Abstract
Autoantibodies target the RNA binding protein Ro60 in systemic lupus erythematosus (SLE) and Sjögren’s syndrome. However, whether Ro60 and its associated RNAs contribute to disease pathogenesis is unclear. We catalogued the Ro60-associated RNAs in human cell lines and found that among other RNAs, Ro60 bound an RNA motif derived from endogenous Alu retroelements. Alu transcripts were induced by type I interferon and stimulated proinflammatory cytokine secretion by human peripheral blood cells. Ro60 deletion resulted in enhanced expression of Alu RNAs and interferon-regulated genes. Anti-Ro60 positive SLE immune complexes contained Alu RNAs, and Alu transcripts were upregulated in SLE whole blood samples compared to controls. These findings establish a link between the lupus autoantigen Ro60, Alu retroelements and type I interferon.
Autoantibodies to the RNA binding protein Ro60/SSA are present in individuals with SLE(1), Sjögren’s Syndrome(2), neonatal lupus with heart block(3–5), and other autoimmune disorders(6). SLE subjects with anti-Ro60 antibodies have an increased prevalence of skin disease, photosensitivity and nephritis, and elevated expression of interferon (IFN)-inducible genes in immune cells and tissues(7, 8). Ro60, encoded by TROVE2, is a component of a ribonucleoprotein complex comprised of the E3 ubiquitin ligase Ro52(9), the Pol-III transcriptional terminator La/SSB, and short noncoding Y RNAs (HY1-4) (10). Ro60 has a specific RNA recognition domain for Y RNAs but binds additional, possibly mis-folded, RNAs through a separate cavity(11). The identity of these other RNA species and their relevance to autoimmune disease is unclear.
To identify RNAs bound to Ro60, we performed iCLIP (individual nucleotide crosslinking and immunoprecipitation)(12–14) followed by high-throughput sequencing in two ENCODE cell lines: GM12878, an EBV transformed B-lymphocyte cell line, and K562, an erythromyeloblastoid leukemia cell line (http://www.genome.gov/encode/) (Fig. S1). Sequencing reads were aligned to the human reference genome (hg19), generating 970,667 and 1,090,671 alignments for GM12878 and K562, respectively. This analysis identified 2,445 Ro60 bound sites in GM12878 and 8,273 sites in K562.
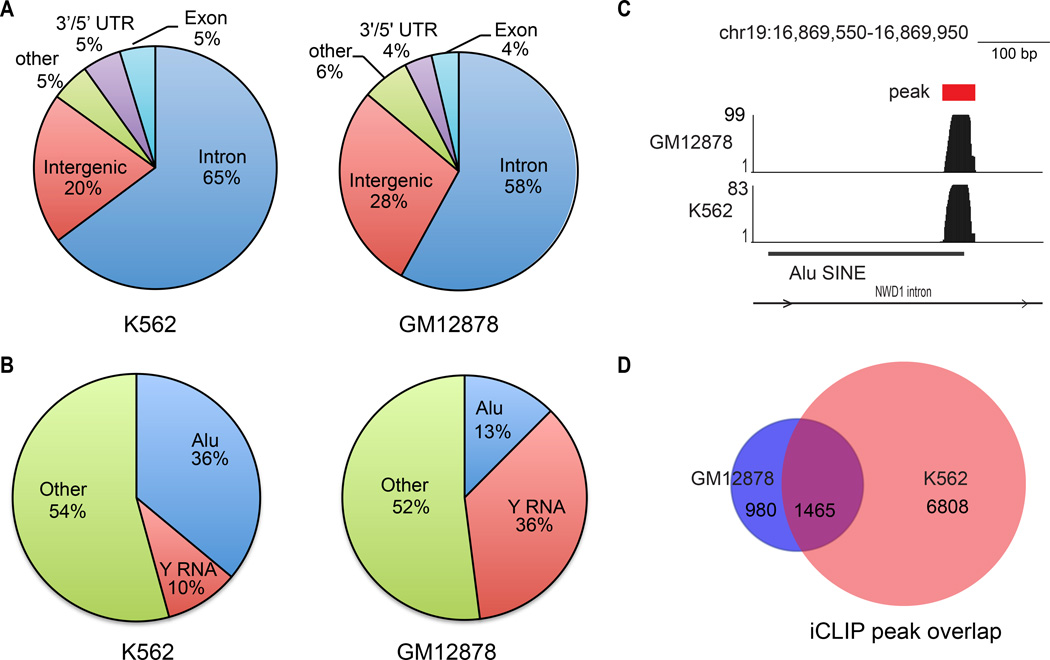
Annotation of all peaks to known genomic features revealed that approximately two-thirds of the iCLIP tags localized to introns in both cell lines (Fig. 1A). As expected, Y RNAs comprised a significant fraction of iCLIP tags in both lines (Fig. 1B). Surprisingly, iCLIP tags within Alu SINEs (short interspersed elements), a common repetitive transposable element (~1.1 M) in the human genome(15), were also represented (Fig. 1B,C). Additional lower frequency tags included 5’ and 3’ untranslated regions, LINEs (long interspersed elements), tRNAs, rRNAs, and simple repeats (Fig. S1). Most of the Ro60 iCLIP peaks identified in GM12878 (1,465/2,445, 60%) were also found in K562 (Fig. 1D, Table S1) indicating significant concordance of iCLIP results between the two cell lines.
