Abstract
The acquisition of mannose 6-phosphate (Man6P) on N-linked glycans of lysosomal enzymes is a structural requirement for their transport from the Golgi apparatus to lysosomes mediated by the mannose 6-phosphate receptors, 300 kDa cation-independent mannose 6-phosphate receptor (MPR300) and 46 kDa cation-dependent mannose 6-phosphate receptor (MPR46). Here we report that the single-chain variable domain (scFv) M6P-1 is a unique antibody fragment with specificity for Man6P monosaccharide that, through an array-screening approach against a number of phosphorylated N-glycans, is shown to bind mono- and diphosphorylated Man6 and Man7 glycans that contain terminal αMan6P(1 → 2)αMan(1 → 3)αMan. In contrast to MPR300, scFv M6P-1 does not bind phosphodiesters, monophosphorylated Man8 or mono- or diphosphorylated Man9 structures. Single crystal X-ray diffraction analysis to 2.7 Å resolution of Fv M6P-1 in complex with Man6P reveals that specificity and affinity is achieved via multiple hydrogen bonds to the mannose ring and two salt bridges to the phosphate moiety. In common with both MPRs, loss of binding was observed for scFv M6P-1 at pH values below the second pKa of Man6P (pKa = 6.1). The structures of Fv M6P-1 and the MPRs suggest that the change of the ionization state of Man6P is the main driving force for the loss of binding at acidic lysosomal pH (e.g. lysosome pH ∼ 4.6), which provides justification for the evolution of a lysosomal enzyme transport pathway based on Man6P recognition.
Keywords: crystal structure, glycan array, lysosome, N-glycosylation, scFv
Introduction
High mannose-type Asn-linked oligosaccharides (N-glycans) terminating in mannose 6-phosphate (Man6P) function as recognition markers for the transport of lysosomal hydrolases from the Golgi to lysosomes (Luzio et al. 2007; Braulke and Bonifacino 2009). Man6P is recognized by the 300 kDa cation-independent mannose 6-phosphate receptor (MPR300) and 46 kDa cation-dependent mannose 6-phosphate receptor (MPR46), which are P-type lectins (Castonguay et al. 2011) that mediate this transport. The MPR300 at the plasma membrane also internalizes extracellular Man6P-containing proteins.
Inherited defects in the phosphotransferase complex (UDP-N-acetylglucosamine-1-phosphotransferase), the key enzyme in the formation of Man6P, cause reduced levels or the complete absence of Man6P residues, leading to the secretion of the lysosomal hydrolases and, as a consequence, dysfunctional lysosomes. The associated lysosomal storage disorders (LSDs) manifest as mucolipidosis type II (MLII also known as I-cell disease) with severe skeletal abnormalities, hepato-splenomegaly, progressive psychomotor retardation and early death, or MLIII (pseudo-Hurler polydystrophy, MLIII) with clinically milder symptoms (Braulke et al. 2013). No treatment is available for MLII or MLIII, whereas some LSDs caused by mutations in genes encoding single lysosomal enzymes are treatable by recombinant enzyme replacement therapy based in most cases on Man6P-dependent uptake mechanisms for proper lysosomal delivery in cells and tissues (Ortolano et al. 2014).
Thus, there is considerable interest in the structural determination and quantification of N-glycans containing Man6P, as well as their positions in lysosomal glycoproteins and the structural characterization of their specific interaction with MPRs. Several studies of the Man6P-binding domains of MPR46 and MPR300 in complex with different ligands have revealed structural insights into the binding of Man6P and provided explanations for the observed pH optimum of binding and distinct trafficking functions in the cellular environment (Dahms et al. 2008; Olson et al. 2008, 2015). Recently, we have isolated a rabbit antibody single-chain variable fragment (scFv) designated M6P-1 by phage display (Figure 1); M6P-1 is specific for Man6P residues and displays low micromolar affinity similar to both MPRs (Müller-Loennies et al. 2010). The scFv M6P-1 was shown to bind to glycoproteins containing Man6P, allowing the direct identification and quantification of Man6P residues on proteins (Schröder et al. 2010; Madhavarao et al. 2014) and the indirect determination of GlcNAc-1-phosphotransferase or Man6P phosphatase activities (Pohl, Tiede, et al. 2010; Makrypidi et al. 2012). Further, these studies have demonstrated that scFv M6P-1 has the potential to be a powerful additional tool for the fast and economical diagnosis of MLII and MLIII by simple western blotting (Müller-Loennies et al. 2010; Pohl, Encarnacão, et al. 2010).
Fig. 1.
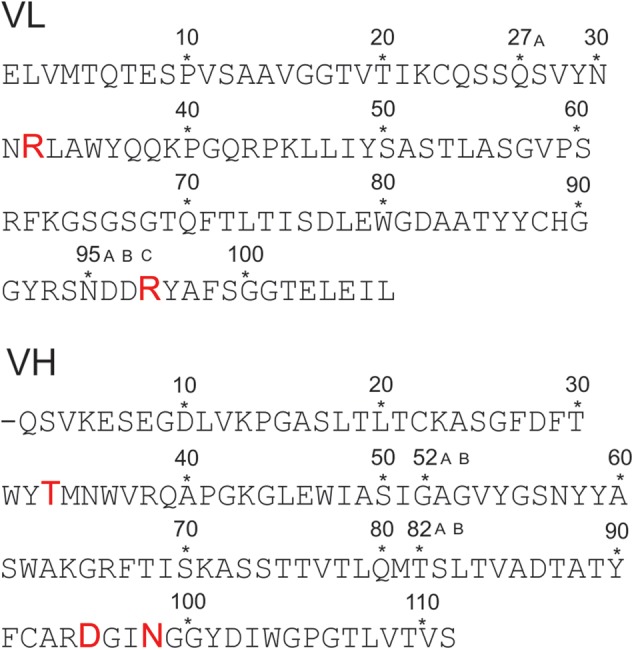
Primary structure of the variable domains of the light (VL) and heavy chain (VH) scFv M6P-1 obtained by phage display from an immunized rabbit. The sequence determination has been published (Müller-Loennies et al. 2010) and residues were numbered using a world wide web interface (http://www.bioinf.org.uk/abs/) applying the Kabat convention (Martin 1996). Amino acids involved in interactions with the ligand in the crystal structure are shown with increased font size. This figure is available in black and white in print and in color at Glycobiology online.
To better understand the specificity of scFv M6P-1 towards phosphorylated high mannose-type N-glycans and to explore its anti-carbohydrate specificity in comparison with MPR P-type lectins, we have determined its reactivity with a large number of high mannose-type N-glycans and used single crystal X-ray diffraction to determine the structure of the Fv in complex with Man6P.
Results
Dependence of scFv M6P-1 binding on pH
The binding of scFv M6P-1 to an immobilized neoglycoconjugate of pentamannose phosphate (PMP) from Hansenula holstii phosphomannan linked to bovine serum albumin (BSA) was investigated by ELISA at pH 5.0, 6.0, 6.4 and 7.4 (Figure 2), as Man6P has a (Hassid and Ballou 1957; Gracy and Noltmann 1968). PMP is a mixture of glycans consisting of α1 → 3-linked Man residues containing Man6P at the non-reducing terminus and a single α1 → 2-linked Man at the reducing end (Parolis et al. 1998; Ferro et al. 2002). The optimum binding was observed at pH 7.4 and 6.4, and it was about 2-fold reduced at pH 6.0. The affinity dramatically decreased with reduction in pH by three orders of magnitude and was essentially lost at pH 5.0. In an ELISA inhibition assay d-mannose 6-sulfate at 100 mM was unable to inhibit the binding of scFv M6P-1 to PMP-BSA.
Fig. 2.
