) signaling in P-Vimentin-positive (green) progenitor cells along ventricular surface. Nuclei counterstained with Hoechst. (E) Schematic depicting potential combinations of daughter cells in pair cell assay (P – progenitor; N – neuron; Tr – transient/intermediate cell type co-expressing Sox2 and Tuj1). (F) LIF stimulates Sox2-positive self-renewal (P-P) of progenitor cells at expense of differentiating cells (P-N; Tr-Tr; identities of pairs of cells represented as mean ± SEM; P-P division: control = 23.3 ± 0.8; LIF = 35.9 ± 1.6; P-N division: control = 18.8 ± 0.9; LIF = 11.6 ± 0.9; Tr-Tr division: control = 38.0 ± 1.9; LIF = 28.8 ± 2.0; N-N division: control = 20.1 ± 2.5; LIF = 23.7 ± 0.6; n=3; p < 0.0001; ANOVA). (G) E10.5 CSF (20%) stimulates Sox2-positive self-renewal (P-P) of progenitor cells at expense of differentiating cells (data presented as above in (F)). P-P division: control = 33.3 ± 2.5; CSF = 40.2 ± 2.2; P-N division: control = 18.2 ± 0.9; CSF = 23.4 ± 3.4; Tr-Tr division: control = 36.8 ± 2.1; CSF = 26.0 ± 0.4; N-N division: control = 12.8 ± 0.8; CSF = 12.2 ± 1.3; n=3; p < 0.05; ANOVA). (H) Interference with LIF signaling in E10.5 CSF (20%) decreases the proportion of P-P division. Data presented as fold change relative to Control IgG (anti-GATA6) (Control IgG = 1.0; LIF neutralization (NAb) = 0.62 ± 0.19; LIFR block = 0.56 ± 0.13; Control vs. LIF NAb, p = n.s.; Control vs. LIFR block, p<0.05; n=3 with approximately 100 pairs counted per condition; Mann-Whitney). (I) Silver staining shows distinct protein patterns in E10.5 CSF collected from lateral vs. fourth ventricles. Arrowheads denote protein bands differentially detected in lateral ventricle CSF (red) vs. fourth ventricle CSF (blue). See also Figure S2.
Lifr-deficient mice are microcephalic by E12.5, a time when LIF is beginning to be produced by the choroid plexus (Gregg and Weiss, 2005). Because the choroid plexus is not yet developed at E10.5, our findings suggest that CSF-LIF must originate from another source to contribute to progenitor proliferation at this earlier developmental stage. While we cannot rule out a contribution by the neuroepithelium (Fig. S2E), LIF is abundantly expressed by the amniotic sac and yolk sac (Fig. S2E). Consistent with CSF drawing fluid from the AF at this stage (Alonso et al., 1998), 10 kDa fluorescein-conjugated dextrans injected into the intra-amniotic space of E10.5 mouse embryos were detected in the CSF 30 minutes following injection (average relative fluorescent units: 10 kDa dextran = 445 and 153.3; Vehicle = 21.3; 33.7; and 50.1). These data suggest that factors available in the AF—possibly including LIF—can reach the CSF by classic diffusion through the neuroepithelium at this age. The zebrafish neuroepithelium is permeable to fluorescent dyes up to 70kDa (Chang and Sive, 2011).
Our proteomic and functional analyses were performed using pooled samples of AF and CSF. However, silver staining analyses of lateral and fourth ventricle CSF revealed that the CSF proteome was regionalized, with distinct protein bands detected in each sample already at E10.5 (Fig. 6I), prior to choroid plexus-driven regionalization that occurs at later ages (Lun et al., 2015a). These data suggest differential diffusion or secretion of proteins into each CSF-filled ventricular sub-compartment, and potentially, differential effects on local progenitor progression.