Figure 1. iCLIP identifies RNAs bound by Ro60.
(A) Pie graph showing annotation of iCLIP peaks to gene structures in K562 and GM12878 cell lines. (B) Pie graph showing annotation of iCLIP peaks to Repeatmasker elements. (C) Genome browser snapshot of a representative Alu iCLIP peak. Y-axis indicates read counts. (D) Venn diagram representation of overlap in Ro60 iCLIP peaks between K562 and GM12878. P value<0.001, hypergeometric test.
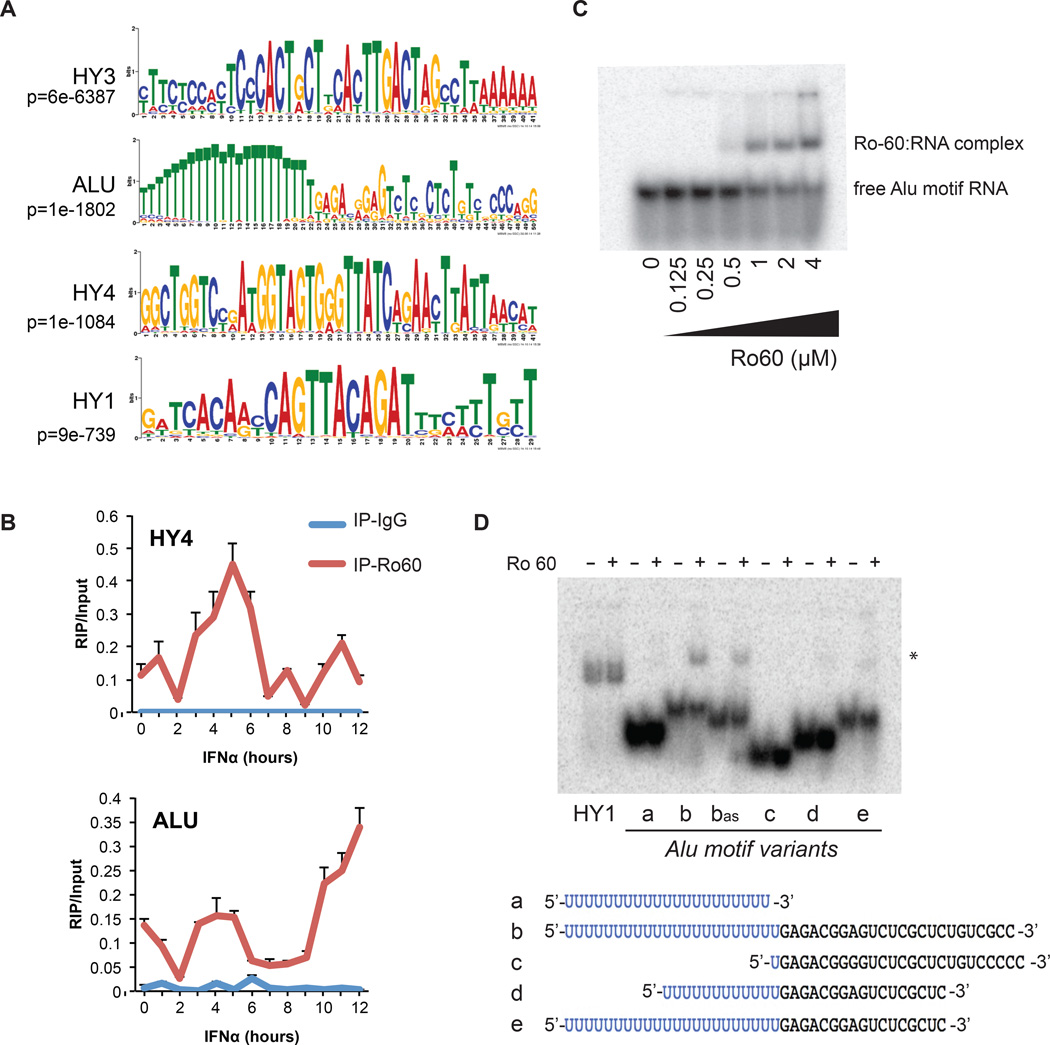
Motifs enriched within shared iCLIP peaks included Y RNA consensus sequences, as well as a U-rich motif that corresponds to the 3’ antisense strand of an Alu element (p<1×10−1800) (Fig. 2A and Fig. S2). The interaction between Ro60 and Alu RNA was confirmed using both RNA immunoprecipitation (RIP)-PCR (Fig. 2B) and RIP-seq (Fig. S3). RIP-PCR over a 12-hour time course of IFNα treatment of GM12878 showed binding of HY4 RNA to Ro60 at baseline, and this association peaked ~5 hours following IFNα treatment (Fig. 2B). Alu RNAs were also bound to Ro60, and showed complex kinetics of association following IFNα with highest levels measured at 12 hours.
Figure 2. Ro60 binds an RNA motif derived from Alu elements.