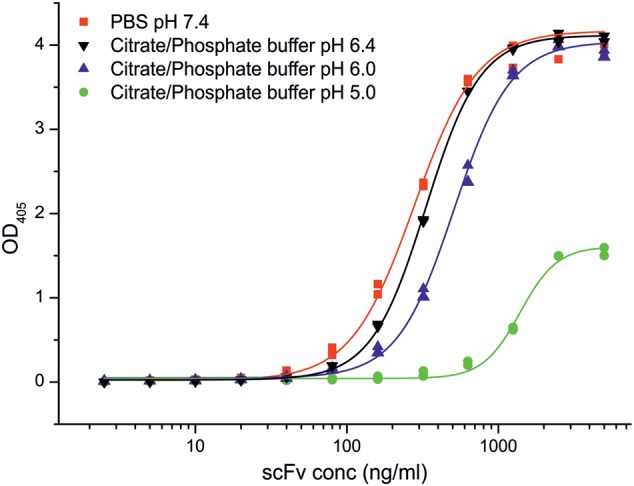
Binding of scFv M6P-1 to immobilized PMP-BSA at different pH in ELISA. ScFv M6P-1 (Mw 28,228 Da) dissolved in the indicated buffers was titrated starting from 5 µg/mL (177 nM) on PMP-BSA neoglycoconjugate immobilized on polystyrene ELISA plates (85 ng/cup ≈ 24 pmol of PMP). The data were fitted by non-linear regression to a logistic function using Origin v. 7.0 SR4 (OriginLab Corp., Northampton). Displayed are the duplicate data points and the fitted curve. This figure is available in black and white in print and in color at Glycobiology online.
Specificity of scFv M6P-1 towards high mannose-type N-glycans
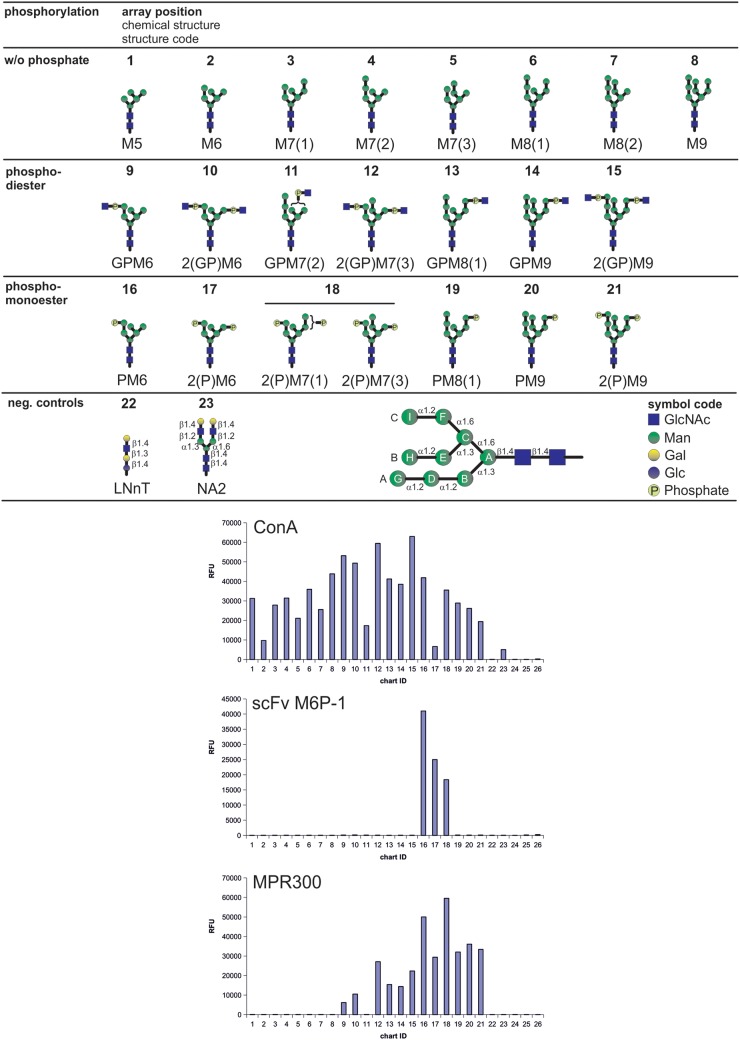
The specificity of scFv M6P-1 towards high mannose-type N-glycans was determined by interrogating a phosphorylated glycan array (Song et al. 2009; Castonguay et al. 2012) with a biotinylated scFv M6P-1 (Figure 3). As positive controls, we measured binding of a soluble form of the MPR300, which was purified from fetal bovine serum by phosphomannan affinity chromatography as described previously (Tong and Kornfeld 1989), and binding by concanavalin A (Con A, a mannose-specific lectin). The glycans were printed on the array and reactivity with Con A at 0.5 µg/mL indicated that all glycans on the array were accessible to binding and present in sufficient amounts. The differential binding observed by Con A is either due to differences in the structures that impact Con A binding or slight differences in the concentration of printed glycan. At higher Con A concentrations (10 µg/mL) binding of the glycans is similar (Castonguay et al. 2012). The binding profile of the MPR300 revealed binding to all phosphorylated glycans, including phosphodiesters and are therefore in agreement with previous results (Song et al. 2009; Castonguay et al. 2012). Taken together, these control studies demonstrate the validity of the phosphorylated glycan array.
Fig. 3.
Binding of scFv M6P-1 to glycans immobilized in an array on glass slides. The glycan microarray used in this analysis contained the schematically depicted glycans that were printed in replicates of four at a concentration of 100 µM with the exception of glycans 3, 7, 11, 12, 15, 18 and 21, which were only available at lower concentrations (<100 µM). Below a structure code is given where G = GlcNAc (blue squares), P = phosphate (light yellow circles) and M = Man (green circles). Glucose and Galactose in controls are depicted as blue and yellow circles, respectively. Numbers indicate the number of residues and isomers are distinguished by numbers in brackets. N-Glycan residues are designated as schematically depicted in the lower panel following the nomenclature as published (Castonguay et al. 2012). The array was interrogated with biotinylated scFv M6P-1 and Con A and affinity purified fetal bovine serum MPR300. Bound scFv M6P-1 and Con A were quantified after incubation with Cy5-labeled streptavidin and MPR300 was assayed using a rabbit polyclonal antibody and a Cy5-labeled anti-rabbit IgG as described (Castonguay et al. 2012). Con A and MPR300 served as positive controls to validate the array. Relative fluorescence units were determined as the average of four replicates, and the glycan numbers correspond to the glycans above. Biotinylated Con A at 0.5 µg/mL (left) was detected with Cy5 Streptavidin (5 µg/mL); biotinylated scFv M6P-1 at 20 µg/mL (middle) detected with Cy5 Streptavidin (5 µg/mL); MPR300 at 5 µg/mL (right) detected with a rabbit polyclonal antibody (1:250 dilution) and Cy5-labeled goat, anti-rabbit IgG at 5 µg/mL. Slides were scanned for Cy5 in a ProScaArray scanner (PerkinElmer) and average fluorescence units were determined using the corresponding ProScanArray software.
The glycan array analysis (Figure 3) using the biotinylated scFv M6P-1 revealed that it bound specifically mono- and diphosphorylated Man6 (PM6 and 2(P)M6, respectively) as well as diphosphorylated Man7 glycans 2(P)M7(1) and 2(P)M7(3) which all contained αMan6P(1 → 2)αMan(1 → 3)αMan, i.e. Man6P in the A-arm. The strongest signal was obtained for PM6. Glycans lacking phosphate and those containing αGlcNAc-1-P-6-αMan phosphodiesters were not bound by the scFv. High mannose-type N-glycans that contained Man6P in the C-arm, i.e. αMan6P(1 → 2)αMan(1 → 6)αMan in structures PM8(1), and PM9 were also not bound by scFv M6P-1. Remarkably, this was also the case for 2(P)M9 despite the presence of Man6P on the A-arm.
