Abstract
Background & aims
Chronic inflammatory liver diseases are associated with estrogen excess and feminization in men, which is thought to be due to compromised liver function to breakdown estrogens. The goal of this study is to determine whether the inflammatory induction of steroid sulfatase (STS), which converts inactive estrogen sulfates to active estrogens, may have contributed to the estrogen excess in chronic liver disease.
Methods
We performed bioinformatic analysis, real-time PCR, immunohistochemistry and UPLC/MS-MS to analyze hepatic STS expression and serum estrogen levels in patients with chronic liver diseases. The crosstalk between NF-κB pathway and STS-regulated estrogen signaling was investigated by electrophoretic mobility shift assay, chromatin immunoprecipitation, luciferase assay and gene knockdown experiments in human hepatocytes.
Results
Hepatic STS was induced in patients with chronic inflammatory liver diseases, which was accompanied by increased circulating estrogen levels. The human STS gene, but not the mouse Sts gene, was induced by inflammatory stimuli in hepatic cells. Mechanistically, STS was established as a novel NF-κB target gene, whose induction facilitated the conversion of inactive estrogen sulfates to active estrogens, and consequently attenuated the inflammatory response. In contrast, genetic or pharmacological inhibition of STS or a direct blockade of estrogen signaling sensitized liver cells to the transcriptional activation of NF-κB and inflammatory response, possibly through the inhibition of IκB kinase (IKK) activation.
Conclusions
Our results suggest a negative feedback loop in chronic inflammatory liver diseases, in which the inflammatory activation of NF-κB induces STS gene expression. The induced STS facilitates the conversion of inactive estrogen sulfates to active estrogens, which in return attenuates the NF-κB-mediated inflammation.
Keywords: steroid sulfatase, estrogens, estrogen metabolism, inflammation, liver disease
Introduction
Abnormal estrogen metabolism in liver disease has been long recognized in the clinic. Concomitants of liver diseases are clinical signs and symptoms like palmar erythema, spider nevus, gynecomastia and infertility due to disturbed homeostasis of steroid hormones, especially the estrogens. Studies have reported increased estrogen levels and signs of endocrine disturbance in patients with chronic liver diseases [1]. The hormone levels are positively correlated to the severity of the liver disease [2], whereas treating patients towards improved liver function resulted in regression of endocrine disturbance [3]. The liver is the primary site of estrogen metabolism through phase I oxidation reactions, which are mainly catalyzed by CYP1A2 and CYP3A4 [4], and phase II conjugation reactions mediated by the estrogen sulfotransferase (EST) [5]. It is thought that damage to the liver impairs its capacity to metabolize and inactivate estrogens, resulting in increased estrogen levels in the circulation [6]. However, there have been reports that changes in steroid hormone levels may occur before the liver functions are compromised [7], suggesting additional mechanisms by which liver disease causes estrogen excess. Although estrogens are known to be the anti-inflammatory hormones, it is unclear whether the estrogen excess can affect the clinical outcome of the underlying liver diseases.
Estrogen sulfation and desulfation represent an important and unique mechanism to control estrogen homeostasis by a reversible metabolic process of conjugation and deconjugation, rather than the destruction of estrogens [8]. Estrogens can be sulfated and deactivated by several sulfotransferases such as EST, SULT1A1 and SULT2A1, with EST being the primary estrogen sulfotransferase at the physiological concentrations [5]. Unlike estrogens, estrogen sulfates cannot bind to the estrogen receptor (ER) and thus are biologically inactive; but they have higher concentrations and prolonged half-life in the circulation, acting as a reservoir for regenerating active estrogens through the STS-mediated desulfation reaction [9]. STS is believed to be the only enzyme responsible for the desulfation of estrogen sulfates.
Consistent with the role of STS in hormonal homeostasis, STS gene deletion or mutation is associated with reproductive manifestations, such as cryptorchidism in males and failed labor progression in females due to disrupted steroid hormone homeostasis [10]. High expression of STS is detected in malignant breast tissues and predicts poor prognosis [11], suggesting an important function of STS in enhancing local estrogen signaling and promoting the development of hormone-dependent breast cancer. Cytokines have been suggested to regulate the expression and activity of STS, but the results have been contradicting. Interleukin-1 decreased the expression and activity of STS in endometrial stromal cells [12]. However, interleukin-6 and tumor necrosis factor α (TNFα) were reported to increase STS activity in breast cancer cells, probably through post-translational mechanisms [13]. It is possible that the effects of cytokines on the expression of STS depend on the cellular context. Nevertheless, little is known about the transcriptional regulation of STS, especially in the liver, the major estrogen-metabolizing organ.