(A) Top four RNA motifs(14) identified from the Ro60 iCLIP peaks shared between K562 and GM12878. T=U in RNA. (B) RIP-PCR for Alu or HY RNA in Ro60 or IgG immunoprecipitates across a time course of IFNα treatment in GM12878 cells. Error bars represent SD of 3 technical replicates, data shown is a representative experiment of 3 (C) EMSA of Alu RNA motif (oligo b from panel D) with indicated concentrations of recombinant Ro60 protein. (D) EMSA of Alu motif variants with recombinant Ro60. a, PolyU sequence alone; b, representative Alu motif; bas, antisense of b; c, deleted polyU; d, partial 5’ and 3’ truncations; e, partial 3’ truncation. *=RNA/Ro60 complex. RNA sequences depicted in bottom panel. See also Fig. S4.
We next tested the binding of radiolabelled Alu motif RNA oligonucleotides to purified recombinant Ro60 protein using electromobility shift assays. Ro60 binding was observed with the full-length Alu motif (Fig. 2C,D, oligo b), and an antisense version of the full-length motif (Fig. 2D, oligo bAS). Reduced (oligos d, e) or no appreciable binding (oligos a, c) to Ro60 was observed for various truncated Alu motifs (Fig. 2D). Several sequence mutants that abrogated predicted secondary structure of the Alu motif did not affect binding to Ro60; however, mutations to the polyU tract disrupted the interaction (Fig. S4).