Quantitative proteomic analysis of maturing AF
We have limited knowledge regarding the mouse AF proteome and how it develops over time. Based upon the observation that E10.5 AF supports the developing olfactory placode (Fig. 3E), we performed quantitative MS to reveal changes in the AF proteome as it develops from E8.5 to E10.5 and E14.5. The composition of E10.5 AF and E14.5 AF were less complex than E8.5 AF (Fig. 7A, Table S2). Only 80 proteins found in the older AF samples were not detected at E8.5 (Fig. 7B, Table S2). Conversely, 476 proteins were exclusive to E8.5 AF (Fig. 7B, Table S2). Unsupervised clustering of expressed proteins followed by enrichment analysis revealed that many proteins enriched in E8.5 AF were ribosomal proteins and translation factors (Fig. 7C, D, Table S2; cluster 1, 515 members). E14.5 AF was enriched in proteins that were secreted (e.g. collagens) or important for cell adhesion (e.g. Ncam1, Vcam1) (Fig. 7C, D, Table S2; cluster 2, 88 members). Cluster 3 (88 members) hosted proteins that were more abundant at later ages than at E8.5 (Fig. 7C, Table S2). Many of these proteins were secreted (e.g. Dkk3, Igf2) or ECM proteins (e.g. collagens, fibronectin; Fig. 7D, Table S2). Only 37 proteins were enriched in E10.5 AF (Fig. 7C, Table S2; cluster 5), and some of these (e.g. Gas1, Mstn, Tgfbr3) were members of canonical signaling pathways.
Figure 7. Mass spectrometry reveals decreasing complexity of AF as embryo develops from E8.5 to E14.5.
(A) Heatmap of normalized spectral counts reveals differential protein availability in AF between E8.5, E10.5 and E14.5. Unsupervised hierarchical clustering grouped biological replicates of each fluid together (E8.5: n=3; E10.5: n=2; E14.5: n=2). Each replicate contains 30μg total AF protein pooled from multiple embryos. Spectral counts were scaled and analyzed as in Fig. 4A. (B) Total of 844 proteins identified across all AF samples are represented as in Fig. 4B. (C) Unsupervised clustering was performed and data are presented as in Fig. 4C. 38 proteins showed less than two-fold difference in availability between the ages, and were not included in the clustering. (D) Functional annotation clustering of proteins in GProX clusters 1, 2 and 3 was performed and is presented as in Fig. 4D. (E) At E10.5, 264 and 14 proteins are exclusive to CSF and AF respectively, and 240 proteins are common to both fluid compartments. (F) By E14.5, 131 and 110 proteins are exclusive to CSF and AF respectively, and 207 proteins are shared between the two fluid compartments. (G) Principal component analysis of protein spectral counts revealed greater degree of variance between fluid compartments than between biological replicates within each compartment. E8.5 AF and E14.5 CSF were the most distinct fluid compartments, whereas E10.5 CSF, E10.5 AF and E14.5 AF were more similar to each other. PC1 and PC2 explained 48.9% and 24.5% of the total variance respectively. Ellipses and lines indicate the 95% confidence interval. See also Table S2.
Despite the expansion of the brain’s ventricles between E8.5-E10.5 (Fig. 1), direct comparison of the AF and CSF proteomes revealed that only 14 proteins found in E10.5 AF were not detected in E10.5 CSF (Fig. 7E). These data are consistent with the model that the CSF draws fluid and proteins from the AF (see above; Alonso et al., 1998; Chang & Sive, 2011). Conversely, many proteins were enriched in the E10.5 CSF vs. E10.5 AF, including Shh (mean Shh concentration [pg/ml] represented as mean ± SEM at E10.5: AF = 50.6 ± 11.9; CSF = 197.6 ± 12.7; n = 3; p < 0.01, t-test; see also Fig. 5A). Importantly, 264 proteins in E10.5 CSF were not detected in E10.5 AF, indicating other sources including the adjacent neuroepithelium (e.g. Marzesco et al., 2005) as major contributors to the nascent CSF proteome (Fig. 7E). By E14.5, the CSF and AF were even more distinct from each other, with 131 proteins exclusively detected in E14.5 CSF and 110 proteins exclusively detected in E14.5 CSF (Fig. 7F), reflecting progressive, age-dependent divergence of the AF and CSF. Principal component analysis showed that E8.5 AF and E14.5 CSF were the most distinct fluid compartments, whereas E10.5 CSF, E10.5 AF and E14.5 AF were more similar to each other (Fig. 7G). Thus, the developing AF and CSF undergo a progressive differentiation with instructive capacities matched to adjacent neurogenic tissues.