Crystal structure analysis: ambiguity in space group assignment and pseudo-merohedral twinning of Fv M6P-1 crystals
The Fv M6P-1 was generated by cleavage of the scFv linker, and subsequently crystallized and soaked with Man6P. Diffraction data were collected to 2.7 Å resolution, and initially indexed, integrated and scaled in a primitive tetragonal Bravais lattice. Systematic absences indicated space group P41212 or P43212. Cell content analysis predicted two molecules in the asymmetric unit (AU); however, molecular replacement (using the coordinates of the variable domain dimer of the only rabbit antigen binding fragment of an antibody (Fab) then deposited in the Protein Data Bank (Berman et al. 2000) (http://www.pdb.org), 4HBC, as a search model initially located only one molecule of the asymmetric unit. An extensively refined partial solution used as a model eventually yielded a full solution in P41212, but refinement stalled with R factors >40% and electron density that did not match entire lengths of the polypeptide chain.
The assumption was made that the space group was incorrect and the data were examined for evidence of twinning. The data were re-indexed into a primitive orthorhombic Bravais lattice with unit cell axes a = 60.6 Å, b = 128.05 Å and c = 127.30 Å. Analysis of data by PHENIX.xtriage (Adams et al. 2010) revealed a significant off-origin peak at coordinates 0.000, 0.473, 0.473, with a height 28.44% of the origin. Twin law tests revealed pseudo-merohedral twinning with the operator –h, l, k and an estimated twinned fraction of 0.419 (Britton analyses), 0.437 (H-test) or 0.435 (maximum likelihood method). The structure was then solved by molecular replacement in space group P212121 with four Fv molecules in the AU. Multiple rounds of model building and refinement yielded a final model with Rwork = 0.228 and Rfree = 0.269 and a final refined twin fraction of 0.480.
Structure of Fv M6P-1 in complex with Man6P
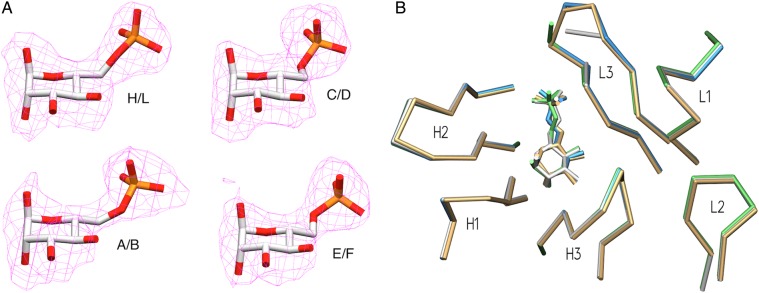
The data collection and refinement statistics for Fv M6P-1 with Man6P are given in Table I. The four crystallographically independent Fv molecules each contain a heavy and light chain, labeled H/L, A/B, C/D and E/F. Excellent electron density was observed for the Man6P monosaccharide in each of the four Fv molecules in the AU (Figure 4A) and for the polypeptide chain generally, although each light chain has regions of weak or interrupted electron density around residues 7–10, 14–16 and 93–95B, opposite the combining site (for numbering of residues see Figure 1). Residues were numbered using a world wide web interface (http://www.bioinf.org.uk/abs/) (Martin 1996) applying the Kabat convention, with heavy and light chain residues designated with an H or L, respectively, and with amino acid insertions designated with capital letters to keep conserved residues properly aligned. Superposition of the four Fv in the AU showed that the complementarity determining regions (CDR) of all four molecules in the AU adopt the same conformations (Figure 4B), with the exception that in two of the four light chains, SerL94 (chain L) or AsnL95A (chain B) of CDR L3 is not modeled due to ambiguous electron density. Each of the four Fv in the AU displays electron density consistent with a buffer-derived sulfate ion linked by hydrogen bonds and a salt bridge to GlnL42 and ArgL43, well outside the combining site. Two of the four Fv display electron density for an additional sulfate ion coordinated to ArgL95C in the combining site (chains D and F), which also maintain their salt bridge interactions to Man6P. The two Fv with this sulfate ion present have somewhat better electron density for residues L93–L95B than the two without.
Table I.
Data collection and refinement statistics for liganded Fv M6P-1
| Fv M6P-1 with Man6P | |
|---|---|
| Resolution (Å) | 30.1–2.72(2.79–2.72)a |
| Space group | P212121 |
| a (Å) | 60.6 |
| b (Å) | 128.1 |
| c (Å) | 127.3 |
| No. of molecules per asymmetric unit | 4 |
| Rsymb | 0.105 (0.392) |
| I/σ(I) | 7.6 (2.4) |
| Completeness (%) | 81.2 (86.3) |
| Redundancy | 2.5 (2.5) |
| Unique reflections | 20805 |
| Refinement | |
| Rwork (%) | 22.6 |
| Rfree (%) | 26.7 |
| No. of waters | 153 |
| r.m.s bonds (Å) | 0.0090 |
| r.m.s. angles (°) | 1.489 |
| Mean B (Å2) | 30.4 |
| Wilson B (Å2) | 33.0 |
| Ramachandran | |
| Favored | 94% |
| Allowed | 5% |
| Outliers | 1% |
aValues in parentheses represent the highest resolution shell.
bRsym = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of each ith redundant observation of reflection hkl, and 〈I(hkl)〉 is the average intensity of a given reflection hkl.
Fig. 4.
Crystal structure of scFv M6P-1 (A). Fo − Fc omit electron density maps contoured to 2σ for Man6P in each of the four Fv molecules in the AU. The refined coordinates of the corresponding Man6P are superposed for clarity. (B) Superposition of M6P-1 binding sites of the four molecules in the asymmetric unit, displaying α-carbon backbones of CDR loops and the Man6P molecule of each unit. Chains H/L in green, A/B white, C/D tan and E/F blue.
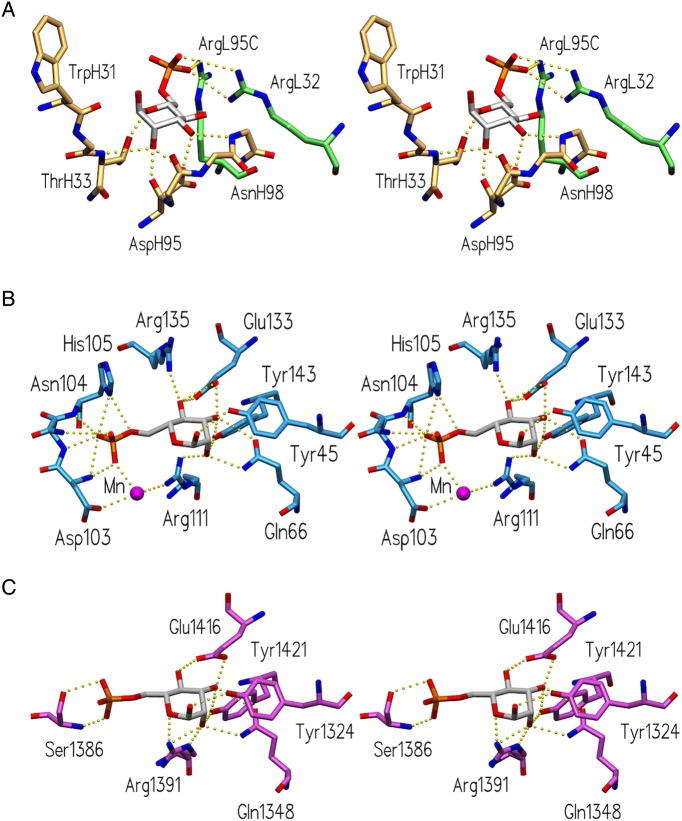
Fv M6P-1 makes four hydrogen bond contacts to the mannose ring of Man6P through CDR H1 and H3 (Figure 5A). The side chain of ThrH33 forms a hydrogen bond with the Man6P ring oxygen, the backbone carbonyl of AspH95 forms a hydrogen bond to OH-2, and the side chain of AspH95 and the backbone amide of AsnH98 form hydrogen bonds to OH-3. There is an additional hydrogen bond seen between ThrH33 main chain N and OH-2, where the distance is observed to lie between 2.99 and 3.45 Å depending on the AU. Fv M6P-1 binds to the phosphate group on Man6P through CDR L1 and CDR L3 where both ArgL32 and ArgL95C form salt bridges. There are small variations in the hydrogen bond distances between the four molecules in the AU due to modest differences in refined orientations of protein side chains and the ligand.