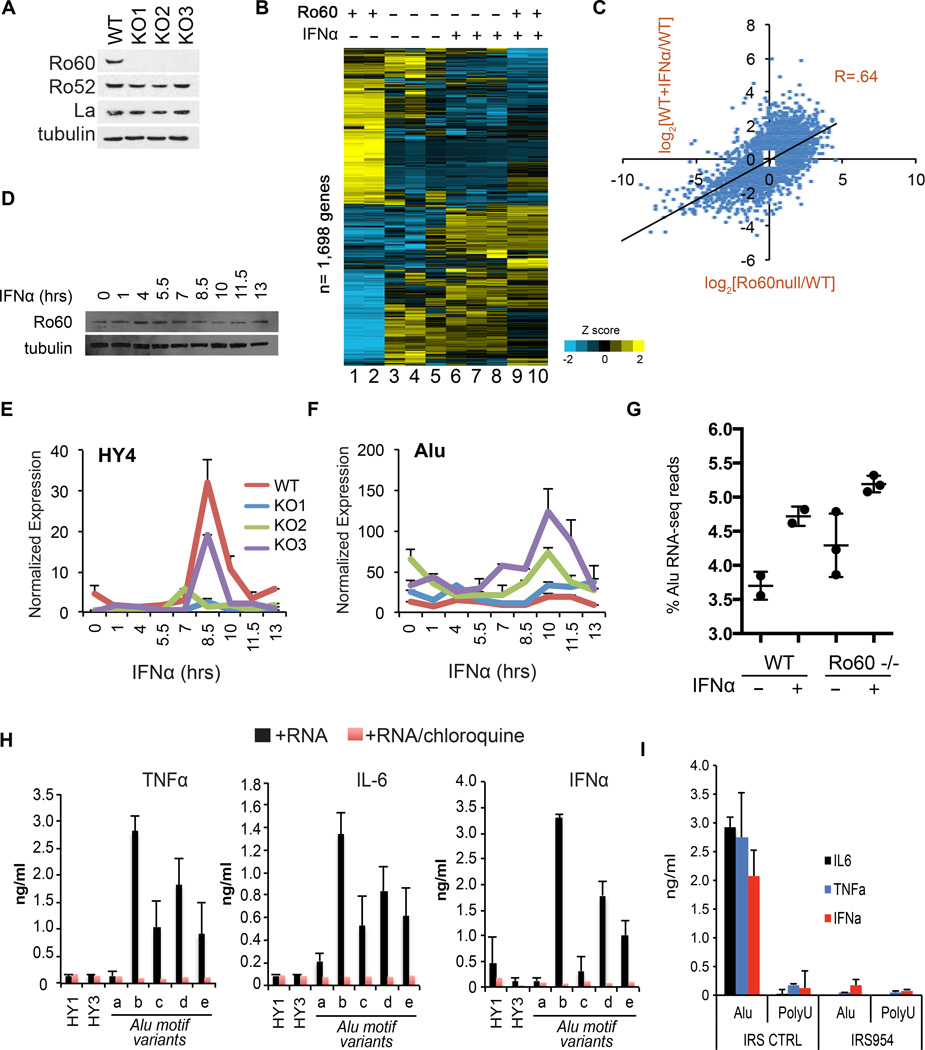
To further explore the functional role of Ro60, we used zinc-finger nuclease technology to generate three GM12878 Ro60 knockout (KO) cell lines, each containing unique bi-allelic frameshift mutations within exon 2 of Ro60/TROVE2 (Fig. S5). The KO lines contained undetectable levels of Ro60 protein (Fig. 3A), and showed markedly reduced retrieval of Alu and Y RNA by Ro60 RIP-PCR (Fig. S6). Surprisingly, RNA-seq at baseline and 6 hours after treatment with IFNα demonstrated that many of the genes altered by IFNα treatment in the parental cells (lanes 9,10) were dysregulated in the Ro60 KOs in the absence of treatment (lanes 3,4,5 Fig. 3B). Gene expression correlated significantly between the unstimulated Ro60 KO lines and the IFNα treated parental cell line (Fig. 3C) (R = 0.64, n=4,520, p<.0001). Genes differentially expressed in the Ro60 KO lines at rest were enriched for a number of immune-related GO terms and Kegg pathways (Fig. S7), suggesting aberrant activation of inflammatory pathways in the absence of Ro60 (see also Table S2 (14)).
Figure 3. Loss of Ro60 in GM12878 cells activates inflammatory gene expression and Alu RNA expression.