DISCUSSION
We present a resource in which we have: 1) developed tools to collect AF and CSF before and after neural tube closure, 2) quantified ventricular expansion in live mouse embryos before and after neural tube closure, 3) systematically documented the dynamic signaling capacity, and protein content of the AF and CSF proteomes as they differentiate over the course of embryonic development, 4) developed robust, heterochronic ex vivo explant systems to assess functional, instructive activities for AF and nascent CSF in promoting neural progenitor cell fate in the developing neural plate, neural tube, and brain as well as the neurogenic cranial placodes, and 5) demonstrated that CSF-based signaling via the LIFR stimulates neuroepithelial cell self-renewal. Paired with the dynamic morphological features of the early developing head and brain, our findings should accelerate future studies investigating the regulation of fluid/tissue interactions and their consequences for signaling during these early, understudied stages of brain development. Since many tissues of the developing embryo, particularly the precursors of the central and peripheral nervous system, are in contact with AF/CSF, our proteomic dataset serves as an important resource for investigators across multiple developmental fields that are interested in studying fluid-tissue interactions and related mechanisms during mid-gestation when embryos are particularly vulnerable to the consequences of disrupted signaling.
Analyzing fluid proteomes and their activities in early embryos
We addressed several challenges in developing this resource for understanding AF and CSF function in mammalian embryos. A number of studies have used proteomics tools to analyze the AF or CSF at older ages or in other species (e.g., Chang et al., 2015; Dislich et al., 2015; Dziegelewska et al., 1981; Lehtinen et al., 2011; Lun et al., 2015a; Parada et al., 2005; Zappaterra et al., 2007). However, to our knowledge, analysis of the murine AF proteome over the course of early development has not been undertaken, likely due to challenges of reliably collecting small amounts of AF and CSF from early mammalian embryos. We minimized variability in AF collection and preparation, with careful dissection, pooling samples across litters, and assessment of contamination by embryonic or maternal tissues. We similarly standardized CSF collection from embryos after neural tube closure and emergence of distinct brain vesicles. While the CSF between ventricles is regionalized by E10.5 (Fig. 6I), analysis of total CSF samples from individual embryos using silver stained gels indicated basic uniformity within and across litters (data not shown). MS analyses showed quantitative differences between pooled AF and CSF samples, which may indicate biological variability independent of technical issues, or subtle differences in embryo stages when fluids are collected. Some variability suggested by MS may not reach a threshold for impacting signaling capacity. Quantitative MS may also be limited in its ability to detect low abundance proteins, proteins that have been processed into small peptides, and proteins with other physicochemical properties (e.g. hydrophobicity). However, our analyses of cardinal signaling activity (e.g. Bmps, RA, Shh) support the model that overall AF and CSF signaling activity is stable among pooled samples. We also detected some variation in AF and CSF activity on neural progenitors in our explant assays; however, these differences may reflect details of the reporter allele (Arnold et al., 2011; Ellis et al., 2004) rather than distinctions of AF or CSF activity on the neuroepithelium. Pair-cell analyses of LIF, a likely AF/CSF signal, indicate stage-specific consistency. Future studies analyzing the influence of AF/CSF on neural stem cell identities in functional assays will further elucidate the biological significance of variability in AF and CSF proteomes.