The development of many chronic inflammatory liver diseases is more common in men than in women. The prognosis of hepatocellular carcinoma is also worse for male than for female patients [14]. These gender differences may be accounted for by sex hormones. Although it is not a classic target organ of sex steroid hormones, the liver has been shown to express functional ER and respond to estrogen stimulation. Since estrogens are known for their anti-inflammatory activities [15], they may provide benefit in inhibiting the progression of chronic inflammatory liver diseases.
In this study, we showed that STS is a novel NF-κB target gene that is induced in the liver of patients with hepatitis and cirrhosis. Our data strongly suggest that the inflammatory induction of STS may have contributed to the estrogen excess in chronic liver disease. Our results also suggested an STS-mediated negative feedback loop to inhibit inflammation, in which the NF-κB responsive activation of STS increases active estrogen levels, which in turn attenuates the NF- κB-mediated inflammation.
Materials and Methods
Microarray and bioinformatic analysis
We previously performed microarray analyses [16] on liver samples from human subjects with normal (< 10 mg/L) or elevated (> 10 mg/L) C-reactive protein (CRP) levels. The data have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) database (GSE32504). The microarray results were verified by TaqMan real-time PCR using the following predesigned TaqMan assays obtained from Life Technologies (Carlsbad, CA): Hs00996676_m1 (STS); Hs00960941_m1 (SULT1E1), and Hs00174103_m1 (IL8). A standard curve was generated for each assay using pooled cDNA from the liver sample collections. With the help of the standard curve, a quantitative measure was calculated which was normalized on the housekeeping gene RPLP0. The Gene Expression Omnibus database was also mined for STS expression in subjects with chronic inflammatory liver disease. The GSE28619 dataset, which contained 15 liver samples from alcoholic hepatitis patients and 9 control liver samples, was analyzed on the Affymetrix Human Genome U133 Plus 2.0 microarrays.
Immunohistochemistry (IHC), real-time PCR analysis, and Western blotting
For IHC analysis, we used a commercial tissue microarray and archived paraffin sections from West China Hospital, Sichuan University, China. The tissue microarray slides of human liver diseases (Cat #LV1201) were purchased from US Biomax (Rockville, MD). The slides were provided in a single core per case with clinical information of sex, age, and pathological diagnosis. Samples contained on the tissue microarray were from autopsy and surgical resection, according to the information provided by the vendor (http://www.biomax.us/faq.php). The tissue microarray contains 66 liver cases, including 22 cases of hepatitis, 30 cases of cirrhosis and 14 cases of controls. One of the cirrhosis cases was excluded because of the lack of hepatocytes. Additional liver tissue sections from 20 patients, including 10 cases of control liver samples and 10 cases of chronic hepatitis liver samples, were collected from surgical resection at West China Hospital, Sichuan University, China. Informed consent in writing was obtained from each patient, and the Ethics Committee of the West China Hospital approved the study. The IHC staining was performed using monoclonal anti-STS antibody (dilution 1:50) purchased from Abcam (Cambridge, MA) following the heat-induced antigen-retrieval procedures. The stained slides were evaluated by a surgical pathologist in a blinded fashion and scored according to the staining intensity.
Real-time PCR analyses were performed on the GSE32504 samples, as well as samples derived from the primary human hepatocytes and the human hepatoma Huh7 cells. For real-time PCR analysis, cDNA was synthesized from total RNA by reverse transcription with random hexamer primers and Superscript RT III enzyme from Invitrogen. SYBR Green-based real-time PCR was performed with the ABI 7300 real-time PCR system. Data were normalized against the GAPDH.
All Western blot analyses were performed on primary human hepatocytes or Huh7 cells. For Western blotting, the anti-STS antibody was used at 1:200 dilution. The anti-IKKα (3G12), anti-IKKβ (D30C6), anti-Phospho-IKKα (Ser176)/IKKβ (Ser177) (C84E11) and anti-p65 (D14E12) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Quantification of the Western blotting was performed using the NIH Image J software.
Human serum samples and estrogen measurement
Patients with alcoholic cirrhosis were eligible for recruitment from University of Louisville Hospitals between January 2013 and November 2014. Blood was drawn from the cubital vein of subjects at fasting state. The serum samples were collected at the time of blood collection and stored at -80°C before analysis. A control group of serum samples were collected concurrently from healthy patients undergoing screening colonoscopy that corresponded to patients by gender and age. We collected 6 alcoholic cirrhosis samples and 3 control samples from male subjects. Informed consent was obtained from all participants, and the study was approved by the Institutional Review Boards at the University of Louisville. To measure the level of serum estrogen and estrogen metabolite, UPLC/MS-MS were carried out with a Waters Acquity UPLC system connected with the Xevo TQ triple quadrupole mass spectrometer as we have previously described [17].