(A) Western analysis of Ro60, Ro52, La and tubulin in three independent GM12878 Ro60 KO clones. (B) RNA-seq heatmap of 1,698 genes >2-fold induced or repressed upon 6 hours of IFNα in parental GM12878 cells. Unsupervised clustering of both genes and conditions. Lanes 1,2: Parental cells at baseline, biologic replicates. Lane 3: KO1 at baseline. Lane 4: KO2 at baseline. Lane 5:KO3 at baseline. Lane 6: KO1+IFNα. Lane 7: KO2 + IFNα. Lane 8: KO3 + IFNα. Lanes 9,10: Parental cell line + IFNα, biologic replicates. See Fig. S7 for global correlation of gene expression between clones. (C) Scatterplot of gene expression changes (log2(fold change)) in Ro60 null clones (x-axis) or wildtype parental cells treated with IFNα (y-axis) compared to baseline wildtype cells. Data represent all genes with ≥1.5 fold induction or repression, n= 4,520. P value=2.2e-16. R value= Pearson’s product-moment correlation. (D) Western blot of whole cell lysates at indicated timepoints following IFNα treatment of the parental cell line. (E, F) RT-PCR of HY4 RNA (E) or Alu RNA (F) at indicated timepoints following IFNα treatment of the parental cell line. Expression values normalized to GAPDH. P values of (E): KO1 vs. WT (p=0.015), KO2 vs. WT (p=0.001), KO3 vs. WT (p=0.001); Student’s t-test (G) Quantitation of Alu reads as a percent of all RNA-seq reads from 6 hr. IFNα treated wildtype (biologic replicates, n=2) or Ro60 null clone (n=3) libraries. Error bars represent SD. (H) ELISA assays in culture supernatants of human PBMCs 15 hours following DOTAP transfection of the indicated RNAs (see Fig. 2D for RNA sequences). Error bars represent SD of 3 technical replicates from a representative experiment (of 3). (I) ELISA assays in culture supernatants of human PBMCs 15 hours following DOTAP cotransfection of the indicated RNAs with TLR7/9 inhibitory oligo IRS954 or a control oligo. Error bars represent technical replicates from a representative experiment (of 4).
We next performed a time course of IFNα treatment to determine whether the widespread changes in gene expression included alterations in Ro60-bound RNAs. In the parental cell line, there were minimal changes in the protein levels of Ro60 during the 12 hour experiment (Fig. 3D). At all timepoints, Y RNAs were expressed at reduced levels in the Ro60 null cells compared to parental cells (Fig. 3E, Fig. S7), while total Alu RNAs were found at significantly higher levels in the null cells (Fig. 3F). We confirmed these differences by quantitation of Alu reads from the RNA-seq data (Fig. 3G). Together, these findings demonstrate elevation of Alu expression and activation of interferon induced genes in the absence of Ro60. A possible interpretation of these data is that aberrant expression of endogenous Alu RNAs stimulates intracellular RNA sensors to induce inflammatory responses.
To test this idea, we transfected synthesized RNAs into PBMCs isolated from healthy human donors. The Alu motif RNAs potently stimulated secretion of cytokines, including IFNα, interleukin (IL)-6 and tumor necrosis factor (TNF)-α (Fig. 3H), consistent with a previous report(16). Importantly, cytokine induction by the Alu RNAs was blocked by co-incubation of the transfected cells with chloroquine, a molecular agent that disrupts signaling of endosome-localized Toll like receptors (TLRs) 3,7 and 8, all of which recognize RNA(17, 18), or by co-transfection of a TLR7/9 antagonist, phosphorothioated oligo IRS954(19, 20) (Fig. 3I). Co-incubation of the transfected PBMCs with BX795, an inhibitor of TBK/IKKe(21) which mediates signaling downstream of cytosolic RNA receptors RIG-I, MDA-5 and TLR3, had no effect on Alu-mediated cytokine production (Fig. S8).
Truncated Alu motifs (see Fig. 2D) elicited sub-maximal cytokine responses. Y RNAs showed no ability to elicit cytokine production in this system. Alu motif sequence variants that retained Ro60 binding in vitro (Fig. S4) also elicited cytokine responses comparable to wildtype (Fig. S9). We conclude that cytokine secretion in Alu-motif transfected human PBMCs is TLR7 dependent.