The Signaling Capacity of the AF and CSF
The AF and nascent CSF have dynamic morphogenetic signaling capacity via Bmps, RA, and Shh. The effective activity of these signals varies in register with patterning in the embryo, including the brain and the neurogenic cranial placodes. The decrease in total Bmp activity during the transition from E8.5 AF to E10.5 CSF may indicate changes in inhibition. For example, Fstl1, a Bmp4 antagonist (Geng et al., 2011), was more abundant in E10.5 CSF than in E8.5 AF, while Bmp4 was identified in each fluid compartment. The timed delivery of fluid-based signals is also demonstrated by the delayed availability of RA in the E14.5 CSF, presumably protecting the developing brain from posteriorizing cues, while permitting local signaling via cranial mesenchymal sources once the neural tube closes (Moody and LaMantia, 2015). Finally, our findings of elevated Shh in E10.5 CSF suggest that the developing dorsal forebrain, as well as the ventral forebrain that is thought to receive Shh signals from local tissue sources (Chiang et al., 1996; Rallu et al., 2002b), is exposed to Shh quite early in embryrogenesis. In all cases, the action of CSF-borne signals on target cells relies on appropriate receptor expression and ability to transduce signals delivered by the CSF.
Since no individual tissue is solely responsible for the production of AF and nascent CSF at the early ages examined (e.g. E8.5 and E10.5; the choroid plexus contributes to CSF production in our E14.5 samples), there is no transcriptome from which to derive the early AF and CSF proteomes. Therefore, our MS approach provides a singular directory of hundreds of differentially available proteins in the maturing AF and CSF over time. The diversity of proteins identified in early AF and CSF includes growth factors and their receptors (e.g. Boc and Gas1 for Shh), axon guidance molecules (e.g. semaphorins, DCC, ephrins), and extracellular matrix components (e.g. reelin, laminin subunits). The presence of prominin-1-containing membrane particles (Dubreuil et al., 2007; Marzesco et al., 2005) in the E10.5 and E14.5 CSF, but not E8.5 AF (Table S1) suggest that the developing neuroepithelium contributes to the CSF proteome. Whether these particles have an active role in CSF-based signaling (Cossetti et al., 2014; Feliciano et al., 2014), or are cellular waste products cleared from the developing neural tissue, remains to be determined. Nevertheless, signaling components of many of the pathways represented in the AF and CSF proteomes have roles in neural tube closure (Wilde et al., 2014), inviting hypotheses for fluid-based regulation of this developmental stage.
AF and CSF proteomes and hypotheses for early mammalian brain development
The resources of AF/CSF proteomes, activities, and embryo-based assays to evaluate signaling capacities also provide an essential foundation for generating and testing hypotheses of mammalian neural development and how it is compromised by genetic and environmental disruption. We found that AF and CSF vary in several dimensions within 48 hours during early to mid-stage mouse embryogenesis. Changes in osmolality and fluid volumes may have substantial consequences for morphogenetic movements that maintain the neural plate, and then support neural tube closure. Importantly, distinctions in AF/CSF protein constituents—especially metabolic and receptor proteins in the fluids at different stages—suggest key changes in cells of embryonic surface tissues that directly contact AF or CSF during the transition from neural plate to forebrain vesicle epithelium. Such changes may be driven by the AF versus CSF proteomes, and may be metabolic indicators of molecular and cellular distinctions in embryonic tissues that influence stem cell progression and early neurogenesis. Despite tremendous focus on neural stem cell progression at later stages of brain development in the spinal cord, hindbrain, and forebrain (e.g. Dias et al., 2014), there has been far less attention paid to the key transition between early neuroectodermal stem cell and neural stem cell identity as the neural tube closes. Mining of our proteomic resource, combined with use of our assays to evaluate stem cell progression will provide important insights into mechanisms that mediate this key period of mammalian brain development.
The early AF and CSF proteomes reflect, in varying degrees, the balance between maternally derived protein constituents and other small molecules, and those derived from the embryo. These proteomes are likely altered dramatically in response to maternal illness, changes in nutrition, substance abuse, and exposure to environmental toxins (Wilde et al., 2014). Thus, the definition of dynamic AF and CSF activities we have provided constitutes an essential resource for understanding risk and consequence of maternal/fetal interactions during first trimester development.