Cell culture and drug treatment
Human primary hepatocytes were obtained through the Liver Tissue Procurement and Distribution System (Pittsburgh, PA). The Huh7 and HEK 293 cell lines were obtained from the American Type Culture Collection (Manassas, VA). In experiments without adding estrogen or estrogen sulfate, cells were cultured in DMEM containing 10% standard fetal bovine serum (FBS) at 37 °C. In experiments in which estrogen, estrogen sulfate or anti-estrogen were exogenously added, the cells were maintained in phenol red free DMEM and dextran coated charcoal stripped FBS before treatment with estrogen or estrogen sulfate. Chemicals used in this study include: estrone sulfate (cat #E0251), estradiol sulfate (cat # E9505), β-estradiol (cat #E8875), STX64 (a STS inhibitor; cat #S1950), Fulvestrant (an ER antagonist, also called ICI 182,780; cat #I4409), LPS 0111:B4 (a Toll-like receptor 4 agonist;cat #L4391), phorbol 12-myristate 13-acetate (PMA, a protein kinase C activator; cat #P8139) and pyrrolidine dithiocarbamate (PFTC, a NF-κB inhibitor;cat #P8765), all from Sigma. The recombinant human TNFα (cat #210-TA) was purchased from R&D Systems. For PDTC treatment, cells were pretreated with PDTC for 30 min prior to the treatment of NF-κB activators.
STS activity assay
The hepatic STS enzymatic activity, normalized to protein concentrations, was determined from cell microsomes by an estrone sulfate conversion assay as described before [18].
Measurement of estrogen levels in cell culture medium
Cells were cultured in phenol red free DMEM and charcoal stripped FBS. Cell culture medium was removed and centrifuged at 15,000g for 10 min. The estradiol content of the supernatant was measured by an ELISA assay kit (cat # 582251) from Cayman Chemicals. The UPLC/MS-MS method was used to measure the estradiol level from culture supernatant of cells treated with exogenously added estradiol sulfate. Estrogen concentrations were normalized by cellular protein concentrations.
Electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) assay EMSA was performed using an in vitro translated p65 protein prepared from T7 Quick Coupled Transcription/Translation System from Promega (Madison, WI) and [32P-dCTP]-labeled DNA probes. The binding reaction mixture consisted of 2 μL of radioactive probe, 4 μL of p65 protein, 1 μL of poly (dI-dC) (cat #P4929; Sigma), 4 μL of 5x binding buffer and sterile water to bring the total volume up to 20 μL. For competition assays, a 200-fold excess of unlabeled oligonucleotide was added to the reactions. For supershift assays, the binding reaction mixture was pre-incubated with 1 μL of anti-p65 antibody for 10 min at room temperature before adding the probe. The binding mixtures were incubated at room temperature for 20 min before being resolved on 5% polyacrylamide gel in 0.5 x Tris borate-EDTA at 4 oC for 1-3 h. ChIP assay was performed in Huh7 cells treated with vehicle or 40 ng/mL TNFα for 6 h. The same anti-p65 antibody was used for ChIP analysis following a two-step cross-linking method as described [19]. See Supplementary Table for the EMSA and ChIP primer sequences.
Plasmid construction, transient transfections, and luciferase assay
The 5′-regulatory sequence of the human STS promoter was inspected for potential NF-κB binding sites by using the JASPAR matrices [20]. The WT (nt −447 to +150) the deletion mutant reporter (nt −56 to +150) was generated by PCR amplification using the human genomic DNA template. The PCR product was digested with KpnI and BglII and inserted into the same enzyme-digested pGL3-basic luciferase reporter plasmid from Promega. Mutations of NF-κB sites within the STS reporter plasmids were generated by PCR-based mutagenesis. See Supplementary Table for the cloning primer sequences. All cloned sequences were verified by DNA sequencing. HEK 293 cells were transfected in 48-well plates using the polyethyleneimine polymer transfection reagent. When necessary, cells were stimulated with drugs for 24 h before the luciferase assay. For experiments in which estrogen and estrogen sulfate were added or estrogen signaling was evaluated, the cells were cultured in phenol red free DMEM and charcoal stripped FBS. Relative reporter activity was calculated by comparing with empty vector-transfected or vehicle-treated cells. All transfections were performed in triplicates. For knockdown experiments, the human STS siRNA (J-009602-09) and control-scrambled siRNA (D-001810-10) were purchased from Dharmacon Research (Lafayette, CO). SignalSilence® NF-κB p65 siRNA II (cat #6534) was purchased from Cell Signaling Technology. Transfection of siRNA was performed using DharmaFECT transfection reagent 4 from Dharmacon Research. After siRNA transfection, the cells were collected at 24 h for mRNA expression analysis, and at 48 h for protein expression analysis.