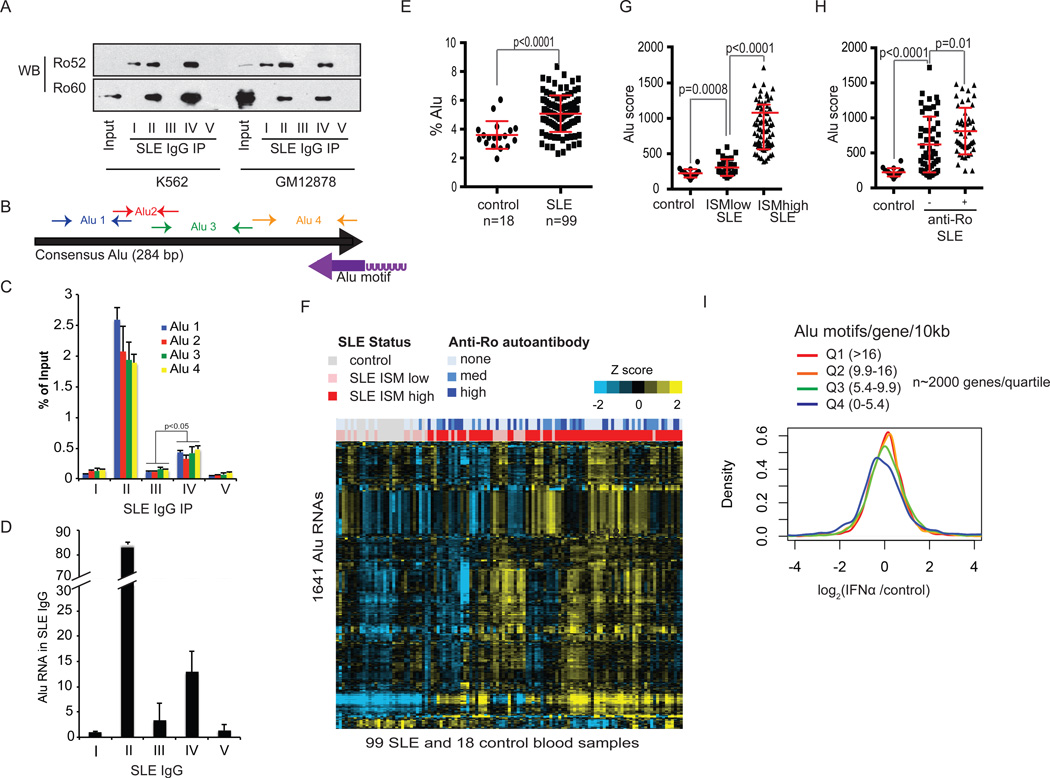
Next, we purified SLE serum IgGs using Protein G beads from subjects representing a spectrum of anti-Ro autoantibodies (Fig. S10), and performed immunoprecipitations from GM12878 and K562 cell lysates. By Western blot, only sera II and IV immunoprecipitated Ro60 (Fig. 4A). Sera II and IV also precipitated significant amounts of Y RNA and POLG RNA (Fig. S10), two of the prominent peaks identified in the Ro60 iCLIP experiments. Using primers spanning the 284 basepair consensus Alu sequence (Fig. 4B), RT-PCR showed that endogenous SLE IgGs also immunoprecipitated full length Alu transcripts (Fig. 4C). Parallel experiments in the Ro60 null cell lines showed reduced immunoprecipitation of Alu and Y RNAs (Fig. S11). Importantly, we found that Alu RNAs were enriched in primary serum IgG immune complexes from anti-Ro60 positive SLE patients (Fig. 4D), suggesting that circulating anti-Ro60 autoantibodies in SLE subjects are pre-loaded with Alu transcripts.
Figure 4. Alu RNAs in Ro60 positive SLE immune complexes and SLE whole blood.
(A) Immunoprecipitation from K562 or GM12878 cell lysates using SLE patient IgGs followed by Western blot of Ro52 or Ro60. Input lane represents 10% total lysate. (B) PCR primer map of Alu elements with respect to the consensus Alu sequence and the identified Ro60 Alu motif from Fig. 2A (purple arrow represents orientation of the motif). (C) SLE IgG immunoprecipitates from GM12878 lysates in (A) were subjected to RT-PCR of Alu RNA using the primers in (B). Error bars represent SD of representative experiment (of 3) (D) RT-PCR of cell-free serum IgGs purified using Protein G beads from 50 ul of SLE lupus sera (I-V). Values represent fold change relative to patient I IgG. Pre-existing Alu RNAs contributed negligibly to the total Alu RNAs retrieved from cell lysates in part (C) (Fig. S11C). (E) Percent of whole blood RNA-seq Alu reads in control or SLE subjects. P value calculated using Student’s t-test. Error bars represent SD (F) Heatmap of the top 1,641 Alu elements expressed in SLE or control subject blood. Unsupervised hierarchical clustering of Alus and subjects. Bars at the top represent anti-Ro titers (blue) or Interferon Signature Metric (ISM) scores (red= ISM high, pink = ISM low). (G, H) Alu score= mean expression of top SLE Alu RNAs(14). (G) Alu scores binned by ISM status; (H) Alu scores binned by anti-Ro status. (I) Distribution of gene expression changes in IFNα treated (6 hr) PBMCs relative to controls; expression is the average of 3 donors. For Alu density calculations, see (14). P value Q4 vs. Q3 <0.001.