EXPERIMENTAL PROCEDURES
Animals
Timed pregnant CD1 dams were obtained from Charles River Laboratories. Sox2:EGFP (Ellis et al., 2004) was shared by L. Pevny. All animal experimentation was carried out under protocols approved by the IACUCs of Boston Children’s Hospital and The George Washington University School of Medicine.
Ventricular system measurements
Uterine horns were placed on Sylgard plates, immersed in PBS, and ultrasound image sequences were captured (Vevo 2100, Visual Sonics; MS550 transducer; step size: 0.076mm). 3D reconstruction of ventricular system was performed for visualization (Vevo 2100) and quantification (Fiji-TrakEM).
AF and CSF collection and measurements
AF was collected by inserting a glass capillary into the intra-amniotic space. CSF was collected by inserting glass capillary into telencephalic ventricles, aqueduct, fourth ventricle or cisterna magna, and processed as described (Zappaterra et al., 2013). Protein concentration (BCA kit) and osmolality (10μl fluid input, Vapro 5600) were measured.
Forebrain neuroectodermal explants
E8.5 anterior neuroectoderm containing the presumptive forebrain, was dissected from the anterior neural ridge to the optic vesicle. E10.5 dorsal telencephalic neuroepithelium was isolated by: 1] dissecting the cortical neuroepithelium caudal to the olfactory bulb at the anterior boundary of the lateral ganglionic eminence (LGE), 2] dissecting caudal to the posterior boundary of the LGE and extending to the medial wall of the developing cortical rudiment, 3] separating the LGE from the developing cortex, and 4] removing the midline at the apex of the neocortex where the lateral cortical surface meets the interhemispheric wall. Following 30 minutes of enzymatic digestion with pancreatin on ice, neuroepithelium was separated from mesenchyme and surface ectoderm using tungsten needles. Explants were placed on polycarbonate membranes (8.0μm pore size; Whatman) in imaging dishes (Mat-Tek), and cultured in 20% AF or CSF in Neurobasal medium supplemented with antibiotics and L-glutamine. Explants were cultured for 24 hours.
Supplementary Material
Acknowledgments
We thank C. Walsh for helpful discussions; E. Paronett, T. Adelita, and A. Malesz for technical assistance; J. Steen for RPS12 and RPL11 antibodies. We are grateful for the following support: NSF Graduate Research Fellowship (KFC), NIH R01 DC011534 (ASL), Pediatric Hydrocephalus Foundation, Eleanor and Miles Shore Fellowship Program for Scholars in Medicine/Boston Children’s Hospital Career Development Award, Alfred P. Sloan Foundation, BCH IDDRC P30 HD18655, NIH K99/R00 NS072192 and R01 NS088566 (MKL).
Footnotes
AUTHOR CONTRIBUTIONS
K.F.C., M.W.S., H.S., A.S.L., and M.K.L. designed research; K.G.B. and H.S. developed proteomic tools and analysis pipeline; K.F.C., M.W.S., K.G.B., H.P., S.T., M.P.L., H.M., T.M., H.S., A.S.L., and M.K.L. performed research; K.F.C., M.W.S., K.G.B., H.P., S.T., T.M., H.S., A.S.L., and M.K.L. analyzed data; K.F.C. and M.K.L. wrote the manuscript; all co-authors edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso MI, Gato A, Moro JA, Barbosa E. Disruption of proteoglycans in neural tube fluid by beta-D-xyloside alters brain enlargement in chick embryos. Anat Rec. 1998;252:499–508. doi: 10.1002/(SICI)1097-0185(199812)252:4<499::AID-AR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Arbeille E, Reynaud F, Sanyas I, Bozon M, Kindbeiter K, Causeret F, Pierani A, Falk J, Moret F, Castellani V. Cerebrospinal fluid-derived Semaphorin3B orients neuroepithelial cell divisions in the apicobasal axis. Nat Commun. 