Statistical analysis
The Student's t-test was used to calculate statistical significance between two group means. One-way ANOVA with Dunnett’s test or Tukey’s test was used to compare multiple group means. Two-way ANOVA with Bonferroni post hoc tests was used to compare two groups with multiple data sets. The association of STS expression with disease category was assessed using the Fisher’s exact test. Spearman rank correlation was used for the gene expression correlation analysis. P< 0.05 was considered as statistically significant.
Results
Frequent induction of STS in chronic inflammatory liver diseases, and increased estrogen levels in patients of alcoholic cirrhosis
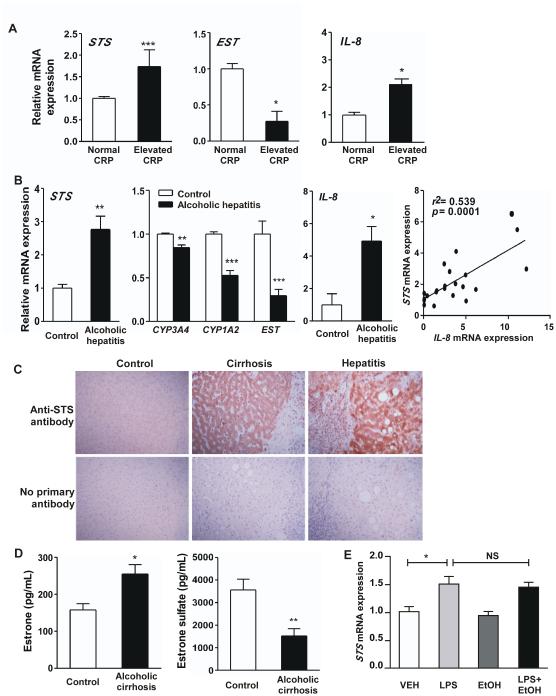
The CRP is an inflammation marker that also predicts disease progression and clinical outcome in patients with liver disease [21]. Microarray analysis on a cohort of human liver samples revealed that the expression of the estrogen re-activating enzyme STS and the estrogen de-activating enzyme EST were respectively induced and reduced in subjects with elevated CRP levels (GSE32504) [16]. The regulation of STS (Fig. 1A, left) and EST (Fig. 1A, middle) was verified by real-time PCR. CRP has been reported to promote inflammation by inducing NF-κB and up-regulation of interleukin-8 (IL-8) [22]. Indeed, our microarray analysis showed that in the same cohort of patients with elevated CRP level, the expression of IL-8 was up-regulated (GSE32504) [16], which was also verified by real-time PCR (Fig. 1A, right).
Figure 1. Frequent induction of STS in chronic inflammatory liver diseases, and increased estrogen levels in patients of alcoholic cirrhosis.
(A) Hepatic mRNA expression of STS (left), EST (middle) and IL-8 (right) in patients with normal or elevated CRP (GSE32504)as determined by real-time PCR. (B) Hepatic mRNA expression of STS (left), CYP3A4, CYP1A2 and EST (middle left) and IL-8 (middle right), and correlation between the expression of IL-8 and STS (right) in control subjects and patients with alcoholic hepatitis (GSE28619). (C) IHC analysis of STS expression. The top panels are representative cases showing STS expression in control, hepatitis and cirrhosis patients. The bottom panels are corresponding controls without the use of primary anti-STS antibody. (D) Serum level of estrone (left) and estrone sulfate (right) in control subjects and patients with alcoholic cirrhosis from the University of Louisville Hospitals. (E) The mRNA expression of STS in human primary hepatocytes treated with vehicle (VEH), LPS (1 μg/mL), Ethanol (EtOH, 100 μM) or LPS + Ethanol for 6 h. *, P<0.05; **, P<0.01; ***, P<0.001; NS, statistically not significant.
In addition, our bioinformatic analysis of the Gene Expression Omnibus database revealed that STS was up-regulated in a cohort of patients with alcoholic hepatitis (GSE28619) [23] (Fig. 1B, left), which is a progressive inflammatory injury to the liver. Concomitant of STS induction was the suppression of phase I and phase II drug-metabolizing enzymes that are involved in estrogen metabolism and de-activation, including CYP1A2, CYP3A4 and EST (Fig. 1B, middle left). Further analysis of this dataset revealed significant inverse correlations between the mRNA expression of STS and CYP1A2 (Spearman R = −0.69; P = 0.0004), CYP3A4 (Spearman R = −0.69; P = 0.0003) and EST (Spearman R = −0.77; P < 0.0001). In patients of alcoholic hepatitis, the expression of IL-8 was also up-regulated (Fig. 1B, middle right), whose expression was positively correlated with the expression of STS (Fig. 1B, right).