Fig. 5.
Comparison of Man6P-binding sites of Fv M6P-1 and bovine MPR. Stereoviews of the binding sites of (A) Fv M6P-1 (light chain green, heavy chain tan), (B) the CRD of MPR46 (pdb code 1M6P; Roberts et al. 1998) and (C) the CRD formed by the N-terminal domains 1–3 of MPR300 (pdb code 1SYO; Olson et al. 2004), showing hydrogen bonds and salt bridges as dashed yellow spheres.
The soaked ligand Man6P is capable of mutarotation, but in each case the electron density of the Man6P molecule is consistent with the α-anomeric form. Superposition of β-mannose in the combining site reveals steric clashes of OH-1 with the backbone of residues TyrH32 and ThrH33.
Fv M6P-1 is a unique anti-carbohydrate antibody in its origin from rabbit. Rabbit antibodies generally have longer and more diverse CDR L3s compared with humans and mice (Lavinder et al. 2014), and Fv M6P-1 follows this trend with a 12-residue CDR L3 [determined with IMGT/V-QUEST (Brochet et al. 2008)]. This does not however appear to be a critical factor in Man6P recognition as CDR L3 is backward leaning (Figure 4B) and makes only one direct interaction from ArgL95C to Man6P (Figure 5A). Fv M6P-1 displays no otherwise unusual features compared with other anti-carbohydrate antibodies (Haji-Ghassemi et al. 2015).
Discussion
ScFv M6P-1 recognizes a single carbohydrate residue
The Man6P-specific scFv M6P-1 was obtained by immunization with PMP, implying a structural similarity between αMan6P(1 → 2)αMan and αMan6P(1 → 3)αMan leading to cross-reaction of the yeast PMP and natural lysosomal glycoproteins with antibodies. Yet, the crystal structure analysis revealed that the lack of specificity for the glycosidic linkage can be explained by a small pocket-type combining site that specifically recognizes only the terminal carbohydrate residue. This is in contrast to almost all other known antibodies against carbohydrates where, despite the dominance of a single residue providing most of the binding energy, multiple residues of the carbohydrate antigen are in contact with the antibody. From early studies, it was estimated that the smallest antibody combining site can accommodate two to three monosaccharide residues (Arakatsu et al. 1966; Bundle et al. 1994). Thus, scFv M6P-1 is one of very few antibodies reported to exhibit high affinity (Kd = 28 µM) (Müller-Loennies et al. 2010) for a single monosaccharide residue.
Two different mechanisms for the antibody recognition of monosaccharides have been observed. First, there exist a number of crystal structures of antibodies specific for various chlamydial oligosaccharide antigens in complex with a 3-deoxy-α-d-manno-oct-2-ulosonic acid (Kdo) monosaccharide (Brooks et al. 2008; Brooks, Blackler, et al. 2010; Brooks, Müller-Loennies et al. 2010; Blackler et al. 2012), which have affinities for the monosaccharide in the range of 3–90 μM. In these cases, monosaccharide binding is achieved through a pocket of conserved sequence containing specific hydrogen bonds and a strong salt bridge to a sugar carboxylate.
Second, and in contrast, antibody S-20-4 has been determined to achieve an affinity of 39 μM for a terminal monosaccharide of the Vibrio cholerae Ogawa O-specific polysaccharide [oligomer of α1 → 2-linked 4-amino-4,6-dideoxy-d-mannose (perosamine) N-acylated with 3-deoxy-l-glycero-tetronic acid] through a significant hydrophobic effect (Wang et al. 1998; Villeneuve et al. 2000).
ScFv M6P-1 achieves specificity for mannose 6-phosphate through targeted hydrogen bonds and charged-residue interactions
ScFv M6P-1 achieves its affinity and specificity through several hydrogen bonds and important salt bridge interactions to the small carbohydrate antigen. ScFv M6P-1 was previously determined not to bind mannose, glucose or glucose 6-phosphate (Glc6P) (Müller-Loennies et al. 2010), and binding to PMP-BSA was not inhibited by mannose 6-sulfate (this study). The crystal structure presents a clear rationale for specific recognition of Man6P by scFv M6P-1, but not mannose, as the scFv makes two salt bridges to the phosphate group from ArgL32 and ArgL95C (Figure 5A). In this sense, M6P-1 is similar to Kdo-binding antibodies that also utilize a salt bridge interaction (Brooks et al. 2008; Brooks, Blackler et al. 2010; Brooks, Müller-Loennies et al. 2010; Blackler et al. 2012).
The lack of measurable affinity for Glc6P can be traced to the hydrogen bonds formed by the axial O2 of Man6P to AspH95 O and ThrH33 N (depending on which molecule in the AU) that could not be formed by the equatorial O2 on Glc. The lack of binding to Man6S (with an ionization state of −1) indicates that strong charged-residue interactions are imperative for binding, and supports the ELISA data that showed that binding is lost at pH below the second pKa of Man6P (pK = 6.1, Hassid and Ballou 1957; Gracy and Noltmann 1968). The scFv M6P-1 was crystallized at a pH of 8.5, which would result in a charge of −2 for the phosphate moiety thus allowing for strong salt bridges to be formed.
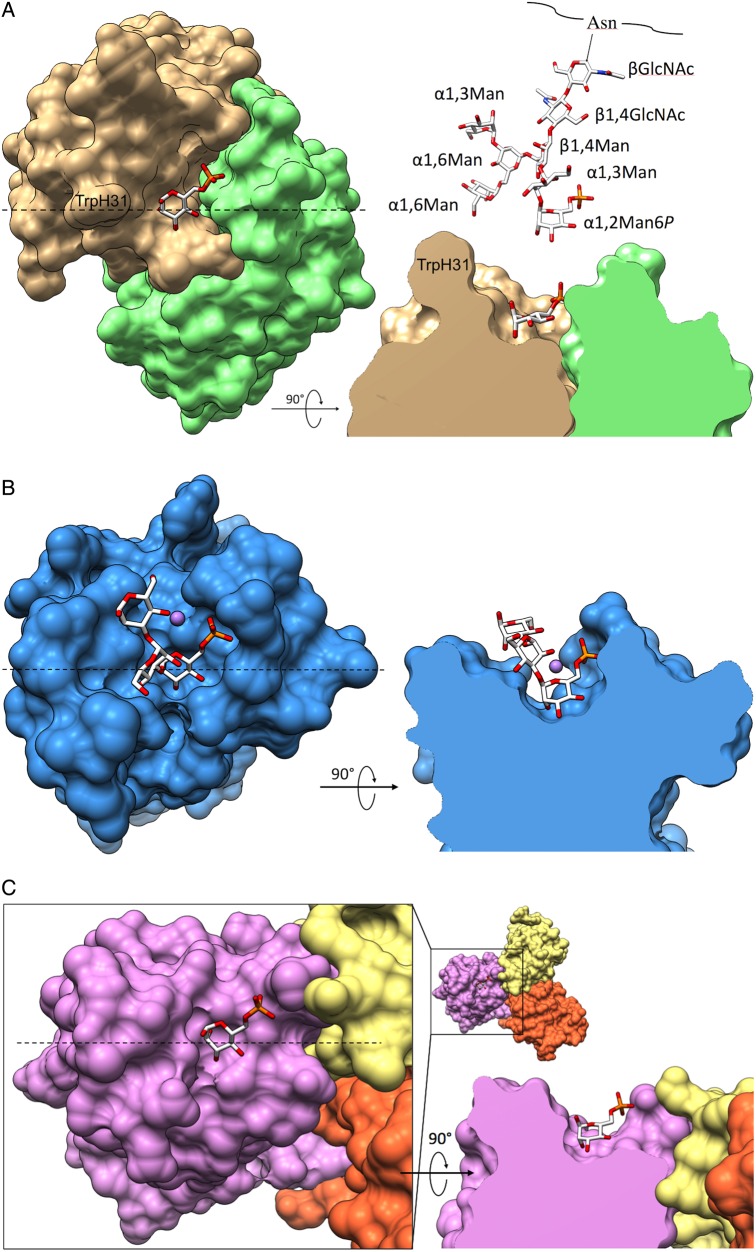
The observed lack of binding to phosphodiesters can be explained by the loss of one negative charge on the phosphate because a charge of −1 is insufficient for binding. In addition, the combining site of the Fv is sterically restricted surrounding the phosphate (Figure 6A) and could only accommodate a phosphodiester with the second sugar directed upwards away from the binding site.