2014;6:6366. doi: 10.1038/ncomms7366. [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson CS, Apostolopoulou M, Tian Y, Temple S. It Takes a Village: Constructing the Neurogenic Niche. Developmental Cell. 2015;32:435–446. doi: 10.1016/j.devcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Sive H. An assay for permeability of the zebrafish embryonic neuroepithelium. J Vis Exp. 2011:e4242. doi: 10.3791/4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Lehtinen MK, Sive H. Zebrafish cerebrospinal fluid mediates cell survival through a retinoid signaling pathway. Dev Neurobiol. 2015 doi: 10.1002/dneu.22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi C, Cunningham TJ, Duester G. Investigation of retinoic acid function during embryonic brain development using retinaldehyde-rescued Rdh10 knockout mice. Dev Dyn. 2013;242:1056–1065. doi: 10.1002/dvdy.23999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Cossetti C, Iraci N, Mercer TR, Leonardi T, Alpi E, Drago D, Alfaro-Cervello C, Saini HK, Davis MP, Schaeffer J, et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56:193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond ME, Jacobson AG. Embryonic brain enlargement requires cerebrospinal fluid pressure. Dev Biol. 1977;57:188–198. doi: 10.1016/0012-1606(77)90364-5. [DOI] [PubMed] [Google Scholar]
- Dias JM, Alekseenko Z, Applequist JM, Ericson J. Tgfβ signaling regulates temporal neurogenesis and potency of neural stem cells in the CNS. Neuron. 2014;84:927–939. doi: 10.1016/j.neuron.2014.10.033. [DOI] [PubMed] [Google Scholar]
- Dislich B, Wohlrab F, Bachhuber T, Mueller S, Kuhn PHH, Hogl S, Meyer-Luehmann M, Lichtenthaler SF. Label-free quantitative proteomics of mouse cerebrospinal fluid detects BACE1 protease substrates in vivo. Mol Cell Proteomics. 2015;14:2550–2563. doi: 10.1074/mcp.M114.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan LT, Javier AL, Furr NM, Nguyen KL, Cho KW, Monuki ES. A Bmp reporter with ultrasensitive characteristics reveals that high Bmp signaling is not required for cortical hem fate. PLoS One. 2012;7:e44009. doi: 10.1371/journal.pone.0044009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Heideveld M, Nieuwkoop PD. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- Dziegielewska KM, Evans CA, Lai PC, Lorscheider FL, Malinowska DH, Møllgård K, Saunders NR. Proteins in cerebrospinal fluid and plasma of fetal rats during development. Dev Biol. 1981;83:193–200. doi: 10.1016/s0012-1606(81)80024-3. [DOI] [PubMed] [Google Scholar]
- Ek CJ, Dziegielewska KM, Saunders NR. Development of the blood-cerebrospinal fluid barrier. In: Zheng W, Chodobski A, editors. The blood-cerebrospinal fluid barrier. Boca Raton: Taylor & Francis; 2005. pp. 3–23. [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Feliciano DM, Zhang S, Nasrallah CM, Lisgo SN, Bordey A. Embryonic cerebrospinal fluid nanovesicles carry evolutionarily conserved molecules and promote neural stem cell amplification. PLoS One. 2014;9:e88810. doi: 10.1371/journal.pone.0088810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foshay KM, Gallicano GI. Regulation of Sox2 by STAT3 initiates commitment to the neural precursor cell fate. Stem Cells Dev. 2008;17:269–278. doi: 10.1089/scd.2007.0098. [DOI] [PubMed] [Google Scholar]
- Gato A, Desmond ME. Why the embryo still matters: CSF and the neuroepithelium as interdependent regulators of embryonic brain growth, morphogenesis and histiogenesis. Dev Biol. 2009;327:263–272. doi: 10.1016/j.ydbio.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Geng Y, Dong Y, Yu M, Zhang L, Yan X, Sun J, Qiao L, Geng H, Nakajima M, Furuichi T, et al. Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc Natl Acad Sci USA. 2011;108:7058–7063. doi: 10.1073/pnas.1007293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Weiss S. CNTF/LIF/gp130 receptor complex signaling maintains a VZ precursor differentiation gradient in the developing ventral forebrain. Development. 2005;132:565–578. doi: 10.1242/dev.01592. [DOI] [PubMed] [Google Scholar]
- Grove EA, Monuki ES. Morphogens, patterning centers, and their mechanisms of action. In: Rubenstein JL, Rakic P, editors. Patterning and cell type specification in the developing CNS and PNS. San Diego: Elsevier, Inc; 2013. pp. 25–44. [Google Scholar]