To validate the STS induction at the protein level, we performed IHC staining of STS on liver sections from control, cirrhosis or chronic hepatitis subjects. The STS protein exhibited a cytoplasmic distribution pattern, whereas the negative control in which the primary antibody was omitted had no detectable staining (Fig. 1C). We categorized patient samples into weak or strong staining groups according to the immunoreactivity of STS. The incidence of chronic hepatitis and cirrhosis was found to be positively associated with the expression of STS (Table 1).
Table 1.
IHC analysis of STS in patients with chronic liver disease.
| STS Staining |
||||
|---|---|---|---|---|
| Total cases | Weak (%) | Strong (%) | P-values | |
| Control liver | 24 | 17 (70.8) | 7 (29.2) | |
| Chronic hepatitis | 32 | 12 (37.5) | 20 (62.5) | 0.017* |
| Cirrhosis | 29 | 7 (24.1) | 22 (75.9) | 0.0009*** |
P<0.05;
P<0.001.
Control liver vs. disease group.
At the functional level, we reasoned the induction of estrogen re-activating STS and the suppression of estrogen-deactivating CYP3A4/CYP1A2/EST might have functioned in concert to increase the estrogen activity in chronic inflammatory liver disease. Indeed, we found that patients with alcoholic cirrhosis showed an increased serum level of estrone and a reduced level of estrone sulfate (Fig. 1D).
Since alcohol consumption has been reported to change estrogen levels and affect estrogen receptor signaling [2, 24], we tested the direct effect of alcohol on STS expression by treating primary human hepatocytes with ethanol in the presence or absence of LPS, an inflammatory inducer. We found that neither the basal nor the LPS-induced STS expression was affected by ethanol (Fig. 1E). The effect of LPS in inducing inflammation was confirmed by the induction of IL-8 (data not shown). However, we cannot exclude the possibility that alcohol may independently affect hepatic STS expression in patients in vivo.
Treatment of human hepatocytes with NF-κB activators induces the expression and activity of STS
The positive correlation between the expression of STS and the level of CRP/IL-8 strongly suggested an association between inflammation and STS gene expression. The NF-κB family of transcription factors mediates various aspects of the inflammatory responses. Chronic NF-κB activation has been implicated in several chronic liver diseases, such as hepatitis, liver fibrosis, cirrhosis and hepatocellular carcinoma. To determine whether activation of NF-κB induced STS expression in liver cells, we treated primary human hepatocytes with LPS, which signals through the toll-like receptors to stimulate NF-κB [25]. Treatment with LPS increased STS mRNA expression (Fig. 2A, left). In the same LPS-treated primary human hepatocytes, the expression of estrogen-deactivating enzymes CYP3A4, CYP1A2 and EST was down-regulated (Fig. 2A, middle), which was consistent with the patient results (Fig. 1A and B). The induction of STS seemed to be human-specific, because treatment of mice with LPS had little effect on the hepatic expression of Sts (Fig. 2A, right). The protein expression (Fig. 2B) and enzymatic activity (Fig. 2C) of STS were also induced by LPS, as well as by two other NF-κB activators TNF-α [26] and PMA [27].
Figure 2. Treatment of human hepatocytes with NF-κB activators induces the expression and activity of STS.
(A) The mRNA expression of STS (left) and CYP3A4, CYP1A2 and EST (middle) in human primary hepatocytes (HPH) treated with VEH or LPS (1 μg/mL) for 24 h, or mouse liver treated with LPS (5 mg/kg) for 24 h. *, P<0.05; ***, P<0.001. (B) Western blotting results. Top panel: the STS protein expression in HPH treated with VEH, LPS (1 μg/ml), or LPS (1 μg/ml) + PDTC (50 μM) for 24 h. Middle panel: the STS protein expression in HPH treated with VEH, PMA (50 ng/mL) or TNFα (40 ng/mL) for 24 h. Bottom panel: the effect of PDTC (50 μM) co-treatment on PMA (50 ng/mL) or TNFα (40 ng/mL) induced STS protein expression in HPH. (C) The STS enzymatic activities in HPH treated with VEH, TNFα (40 ng/mL), PMA (50 ng/mL), or LPS (1 μg/ml). *, P<0.05; **, P<0.01. (D to F) The mRNA expression (D), protein expression (E) and enzymatic activities (F) of STS in Huh7 cells treated with vehicle, PMA (50 ng/mL) or TNFα (40 ng/mL) for 24 h. When applicable, the cells were co-treated with PDTC (50 μM). (G) The protein expression of p65 and STS in PMA (50 ng/mL)-treated Huh7 cells that were transfected with control siRNA or sip65. *, P<0.05; ***, P<0.001.