Fig. 6.
Binding surfaces of Fv M6P-1 and MPRs. Top-down and side-view surface representations of Fv M6P-1, MPR46 (PDB code 1C39) and MPR300 domains 1–3 (PDB code 1SZ0). In each side-view, the model is clipped through the binding pocket. The dashed line in the top-down view indicates the location of the clipping plane. (A) Fv Man6P in complex with Man6P. Displayed above the side-view is a model of array glycan #16 for size comparison. (B) MPR46 in complex with PMP, showing only αMan6P(1 → 3)αMan(1 → 3)αMan as observed in that structure. Manganese ion is displayed as a mauve sphere. (C) MPR300 domains 1–3, sequentially colored yellow, orange, and mauve, in complex with Man6P.
Comparison of scFv M6P-1 and P-type lectin Man6P recognition strategy
The MPR46 and MPR300 are well-studied P-type lectins that bind high mannose-type glycans containing Man6P (reviewed in Dahms et al. 2008) allowing a comparison of the antibody combining site with the lectin carbohydrate recognition domains (CRDs). MPR46 forms stable homodimers with a single Man6P CRD per monomer and binds almost exclusively phosphomonoesters (Distler et al. 1991; Olson et al. 2008; Song et al. 2009). In contrast, the MPR300 contains four independent Man6P-binding sites and binds strongly to almost all phosphomono- as well as phosphodiesters, such as Man-P-GlcNAc (Song et al. 2009) (Figure 5). It contains two high-affinity Man6P-binding sites and a low-affinity-binding site that has been shown to bind Man-P-GlcNAc with higher affinity than Man6P (Chavez et al. 2007; Olson et al. 2010).
The crystal structures of the bovine MPR46 in complex with pentamannosyl phosphate (PMP, PDB code 1C39) (Olson et al. 1999) and of the N-terminal domains 1–3 of MPR300 in complex with Man6P (PDB code 1SZ0) (Olson et al. 2004) have revealed in both MPRs five conserved interactions from Tyr, Gln, Arg, Tyr and Glu to the pyranose ring (Figure 5). A total of 11 possible hydrogen bonds to the pyranose ring lead to a high-affinity-binding site.
Surprisingly, the phosphate is bound by the high-affinity-binding site in domains 1–3 of the MPR300 only by two hydrogen bonds without any salt bridge interaction, yet it has been reported to exhibit an affinity for Man6P similar to MPR46 (Tong et al. 1989; Tong and Kornfeld 1989), which binds the phosphate through one salt bridge (His105) and three hydrogen bonds (Asn103, Asn104) and loses binding to Man6P upon His105 deprotonation (Olson et al. 2008).
In contrast to the lectin MPR structures, the interaction between the antibody Fv M6P-1 and Man6P is dominated by the phosphate moiety, which forms mono- and bidentate salt bridges through two arginine side chains (ArgL32, ArgL95C). The combining site of Fv M6P-1 displays only four hydrogen bonds to the pyranose ring of Man6P. Two of these bonds are from the protein main chain atoms in Fv M6P-1, whereas all hydrogen bonds in the MPRs to the pyranose ring are from side chains.
The combining site of Fv M6P-1 is similar to the MPR46 binding site with steric restriction about the phosphate moiety (Figure 6A and B), consistent with its inability to accommodate phosphodiesters. The MPR300-binding site formed by domains 1–3 is less restricted than that of MPR46 (Figure 6C); however, a Man-P-GlcNAc phosphodiester-binding site is also located in domain 5 (Bohnsack et al. 2009). Determination of the solution NMR structure of domain 5 revealed a binding site less sterically crowded than MPR46 (Olson et al. 2010).
ScFv M6P-1 is specific for glycans containing terminal αMan6P(1 → 2)αMan(1 → 3)αMan
In addition to its inability to bind non-phosphorylated or phosphodiester glycans, scFv M6P-1 differentiates between phosphomonoester glycans with specificity for those containing αMan6P(1 → 2)αMan(1 → 3)αMan (Figure 3, structures 16–18). The lack of scFv M6P-1 binding to glycans PM8(1), PM9 and 2(P)M9 showed that it does not cross-react with αMan6P(1 → 2)αMan(1 → 6)αMan and indicated that glycans with two consecutive terminal α(1 → 2)-linkages as in αMan6P(1 → 2)αMan(1 → 2)αMan(1 → 3)αMan are not bound. Therefore, the observed cross-reaction between PMP and N-glycans is due to the fact that the scFv is permissive for the change from α(1 → 3) to α(1 → 2) only in the terminal linkage but not in the penultimate position.
While Man6P monosaccharide is capable of mutarotation, each of the Man6P molecules is observed clearly bound in the α-configuration, which points the anomeric oxygen away from the binding pocket (Figure 6A). The CDR loops do not extend significantly beyond the level of Man6P in this pocket nor do many side chains protrude. With the exception of TrpH31, which could possibly stack against additional mannose residues, there is little potential for steric hindrance of glycans displaying a terminal αMan6P. However, when αMan6P(1 → 3)αMan(1 → 3)αMan of PMP from the complex structure with MPR46 (PDB 1C39) is aligned with bound Man6P in the M6P-1 structure, there are significant clashes with the second and third mannose residues, with the third completely overlapping the protein. Yet, as M6P-1 was generated using PMP, alternate conformations of this ligand must exist.
Considering ligand flexibility, the shallow-binding site and the dominance of the phosphate in the interaction, binding of Fv M6P-1 to αMan6P(1 → 2)αMan(1 → 2)αMan(1 → 3)αMan or αMan6P(1 → 2)αMan(1 → 6)αMan appeared feasible, in principal. An alternative explanation for the lack of binding may therefore be a conformational change induced in the antigen due to the presence of Man (Figure 3, residue I) inducing steric crowding which renders not only Man6P on the C-arm but also on the A-arm inaccessible due to backfolding of the C-arm towards the core GlcNAc. Such conformational rearrangements affecting the whole triantennary structure have been predicted for higher N-glycans from molecular dynamics simulations (Balaji et al. 1994; Qasba et al. 1994) and nuclear magnetic resonance experiments (Vliegenthart et al. 1983; Kamiya et al. 2013). Nevertheless, the binding to PM8 and PM9 by MPR300 domain 9 (Bohnsack et al. 2009) showed that the Man6P on the C-arm is in fact accessible to protein binding. Therefore, the surface of M6P-1 adjacent the Man6P pocket must clash with the PM9, 2(P)M9 and PM8(1) epitopes bound by MRP300.