Finally, we tested the levels of Alu transcripts in blood cells of SLE patients and controls(22) using RNAseq (99 active SLE, 18 healthy controls; Fig. S12). RNA-seq reads mapping to Alu elements were found at significantly higher levels in SLE subjects than controls (p=6.5E-6), Fig. 4E). Hierarchical clustering of the most highly expressed Alu RNAs (Fig. S13) segregated Interferon Signature Metric (ISM)-high SLE subjects from control and ISM-low patients (Fig. 4F). Alu scores(14) were higher in ISM-low SLE than in controls, and were highest in ISM-high SLE (Fig. 4G). Alu scores were also significantly elevated in anti-Ro autoantibody positive SLE compared to SLE lacking anti-Ro (p=0.01) (Fig. 4H). A subset of ISM-high/Alu score-high SLE subjects lacked anti-Ro antibodies, indicating that there are other mechanisms for induction of IFN and Alu RNA in SLE, likely including autoantibodies to other RNA binding proteins(23).
Endogenous retroelements were recently shown to activate humoral immune responses in mice(24), and aberrant accumulation of retroelements contributes to two human genetic diseases with features of SLE - Aicardi-Goutieres syndrome and familial chiblain lupus(25). Our investigation of the Alu RNAs bound to Ro60 support the hypothesis that retroelements contribute to human autoimmunity, and suggest a model that links Ro60 autoantibody responses and Alu retroelement stimulation of intracellular RNA sensors, including TLR7, with development of SLE, Sjögren’s Syndrome, and congenital heart disease in the offspring of anti-Ro60 mothers(4) (see Fig. S14). We speculate that interferon-stimulated modifications to Alu RNA, such as N6-Methyladenosine (m6A), may mediate this interaction (Fig. S14B). These findings also provide new insights into the lupus-like disorder described in Ro60 knockout mice(26), suggesting that autoimmunity in this model may be mediated by dysregulation of murine retroelements.
Finally, this work may help to explain the maintenance of Alu sequences in the genome over the course of primate evolution. There are more than 1 million copies of the Alu retroelement in the human genome, and it has been postulated that the retention of Alu sequences in gene-rich regions and their transcription in response to stress might provide an evolutionary advantage(27). When we binned all coding genes in quartiles from high to low density of the Alu motif, and plotted gene expression changes upon IFNα treatment, we observed that genes with higher densities of the Alu motif showed increased IFN-inducible gene expression (p<0.001, Fig. 4I, Fig. S14A). These data support a model where the transcription of Alu-containing genes upon pathogen infection upregulates endogenous Alu RNA ‘adjuvants’, which serve to amplify immune responses via stimulation of intracellular RNA sensors, possibly by temporarily overwhelming the ability of endogenous Ro60 to target these RNAs for destruction. Thus, Alu sequences may have persisted in the genome by virtue of their beneficial effects for defense against infectious agents, while at the same time predisposing to anti-Ro60 autoantibody production in the setting of systemic autoimmunity.
Supplementary Material
Acknowledgments
We thank J. Lowe, E. Brown and V. Dixit for their critical review of the manuscript. This work was partially supported by NIH grants HG004659, NS075449 and HG007005 to GWY. GWY is a recipient of the Alfred P Sloan Research Fellowship. GP is a recipient of the National Science Foundation Graduate Research Fellowship Grant No. DGE-1144086.
Footnotes
The data presented in this manuscript are tabulated in the main paper and in the supplementary materials.
An MTA is required for access to the GM12878 Ro60 KO cell lines. Sequencing data available at NCBI GEO, accession number GSE72420.