To determine whether activation of NF-κB was necessary for the STS induction, we analyzed the STS induction in the presence of a selective NF-κB inhibitor PDTC [28]. As shown in Fig. 2B, pretreatment of primary hepatocytes with PDTC abolished the protein induction of STS by all three NF-κB activators, suggesting the induction was NF-κB dependent. The NF-κB-dependent regulation of STS at the mRNA (Fig. 2D), protein (Fig. 2E) and enzymatic (Fig. 2F) levels was also observed in the human hepatoma Huh7 cell line. Since PDTC has been reported to inhibit 11β-hydroxysteroid dehydrogenase [29] and thus may elicit off-target effect, we also used p65 siRNA to study the role of NF-κB in STS induction. Consistent with the pharmacological inhibition by PDTC, genetic knockdown of p65 by siRNA also attenuated the expression of STS in PMA-treated Huh7 cells (Fig. 2G), further suggesting that the STS induction was NF-κB dependent.
Promoter analysis establishes STS as a NF-κB target gene
To determine whether STS is a direct NF-κB target gene, we cloned the human STS gene promoter and analyzed its regulation by NF-κB. Inspection of the 5′-flanking region of the STS gene revealed three potential NF-κB binding motifs located between nucleotides −50 and −500 (Fig. 3A). EMSA was used to assess the ability of these binding sites to form complexes with the p65 protein, a major subunit of the NF-κB transcription complex. We radiolabeled STS probes containing an individual NF-κB binding site and their mutant variants. As shown in Fig. 3B, the radiolabeled WT probes formed complexes with p65 with mobilities similar to that of the complex formed with the consensus NF-κB binding site derived from the MHC class II-associated invariant chain gene [30]. All three NF-κB binding sites were able to bind to p65, with NF-κB2 and NF-κB3 showing the strongest and weakest binding, respectively. Mutations of the putative NF-κB binding sites attenuated or eliminated the p65 binding, demonstrating the site-specific binding. The binding specificity was further confirmed by competition experiments, in which excess unlabeled WT probes, but not mutant probes, out-competed the p65 binding with the radiolabeled WT probes. To verify the identity of the protein bound to the DNA, the p65 antibody was included in the binding reaction, which decreased the mobility of the protein-DNA complex with a supershift. ChIP assay on TNFα-treated Huh7 cells was used to assess the in situ recruitment of p65 to the three NF-κB sites. As shown in Fig. 3C, the p65 protein was significantly recruited to NF-κB1 and NF-κB2, but not NF-κB3, upon the TNFα stimulation. The recruitment of p65 to a classical NF-κB site in the IL-8 gene promoter was included as a positive control.
Figure 3. STS is a transcriptional target of NF-κB.
(A) Schematic diagram of the 5′-flanking region of the human STS gene and the three putative NF-κB binding sites. Sequences of the WT and mutant NF-κB binding sites and a consensus NF-κB binding site are shown. (B) The binding of p65 to the three STS NF-κB binding sites was evaluated by EMSA. (C) The recruitment of p65 to the three STS NF-κB binding sites was evaluated by ChIP assay on Huh7 cells treated with VEH or TNFα using an anti-p65 antibody. The recruitment of p65 to a NF-κB binding site on the IL-8 gene promoter was included as a positive control. (D) The transactivation of the WT and mutant STS promoter by p65 was evaluated by transient transfection and luciferase reporter assay. Results are fold induction over the luciferase activity of the empty vector pGL-3 from triplicate assays. *, P<0.05; **, P<0.01.
To functionally test the STS gene promoter and the three NF-κB binding sites, we cloned the WT (nt −447 to +150), mutant and deletion variant of the STS promoter sequences and placed them upstream of the luciferase reporter gene. The luciferase reporters were transfected into human HEK293 cells. As shown in Fig. 3D, co-transfection of p65 induced the luciferase reporter gene driven by the WT STS promoter, but not by the deletion mutant lacking all three NF-κB binding sites or when all three sites were mutated. When the NF-κB binding sites were mutated individually, mutations of NF-κB1 or NF-κB2 alone abolished the transactivation by p65. However, mutation of NF-κB3 alone failed to abolish the p65 effect, consistent with its weak p65 binding in EMSA and lack of p65 recruitment in ChIP assay.