ScFv M6P-1 will recognize several glycoproteins possessing terminal Man6P residues, which facilitates the diagnosis of MLII and MLIII by western blotting demonstrating the loss or significantly lowered levels of several Man6P-containing proteins, respectively, in extracts of cultured fibroblast of patients compared with healthy individuals (Müller-Loennies et al. 2010; Pohl, Encarnacão, et al. 2010). The lack of scFv M6P-1 binding to higher Man8 or Man9 N-glycan structures that are phosphorylated on the C-arm will certainly lead to an underestimation of the total Man6P content of glycoproteins based on western blot analysis. However, structural analyses by mass spectrometry indicate that the glycan structures containing <8 mannose residues are by far the most abundant on lysosomal enzymes, e.g. arylsulfatase A (Schröder et al. 2010).
pH-dependent binding of scFv M6P-1 and MPRs stems from ionization of phosphate
The scFv M6P-1, MPR46 and MPR300 all have similar observed affinities for Man6P with KD values in the range of 7–28 µM (Tong and Kornfeld 1989; Müller-Loennies et al. 2010). The common theme is the presence of charged residues making ion-dipole and/or salt bridge interactions. Neither MPR binds Man6P at pH <5, which has been explained in MPR46 by the disruption of electrostatic inter-subunit interactions and the protonation of E133, one of the critical Man6P-binding residues (Waheed et al. 1990; Waheed and von Figura 1990; Olson et al. 2008). Interestingly, the strongest binding of scFv M6P-1 to PMP was seen at pH 6.4 and 7.4, with ∼2-fold reduced binding at pH 6.0 and no observed binding at pH 5.0, yet the binding site possesses no residues with pKa in this range. It is therefore likely that the loss of affinity at pH 5.0 is mainly driven by a change of the protonation state of the phosphate moiety of Man6P (, Hassid and Ballou 1957; Gracy and Noltmann 1968).
While the effect of phosphate ionization on the loss of Man6P binding by MPR46 at acidic pH was not discussed for the release of cargo into the endosomal compartment (Olson et al. 2008), it is likely a critical factor in pH-dependent binding (Tong and Kornfeld 1989) given the extensive charge coordination in the binding site (Figure 5B). Although there are no salt bridges to Man6P observed in the structure of MPR300 domains 1–3, there are potential long-range charge interactions from Arg391, Lys358 and Lys125 (between 4.8 and 6.9 Å) that may be weakened by Man6P protonation. The sequence alignment of MPR300 domains 3, 5 and 9 with MPR46 reveals histidine and lysine residues of MPR300 domain 9 in a similar position to His105 of MPR46 that forms a salt bridge to Man6P (Reddy et al. 2004). Therefore, Man6P protonation may also explain the mechanism by which the MPR300 releases its cargo into the acidic lumen of endosomes, for which structural changes of the protein have not been observed, and provides an explanation why a system based on Man6P recognition has evolved for shuttling glycoproteins to the lysosome.
Conclusions
The Fv M6P-1 is shown to distinguish N-glycan phosphorylation in a branch-specific manner, with specificity for A-branch terminal αMan6P(1 → 2)αMan(1 → 3)αMan present on glycans PM6, 2(P)M6, 2(P)M7(1) and 2(P)M7(3). This specificity and the predominance of glycans with fewer than 8 Man residues on lysosomal enzymes support the potential utility of scFv M6P-1 in the diagnosis of MLII and MLIII or in protein purification for enzyme replacement therapy. The crystal structure of Fv M6P-1 in complex with Man6P reveals that the basis for its ability to react with terminal αMan6P is a shallow monosaccharide-binding pocket with multiple hydrogen bonds to the pyranose ring and two salt bridges to the phosphate moiety. The loss of Man6P binding by scFv M6P-1 <pH 6.0 is found to be due to Man6P protonation and the associated weakening of salt bridge interactions, and a similar effect occurring in the MPRs with Man6P protonation rationalizes the evolution of the Man6P-based lysosomal enzyme transport pathway.
Materials and methods
Preparation of Fv M6P-1
The generation of scFv M6P-1 by immunization and phage display and its expression and purification have been described previously (Müller-Loennies et al. 2010). Fv was produced from M6P-1 scFv by incubation with subtilisin at a ratio of 1:300 (w/w) to digest the flexible linker so as to prevent oligomerization. The digest was performed at room temperature in 20 mM HEPES, pH 7.2, with 150 mM NaCl for 3 h and quenched by the addition of 1 mM PMSF (G-biosciences). The Fv was further purified by size-exclusion chromatography (Phenomenex BioSep SEC-s3000 column) using HEPES, pH 7.2, with 150 mM NaCl as the mobile phase.
Preparation of mannose-6-phosphate
Man6P was prepared from benzyl 2,3,4-tri-O-benzyl α-d-mannoside (Lu et al. 2005). In brief, benzyl 2,3,4-tri-O-benzyl β-d-mannoside was phosphorylated with dibenzyl N,N-diethylphosphoramidite and treated with mCPBA to give the phosphate triester. Hydrogenolysis of the resulting benzyl 2,3,4-tri-O-benzyl-1-O-dibenzylphosphoryl β-d-mannoside over Pd/C and purification using ion exchange resin (Dowex® Na+ form) gave the disodium salt of Man6P.
Biotinylation and ELISA-binding assays
The scFv M6P-1 was biotinylated using sulfo-NHS-LC-biotin (Thermo Fisher Scientific, Bonn, Germany). The reagent was dissolved to 10 mM in water and 0.05 mL added to 0.7 mL of purified scFv (1.0 mg/mL) in PBS, pH 7.4. After 2 h on ice excess reagent was removed by dialysis against PBS. The activity of the biotinylated scFv was verified by ELISA against PMP-BSA using a secondary antibody (ascites from mAb 9E10) against the c-myc tag as described (Müller-Loennies et al. 2010) and an HRP-conjugate of streptavidin (Dianova, Germany).
For the ELISA-binding assays at different pH, the scFv (1 mg/mL in PBS, pH 7.4) was diluted to the starting concentration of 0.005 mg/mL in citrate-phosphate buffer containing 150 mM NaCl with a pH adjusted to 5.0, 6.0 or 6.4. The binding curves were determined as described (Müller-Loennies et al. 2010) in these buffers and compared with the binding curve of scFv in PBS, pH 7.4.
ELISA inhibition was performed as described (Müller-Loennies et al. 2010) using mannose 6-sulfate (sodium salt, Dextra), Man6P and Man at starting concentrations of 100 mM.
Glycan microarray analysis
The phosphorylated glycan array was produced and the analyses were performed as previously described, and the data were reported as histograms of average Relative Fluorescent Units corresponding to glycan structures identified by chart ID (Song et al. 2009; Castonguay et al. 2012). The carbohydrate structures are depicted in Figure 3. As negative controls lacto-N-neotetraose (#22), NA2 (#23), PBS (#24 and 25) and biotin (#26) were included. The positive controls were carried out with biotinylated concanavalin A (Vector Labs) at 0.5 µg/mL detected with Cy5-labeled Streptavidin (Invitrogen) and a soluble form of the MPR300 at 5 µg/mL detected with a rabbit anti-MPR300 polyclonal antibody (B14.5 at 1:250 dilution) and Cy5-labeled anti-rabbit IgG (Invitrogen) at 5 µg/mL. The MPR and B14.5 antibody were kindly provided by Nancy Dahms (Medical College of Wisconsin, Milwaukee, WI). The biotinylated scFv M6P-1 was analyzed on the CFG glycan microarray and those data, which are available on the CFG website (http://www.functionalglycomics.org), indicated that it bound only mannose-6-phosphate among the 610 glycans printed on v5.1 of the CFG array. The biotinylated scFv M6P-1 was applied to the phosphorylated glycan array at 200, 20 and 2 µg/mL in buffer containing EDTA and detected with Cy5-labeled Streptavidin.
Crystallization of Fv M6P-1
Purified Fv was concentrated to 14 mg/mL using Amicon Ultra centrifugal filters. Crystallization screens were set up using an Art Robbins Instruments Gryphon robot. Small needle-like crystals appeared in Hampton Index screen (Hampton Research, Aliso Viejo, CA) condition #6 (0.1 M Tris, pH 8.5, with 2 M ammonium sulfate) after 4 days.