- Haskell GT, LaMantia AS. Retinoic acid signaling identifies a distinct precursor population in the developing and adult forebrain. J Neurosci. 2005;25:7636–7647. doi: 10.1523/JNEUROSCI.0485-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta T, Matsumoto A, Ono A, Udagawa J, Nimura M, Hashimoto R, Otani H. Quantitative analyses of leukemia inhibitory factor in the cerebrospinal fluid in mouse embryos. Neuroreport. 2006;17:1863–1866. doi: 10.1097/WNR.0b013e3280113962. [DOI] [PubMed] [Google Scholar]
- Hatta T, Moriyama K, Nakashima K, Taga T, Otani H. The Role of gp130 in cerebral cortical development: in vivo functional analysis in a mouse exo utero system. J Neurosci. 2002;22:5516–5524. doi: 10.1523/JNEUROSCI.22-13-05516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Guo J, Yokota Y, Umberger N, Su C, Li J, Verma N, Hirt J, Ghukasyan V, Caspary T, Anton ES. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci. 2013;16:1000–1007. doi: 10.1038/nn.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ketova T, Fleming JT, Wang H, Dey SK, Litingtung Y, Chiang C. Sonic hedgehog signaling regulates a novel epithelial progenitor domain of the hindbrain choroid plexus. Development. 2009b;136:2535–2543. doi: 10.1242/dev.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc Natl Acad Sci USA. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KE, Hatten ME. Radial glial cell transformation to astrocytes is bidirectional: regulation by a diffusible factor in embryonic forebrain. Proc Natl Acad Sci USA. 1995;92:2061–2065. doi: 10.1073/pnas.92.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352:40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Colbert MC, Linney E. Retinoic acid induction and regional differentiation prefigure olfactory pathway formation in the mammalian forebrain. Neuron. 1993;10:1035–1048. doi: 10.1016/0896-6273(93)90052-s. [DOI] [PubMed] [Google Scholar]
- Layton MJ, Cross BA, Metcalf D, Ward LD, Simpson RJ, Nicola NA. A major binding protein for leukemia inhibitory factor in normal mouse serum: identification as a soluble form of the cellular receptor. Proc Natl Acad Sci USA. 1992;89:8616–8620. doi: 10.1073/pnas.89.18.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays. 2009;31:446–458. doi: 10.1002/bies.200800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun MP, Johnson MB, Broadbelt KG, Watanabe M, Kang YJ, Chau KF, Springel MW, Malesz A, Sousa AMM, Pletikos M, et al. Spatially Heterogeneous Choroid Plexus Transcriptomes Encode Positional Identity and Contribute to Regional CSF Production. J Neurosci. 2015a;35:4903–4916. doi: 10.1523/JNEUROSCI.3081-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat Rev Neurosci. 2015b;16:445–457. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–339. doi: 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118:2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- Massarwa R, Niswander L. In toto live imaging of mouse morphogenesis and new insights into neural tube closure. Development. 2013;140:226–236. doi: 10.1242/dev.085001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- Moody SA, LaMantia AS. Transcriptional regulation of cranial sensory placode development. Curr Top Dev Biol. 2015;111:301–350. doi: 10.1016/bs.ctdb.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Onishi K, Zandstra PW. LIF signaling in stem cells and development. Development. 2015;142:2230–2236. doi: 10.1242/dev.117598. [DOI] [PubMed] [Google Scholar]
- Parada C, Gato A, Bueno D. Mammalian embryonic cerebrospinal fluid proteome has greater apolipoprotein and enzyme pattern complexity than the avian proteome. J Proteome Res. 2005;4:2420–2428. doi: 10.1021/pr050213t. [DOI] [PubMed] [Google Scholar]