The inflammatory induction of STS induces estrogen activity in liver cells
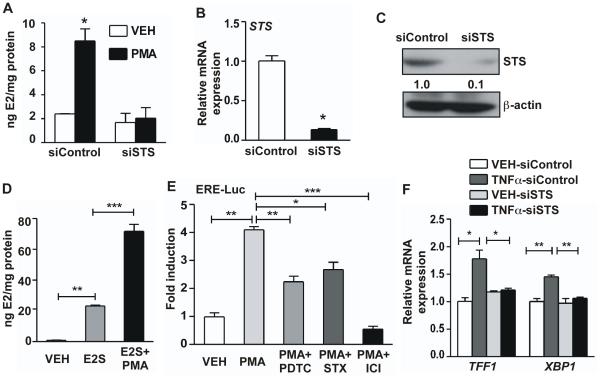
Estrogen sulfates are the preferred STS substrates that exist in high concentrations in vivo [9]. We then hypothesized that the induction of STS by NF-κB activators may convert the inactive estrogen sulfates to active estrogens, and thus enhance estrogen signaling in the liver cells. Indeed, treatment of Huh7 cells cultured in charcoal stripped FBS with PMA increased the estrogen level in the cell culture supernatant, which was abolished upon siSTS knockdown (Fig. 4A). The STS knockdown efficiency was confirmed at both the mRNA and protein levels (Fig. 4B and 4C). The estrogen may have been converted from serum estrogen sulfate in the charcoal stripped FBS, which contained high levels of estrogen sulfate but little estrogen (data not shown). Indeed, treatment of Huh7 cells with exogenously added estradiol sulfate increased the culture supernatant level of estradiol likely due to the expression of endogenous STS, whereas co-treatment with the STS-inducing PMA further increased the estradiol level (Fig. 4D). Consistent with the increased estrogen levels, the estrogen activity as measured by the activation of the estrogen-responsive luciferase reporter gene ERE-Luc, was enhanced by the PMA treatment (Fig. 4E). The induction of ERE-Luc by PMA was both NF-κB and STS dependent, because pretreatment of cells with PDTC or the STS inhibitor STX64 [31] attenuated the PMA effect in inducing the reporter activity (Fig. 4E). The PMA effect on the ERE-Luc reporter gene was also abolished in cells treated with the ER antagonist ICI (Fig. 4E), suggesting that the PMA effect on the ERE-Luc reporter was estrogen/ER dependent. When the expression of endogenous estrogen-responsive genes TFF1 and XBP1 [32, 33] was measured, we found the expression of both genes was induced by TNFα, and this induction was abolished when STS was knocked-down by siSTS (Fig. 4F).
Figure 4. The inflammatory induction of STS enhances estrogen activity in Huh7 cells.
(A to C) The E2 levels in cell culture supernatant from control siRNA or siSTS-transfected Huh7 cells treated with either VEH or PMA (50 ng/mL) for 24 h (A). The knockdown efficiency of siSTS as determined by real-time PCR (B) and Western blotting (C). (D) The level of E2 in cell culture supernatant of Huh7 cells treated with VEH, E2S (10 μM), or E2S (10 μM) + PMA (50 ng/mL) for 24 h. (E) The effects of PDTC (25 μM), STX (10 μM) or ICI (100 nM) on PMA (50 ng/mL)-induced ERE-luciferase reporter activity. (F) TNFα (40 ng/mL)-induced expression of estrogen-responsive genes in Huh7 cells transfected with control siRNA or siSTS.
The inflammation-responsive and STS-mediated induction of estrogen activity attenuates the inflammatory response in liver cells, possibly through the inhibition of IκB kinase (IKK) activation
Given the known anti-inflammatory activity of estrogens [15], we further hypothesized that the induction of STS in response to NF-κB activation may help to attenuate the NF-κB response by activating the estrogen signaling pathway. Using an NF-κB responsive luciferase reporter gene, we showed the PMA-induced activation of NF-κB was attenuated by the treatment of either estrone sulfate or estradiol (Fig. 5A). The effect of estrone sulfate may have resulted from its conversion to estrone by the endogenous STS. In contrast, the PMA-induced NF-κB activation was enhanced by STX64 and ICI (Fig. 5B), presumably by inhibiting the STS-mediated estrogen formation and a direct inhibition of estrogen activity, respectively. When the expression of endogenous inflammatory/NF-κB target genes was measured, we found the TNFα-responsive activation of IL-8 and MCP-1 or VCAM was enhanced when STS was pharmacologically inhibited by STX64 (Fig. 5C) or genetically knocked down by siSTS (Fig. 5D). The pattern of the STX64 and siSTS effect was recapitulated in cells co-treated with ICI (Fig. 5E), suggesting the effect of STX64 and siSTS was achieved through the inhibition of estrogen/ER signaling.