References and notes
- 1.Clark G, Reichlin M, Tomasi TB., Jr Characterization of a soluble cytoplasmic antigen reactive with sera from patients with systemic lupus erythmatosus. J Immunol. 1969 Jan;102:117. [PubMed] [Google Scholar]
- 2.Alspaugh M, Maddison P. Resolution of the identity of certain antigen-antibody systems in systemic lupus erythematosus and Sjogren's syndrome: an interlaboratory collaboration. Arthritis and rheumatism. 1979 Jul;22:796. doi: 10.1002/art.1780220719. [DOI] [PubMed] [Google Scholar]
- 3.Lee LA. Transient autoimmunity related to maternal autoantibodies: neonatal lupus. Autoimmunity reviews. 2005 Apr;4:207. doi: 10.1016/j.autrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Clancy RM, et al. Ro60-associated single-stranded RNA links inflammation with fetal cardiac fibrosis via ligation of TLRs: a novel pathway to autoimmune-associated heart block. J Immunol. 2010 Feb 15;184:2148. doi: 10.4049/jimmunol.0902248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clancy RM, et al. Impaired clearance of apoptotic cardiocytes is linked to anti-SSA/Ro and -SSB/La antibodies in the pathogenesis of congenital heart block. The Journal of clinical investigation. 2006 Sep;116:2413. doi: 10.1172/JCI27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulte-Pelkum J, Fritzler M, Mahler M. Latest update on the Ro/SS-A autoantibody system. Autoimmunity reviews. 2009 Jun;8:632. doi: 10.1016/j.autrev.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004 Sep;16:541. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman IE, et al. Specific antinuclear antibodies are associated with clinical features in systemic lupus erythematosus. Annals of the rheumatic diseases. 2004 Sep;63:1155. doi: 10.1136/ard.2003.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Chetrit E, Chan EK, Sullivan KF, Tan EM. A 52-kD protein is a novel component of the SS-A/Ro antigenic particle. The Journal of experimental medicine. 1988 May 1;167:1560. doi: 10.1084/jem.167.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maraia RJ, Sasaki-Tozawa N, Driscoll CT, Green ED, Darlington GJ. The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic acids research. 1994 Aug 11;22:3045. doi: 10.1093/nar/22.15.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005 May 20;121:529. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010 Jul;17:909. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppertz I, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014 Feb;65:274. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Materials and methods are available as supplementary materials on Science Online.
- 15.Deininger P. Alu elements: know the SINEs. Genome biology. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green NM, Moody KS, Debatis M, Marshak-Rothstein A. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. The Journal of biological chemistry. 2012 Nov 16;287:39789. doi: 10.1074/jbc.M112.383000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutz M, et al. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. European journal of immunology. 2004 Sep;34:2541. doi: 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- 18.Hart OM, Athie-Morales V, O'Connor GM, Gardiner CM. TLR7/8-mediated activation of human NK cells results in accessory cell-dependent IFN-gamma production. J Immunol. 2005 Aug 1;175:1636. doi: 10.4049/jimmunol.175.3.1636. [DOI] [PubMed] [Google Scholar]
- 19.Barrat FJ, Meeker T, Chan JH, Guiducci C, Coffman RL. Treatment of lupus-prone mice with a dual inhibitor of TLR7 and TLR9 leads to reduction of autoantibody production and amelioration of disease symptoms. European journal of immunology. 2007 Dec;37:3582. doi: 10.1002/eji.200737815. [DOI] [PubMed] [Google Scholar]
- 20.Guiducci C, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010 Jun 17;465:937. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. The Journal of biological chemistry. 2009 May 22;284:14136. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride JM, et al. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis and rheumatism. 2012 Nov;64:3666. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 23.Migliorini P, Baldini C, Rocchi V, Bombardieri S. Anti-Sm and anti-RNP antibodies. Autoimmunity. 2005 Feb;38:47. doi: 10.1080/08916930400022715. [DOI] [PubMed] [Google Scholar]
- 24.Zeng M, et al. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014 Dec 19;346:1486. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nature immunology. 2014 May;15:415. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue D, et al. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proceedings of the National Academy of Sciences of the United States of America. 2003 Jun 24;100:7503. doi: 10.1073/pnas.0832411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409:860. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.