- Pitman M, Emery B, Binder M, Wang S, Butzkueven H, Kilpatrick TJ. LIF receptor signaling modulates neural stem cell renewal. Mol Cell Neurosci. 2004;27:255–266. doi: 10.1016/j.mcn.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Rallu M, Corbin JG, Fishell G. Parsing the prosencephalon. Nat Rev Neurosci. 2002a;3:943–951. doi: 10.1038/nrn989. [DOI] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002b;129:4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- Rawson NE, Lischka FW, Yee KK, Peters AZ, Tucker ES, Meechan DW, Zirlinger M, Maynard TM, Burd GB, Dulac C, et al. Specific mesenchymal/epithelial induction of olfactory receptor, vomeronasal, and gonadotropin-releasing hormone (GnRH) neurons. Dev Dyn. 2010;239:1723–1738. doi: 10.1002/dvdy.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Vanselow JT, Blagoev B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics. 2011;10:O110. doi: 10.1074/mcp.O110.007450. 007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JL, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shimazaki T, Shingo T, Weiss S. The ciliary neurotrophic factor/leukemia inhibitory factor/gp130 receptor complex operates in the maintenance of mammalian forebrain neural stem cells. J Neurosci. 2001;21:7642–7653. doi: 10.1523/JNEUROSCI.21-19-07642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell. 2009;139:597–609. doi: 10.1016/j.cell.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridis MP, Collins BJ, Storey KG. Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development. 2010;137:881–890. doi: 10.1242/dev.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida M. Structural and functional studies on the leukemia inhibitory factor receptor (LIF-R): gene and soluble form of LIF-R, and cytoplasmic domain of LIF-R required for differentiation and growth arrest of myeloid leukemic cells. Leuk Lymphoma. 2000;37:517–525. doi: 10.3109/10428190009058503. [DOI] [PubMed] [Google Scholar]
- Tong CK, Han YG, Shah JK, Obernier K, Guinto CD, Alvarez-Buylla A. Primary cilia are required in a unique subpopulation of neural progenitors. Proc Natl Acad Sci USA. 2014;111:12438–12443. doi: 10.1073/pnas.1321425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, Lamantia ASS. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137:2471–2481. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ES, Segall S, Gopalakrishna D, Wu Y, Vernon M, Polleux F, Lamantia AS. Molecular specification and patterning of progenitor cells in the lateral and medial ganglionic eminences. J Neurosci. 2008;28:9504–9518. doi: 10.1523/JNEUROSCI.2341-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Gallego Llamas J, Fraulob V, Niederreither K, Chambon P, Dollé P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, McCaffery P, Drager UC. Influence of the choroid plexus on cerebellar development: analysis of retinoic acid synthesis. Brain Res Dev Brain Res. 1996;93:182–190. doi: 10.1016/0165-3806(96)00038-7. [DOI] [PubMed] [Google Scholar]
- Yeh C, Li A, Chuang JZ, Saito M, Caceres A, Sung CH. IGF-1 activates a cilium-localized noncanonical Gbetagamma signaling pathway that regulates cell-cycle progression. Dev Cell. 2013;26:358–368. doi: 10.1016/j.devcel.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MW, LaMantia AS, Walsh CA, Lehtinen MK. Isolation of cerebrospinal fluid from rodent embryos for use with dissected cerebral cortical explants. J Vis Exp. 2013:e50333. doi: 10.3791/50333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MW, Lehtinen MK. The cerebrospinal fluid: regulator of neurogenesis, behavior, and beyond. Cell Mol Life Sci. 2012;69:2863–2878. doi: 10.1007/s00018-012-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MD, Lisgo SN, Lindsay S, Gygi SP, Walsh CA, Ballif BA. A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J Proteome Res. 2007;6:3537–3548. doi: 10.1021/pr070247w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.