Ligand soaking, data collection, structure determination and refinement
Crystals were soaked in mother liquor with 2 mM Man6P and flash frozen to −160°C with an Oxford Cryostream 700 crystal cooler (Oxford Cryosystems, Devens, MA) using CryoOil (MiTeGen, Ithaca, NY) as cryoprotectant. Data were collected on beamline CMCF-ID at the Canadian Light Source synchrotron (Saskatoon, SK) and processed using HKL2000 (HKL Research, Charlottesville, VA). Data quality were analyzed using PHENIX.xtriage (Adams et al. 2010). The structure of liganded Fv M6P-1 was solved by molecular replacement using PHASER (McCoy et al. 2007) with the variable domain of the antibody heavy chain (VH) and variable domain of the antibody light chain (VL) of a rabbit Fab (PDB code 4HBC) as a model. Manual fitting of Fo − Fc and 2Fo − Fc electron density maps was carried out with Coot (Emsley et al. 2010) and SetoRibbon (S. V. Evans, unpublished). TLS and restrained amplitude-based twin refinement allowing isotropic thermal motion was carried out with REFMAC5 as implemented in CCP4 (Winn et al. 2011). The final refinement and model statistics are given in Table I.
Visualization and graphics
Crystal structure figures were produced using SetoRibbon (S. V. Evans, unpublished) and UCSF Chimera (Pettersen et al. 2004). Chimera is developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Sequence analysis
The sequence of Fv M6P-1 was analyzed with UCL AbCheck and the structure was numbered according to the Kabat method (http://www.bioinf.org.uk/abs/; Martin 1996).
Funding
This work was supported in part by grants from the Natural Resources and Engineering Research Council of Canada and the Canadian Institute for Health Research to S.V.E . S.V.E. is a recipient of a senior scholarship from the Michael Smith Foundation for Health Research. R.J.B. is a recipient of an NSERC CGSD Award. X.L. thanks support from NIH (P50CA121973). A Grant P41GM103694 (R.D.C.) to the National Center for Functional Glycomics also supported work.
Conflict of interest statement
S.M.-L. and T.B. hold a patent on the application of scFv M6P-1.
Abbreviations
AU, asymmetric unit; BSA, bovine serum albumin; CDR, complementarity determining region; CRD, carbohydrate recognition domain; Fab, antigen binding fragment of an antibody; Fv, variable fragment of an antibody (scFv-fragment after enzymatic cleavage of the linker); Glc, glucose; Glc6P, glucose 6-phosphate; GlcNAc-1-phosphotransferase, UDP-N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase; Man, mannose; Man6P, mannose 6-phosphate; ML, mucolipidosis; MPR46, 46 kDa cation-dependent mannose 6-phosphate receptor; MPR300, 300 kDa cation-independent mannose 6-phosphate receptor; PMP, pentamannose phosphate from Hansenula holstii; scFv, single-chain fragment variable; VH, variable domain of the antibody heavy chain; VL, variable domain of the antibody light chain.
Acknowledgments
We thank Nadine Harmel, Christine Schneider and Ute Agge for their technical assistance. We also thank Dr. C.R. MacKenzie from the Institute of Biological Sciences at the National Research Council of Canada (Ottawa) for the kind gift of mAb 9E10 ascites. The authors acknowledge The Consortium for Functional Glycomics funded by the NIGMS GM62116 and GM98791 for services provided by the Glycan Array Synthesis Core (The Scripps Research Institute, LaJolla, CA) that produced the mammalian glycan microarray and the Protein-Glycan Interaction Core (Emory University School of Medicine, Atlanta, GA) that assisted with analysis of samples on the array.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW et al. . 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakatsu Y, Ashwell G, Kabat EA. 1966. Immunochemical studies on dextrans. V. Specificity and cross-reactivity with dextrans of the antibodies formed in rabbits to isomaltonic and isomaltotrionic acids coupled to bovine serum albumin. J Immunol. 97:858–866. [PubMed] [Google Scholar]
- Balaji PV, Qasba PK, Rao VS. 1994. Molecular dynamics simulations of high-mannose oligosaccharides. Glycobiology. 4:497–515. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackler RJ, Müller-Loennies S, Brade L, Kosma P, Brade H, Evans SV. 2012. Antibody recognition of chlamydia LPS: Structural insights of inherited immune responses. In: Kosma P, Müller-Loennies S, editors. Anticarbohydrate Antibodies. Vienna: Springer; p. 75–120. [Google Scholar]
- Bohnsack RN, Song X, Olson LJ, Kudo M, Gotschall RR, Canfield WM, Cummings RD, Smith DF, Dahms NM. 2009. Cation-independent mannose 6-phosphate receptor: A composite of distinct phosphomannosyl binding sites. J Biol Chem. 284:35215–35226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braulke T, Bonifacino JS. 2009. Sorting of lysosomal proteins. Biochim Biophys Acta. 1793:605–614. [DOI] [PubMed] [Google Scholar]
- Braulke T, Raas-Rothschild A, Kornfeld S. 2013. I-cell disease and pseudo-Hurler polydystrophy: Disorders of lysosomal enzyme phosphorylation and localization. In: Valle D, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Scriver CR, Sly WS, Bunz F, Gibson KM, Mitchell G, editors. The Online Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill. [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Blackler RJ, Sixta G, Kosma P, Müller-Loennies S, Brade L, Hirama T, MacKenzie CR, Brade H, Evans SV. 2010. The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen. Glycobiology. 20:138–147. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Müller-Loennies S, Borisova SN, Brade L, Kosma P, Hirama T, Mackenzie CR, Brade H, Evans SV. 2010. Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition. Biochemistry. 49:570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Müller-Loennies S, Brade L, Kosma P, Hirama T, MacKenzie CR, Brade H, Evans SV. 2008. Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes. J Mol Biol. 377:450–468. [DOI] [PubMed] [Google Scholar]
- Bundle DR, Eichler E, Gidney MA, Meldal M, Ragauskas A, Sigurskjold BW, Sinnott B, Watson DC, Yaguchi M, Young NM. 1994. Molecular recognition of a Salmonella trisaccharide epitope by monoclonal antibody Se155-4. Biochemistry. 33:5172–5182. [DOI] [PubMed] [Google Scholar]
- Castonguay AC, Lasanajak Y, Song X, Olson LJ, Cummings RD, Smith DF, Dahms NM. 2012. The glycan-binding properties of the cation-independent mannose 6-phosphate receptor are evolutionary conserved in vertebrates. Glycobiology. 22:983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay AC, Olson LJ, Dahms NM. 2011. Mannose 6-phosphate receptor homology (MRH) domain-containing lectins in the secretory pathway. Biochim Biophys Acta. 1810:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez CA, Bohnsack RN, Kudo M, Gotschall RR, Canfield WM, Dahms NM. 2007. Domain 5 of the cation-independent mannose 6-phosphate receptor preferentially binds phosphodiesters (mannose 6-phosphate N-acetylglucosamine ester). Biochemistry. 46:12604–12617. [DOI] [PubMed] [Google Scholar]
- Dahms NM, Olson LJ, Kim JJ. 2008. Strategies for carbohydrate recognition by the mannose 6-phosphate receptors. Glycobiology. 18:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler JJ, Guo JF, Jourdian GW, Srivastava OP, Hindsgaul O. 1991. The binding specificity of high and low molecular weight phosphomannosyl receptors from bovine testes. Inhibition studies with chemically synthesized 6-O-phosphorylated oligomannosides. J Biol Chem. 266:21687–21692. [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro V, Li C, Fewings K, Palermo MC, Linhardt RJ, Toida T. 2002. Determination of the composition of the oligosaccharide phosphate fraction of Pichia (Hansenula) holstii NRRL Y-2448 phosphomannan by capillary electrophoresis and HPLC. Carbohydr Res. 337:139–146. [DOI] [PubMed] [Google Scholar]
- Gracy RW, Noltmann EA. 1968. Studies on phosphomannose isomerase. 3. A mechanism for catalysis and for the role of zinc in the enzymatic and the nonenzymatic isomerization. J Biol Chem. 243:5410–5419. [PubMed] [Google Scholar]
- Haji-Ghassemi O, Blackler RJ, Martin Young N, Evans SV. 2015. Antibody recognition of carbohydrate epitopes. Glycobiology. 25:920–952. [DOI] [PubMed] [Google Scholar]
- Hassid WZ, Ballou CE. 1957. Phosphate esters. In: Pigman WW, editor. The Carbohydrates. Chemistry, Biochemistry, Physiology. New York: Academic Press; p. 172–187. [Google Scholar]
- Kamiya Y, Yanagi K, Kitajima T, Yamaguchi T, Chiba Y, Kato K. 2013. Application of metabolic 13C labeling in conjunction with high-field nuclear magnetic resonance spectroscopy for comparative conformational analysis of high mannose-type oligosaccharides. Biomolecules. 3:108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. 2014. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS ONE. 9:e101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Navidpour L, Taylor SD. 2005. An expedient synthesis of benzyl 2,3,4-tri-O-benzyl-beta-D-glucopyranoside and benzyl 2,3,4-tri-O-benzyl-beta-D-mannopyranoside. Carbohydr Res. 340:1213–1217. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. 2007. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol. 8:622–632. [DOI] [PubMed] [Google Scholar]
- Madhavarao CN, Agarabi CD, Wong L, Müller-Loennies S, Braulke T, Khan M, Anderson H, Johnson GR. 2014. Evaluation of butyrate-induced production of a mannose-6-phosphorylated therapeutic enzyme using parallel bioreactors. Biotechnol Appl Biochem. 61:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrypidi G, Damme M, Müller-Loennies S, Trusch M, Schmidt B, Schluter H, Heeren J, Lübke T, Saftig P, Braulke T. 2012. Mannose 6 dephosphorylation of lysosomal proteins mediated by acid phosphatases Acp2 and Acp5. Mol Cell Biol. 32:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC. 1996. Accessing the Kabat antibody sequence database by computer. Proteins. 25:130–133. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr. 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Loennies S, Galliciotti G, Kollmann K, Glatzel M, Braulke T. 2010. A novel single-chain antibody fragment for detection of mannose 6-phosphate-containing proteins: Application in mucolipidosis type II patients and mice. Am J Pathol. 177:240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Castonguay AC, Lasanajak Y, Peterson FC, Cummings RD, Smith DF, Dahms NM. 2015. Identification of a fourth mannose 6-phosphate binding site in the cation-independent mannose 6-phosphate receptor. Glycobiology. 25:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Dahms NM, Kim JJ. 2004. The N-terminal carbohydrate recognition site of the cation-independent mannose 6-phosphate receptor. J Biol Chem. 279:34000–34009. [DOI] [PubMed] [Google Scholar]
- Olson LJ, Hindsgaul O, Dahms NM, Kim JJ. 2008. Structural insights into the mechanism of pH-dependent ligand binding and release by the cation-dependent mannose 6-phosphate receptor. J Biol Chem. 283:10124–10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Peterson FC, Castonguay A, Bohnsack RN, Kudo M, Gotschall RR, Canfield WM, Volkman BF, Dahms NM. 2010. Structural basis for recognition of phosphodiester-containing lysosomal enzymes by the cation-independent mannose 6-phosphate receptor. Proc Natl Acad Sci USA. 107:12493–12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Zhang J, Lee YC, Dahms NM, Kim JJ. 1999. Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor. J Biol Chem. 274:29889–29896. [DOI] [PubMed] [Google Scholar]
- Ortolano S, Vieitez I, Navarro C, Spuch C. 2014. Treatment of lysosomal storage diseases: Recent patents and future strategies. Recent Pat Endocr Metab Immune Drug Discov. 8:9–25. [DOI] [PubMed] [Google Scholar]
- Parolis LA, Parolis H, Kenne L, Meldal M, Bock K. 1998. The extracellular polysaccharide of Pichia (Hansenula) holstii NRRL Y-2448: The phosphorylated side chains. Carbohydr Res. 309:77–87. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera – A visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pohl S, Encarnacão M, Castrichini M, Müller-Loennies S, Muschol N, Braulke T. 2010. Loss of N-acetylglucosamine-1-phosphotransferase gamma subunit due to intronic mutation in GNPTG causes mucolipidosis type III gamma: Implications for molecular and cellular diagnostics. Am J Med Genet A. 152A:124–132. [DOI] [PubMed] [Google Scholar]
- Pohl S, Tiede S, Marschner K, Encarnacao M, Castrichini M, Kollmann K, Muschol N, Ullrich K, Müller-Loennies S, Braulke T. 2010. Proteolytic processing of the gamma-subunit is associated with the failure to form GlcNAc-1-phosphotransferase complexes and mannose 6-phosphate residues on lysosomal enzymes in human macrophages. J Biol Chem. 285:23936–23944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba PK, Balaji PV, Rao VS. 1994. Molecular dynamics simulations of oligosaccharides and their conformation in the crystal structure of lectin-carbohydrate complex: Importance of the torsion angle psi for the orientation of alpha 1,6-arm. Glycobiology. 4:805–815. [DOI] [PubMed] [Google Scholar]
- Reddy ST, Chai W, Childs RA, Page JD, Feizi T, Dahms NM. 2004. Identification of a low affinity mannose 6-phosphate-binding site in domain 5 of the cation-independent mannose 6-phosphate receptor. J Biol Chem. 279:38658–38667. [DOI] [PubMed] [Google Scholar]
- Roberts DL, Weix DJ, Dahms NM, Kim JJ. 1998. Molecular basis of lysosomal enzyme recognition: Three-dimensional structure of the cation-dependent mannose 6-phosphate receptor. Cell. 93:639–648. [DOI] [PubMed] [Google Scholar]
- Schröder S, Matthes F, Hyden P, Andersson C, Fogh J, Müller-Loennies S, Braulke T, Gieselmann V, Matzner U. 2010. Site-specific analysis of N-linked oligosaccharides of recombinant lysosomal arylsulfatase A produced in different cell lines. Glycobiology. 20:248–259. [DOI] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Olson LJ, Boonen M, Dahms NM, Kornfeld S, Cummings RD, Smith DF. 2009. Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. J Biol Chem. 284:35201–35214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong PY, Gregory W, Kornfeld S. 1989. Ligand interactions of the cation-independent mannose 6-phosphate receptor. The stoichiometry of mannose 6-phosphate binding. J Biol Chem. 264:7962–7969. [PubMed] [Google Scholar]
- Tong PY, Kornfeld S. 1989. Ligand interactions of the cation-dependent mannose 6-phosphate receptor. Comparison with the cation-independent mannose 6-phosphate receptor. J Biol Chem. 264:7970–7975. [PubMed] [Google Scholar]
- Villeneuve S, Souchon H, Riottot MM, Mazie JC, Lei P, Glaudemans CP, Kovac P, Fournier JM, Alzari PM. 2000. Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: Molecular basis for serotype specificity. Proc Natl Acad Sci USA. 97:8433–8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliegenthart JFG, Dorland L, Halbeek Hv. 1983. High-resolution, 1H-nuclear magnetic resonance spectroscopy as a tool in the structural analysis of carbohydrates related to glycoproteins. In: Tipson RS, Derek H, editors. Advances in Carbohydrate Chemistry and Biochemistry. Academic Press; p. 209–374. [Google Scholar]
- Waheed A, Hille A, Junghans U, von Figura K. 1990. Quaternary structure of the Mr 46,000 mannose 6-phosphate specific receptor: Effect of ligand, pH, and receptor concentration on the equilibrium between dimeric and tetrameric receptor forms. Biochemistry. 29:2449–2455. [DOI] [PubMed] [Google Scholar]
- Waheed A, von Figura K. 1990. Rapid equilibrium between monomeric, dimeric and tetrameric forms of the 46-kDa mannose 6-phosphate receptor at 37 degrees C. Possible relation to the function of the receptor. Eur J Biochem. 193:47–54. [DOI] [PubMed] [Google Scholar]
- Wang J, Villeneuve S, Zhang J, Lei P, Miller CE, Lafaye P, Nato F, Szu SC, Karpas A, Bystricky S et al. . 1998. On the antigenic determinants of the lipopolysaccharides of Vibrio cholerae O:1, serotypes Ogawa and Inaba. J Biol Chem. 273:2777–2783. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A et al. . 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]