Figure 5. The inflammation-responsive and STS-mediated induction of estrogen activity attenuates the inflammatory response in Huh7 cells.
(A) Treatment of Huh7 cells with E1S (100 nM) or E2 (10 nM) attenuated the PMA (50 ng/mL)-induced NF-κB luciferase reporter activity. (B) The effect of STX (10 μM) or ICI (100 nM) on PMA (50 ng/mL)-induced NF-κB reporter activity. (C to E) The effect of STX (10 μM) (C), siSTS (D), or ICI (100 nM) (E) on TNFα (40 ng/mL)-induced expression of endogenous NF-κB target genes. (F and G) The effects of E2 (F) and ICI and STX (G) on PMA-induced phosphorylation of IKK α and β as shown by Western blotting.
To understand the molecular mechanism by which estrogens inhibited NF-κB, we analyzed the activation of IKK, which is an upstream kinase for NF-κB activation [34]. Pretreatment of Huh7 cells with estradiol attenuated the PMA-induced phosphorylation of IKK α and β (Fig. 5F). In contrast, the PMA-induced IKK α and β phosphorylation was enhanced by the co-treatment of STX64 or ICI (Fig. 5G). These results suggested that the estrogen/ER signaling may inhibit NF-κB activation and inflammation by blocking the IKK phosphorylation.
Discussion
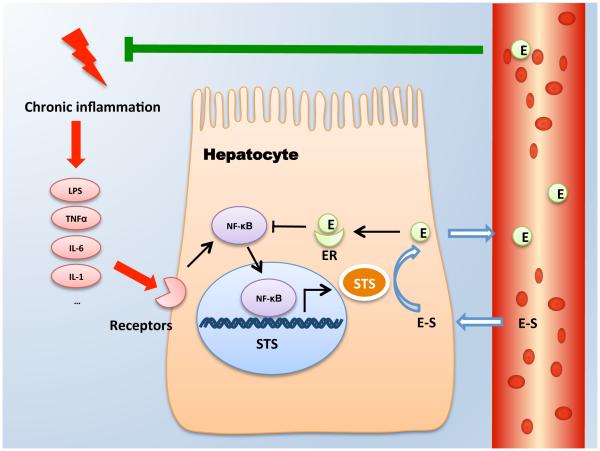
In this study, we have uncovered a novel endocrine basis for the estrogen excess in chronic liver diseases. Based on our results, we propose a negative feedback loop in chronic inflammatory liver diseases that is triggered initially by inflammation and activation of NF-κB, which then induces STS gene expression in hepatocytes and converts inactive estrogen sulfates into active estrogens. The resulting estrogens in return, inhibit the NF-κB-mediated inflammation via the estrogen/ER signaling, thereby completing the negative feedback loop (Fig. 6).
Figure 6. A proposed STS-mediated negative feedback loop in chronic inflammatory liver disease that limits the activity of NF-κB and attenuates inflammation.
During chronic inflammation, inflammatory mediators such as LPS and TNFα act on hepatocytes to elicit NF-κB activation. NF-κB then induces the expression of STS, which converts inactive estrogen sulfates to active estrogens and increases estrogen level in the liver and circulation. The estrogens/ER signaling in return suppresses the NF-κB response and inhibits inflammation.
Chronic inflammatory liver diseases are commonly associated with estrogen excess. The conventional explanation is that liver damage weakens the liver's ability to metabolically deactivate estrogens. Indeed, in both cohorts of patients we analyzed, the expression of estrogen deactivating enzymes CYP1A2, CYP3A4 and EST was decreased. In the current study, we presented comprehensive evidence demonstrating that in addition to the down-regulation of estrogen deactivating enzymes, the transcriptional activation of the estrogen-generating STS may have also contributed to the estrogen excess. Consistent with our hypothesis, patients with alcoholic cirrhosis showed decreased estrogen sulfate and increased estrogen levels in their serum. Considering the high circulating level and longer half-life of estrogen sulfates, it is even possible that the induction of STS may have played a dominating role in causing the estrogen excess in patients of chronic inflammatory liver diseases. Our results also provided an explanation why changes in steroid hormone levels in patients can occur before the liver functions are compromised [7]. However, we cannot exclude the possibilities that other mechanisms may also contribute to the estrogen excess in liver disease, such as the alcohol responsive increase of conversion of androgens to estrogens, and the relative hyperestrogenism due to alcohol induced gonadal toxicity as seen in some patients with alcoholic liver disease [35].
One of our most interesting and surprising findings is the inflammatory induction of STS and the establishment of STS as a NF-κB target gene. This finding is intriguing because most drug- metabolizing enzymes as well as transporters are suppressed by inflammation and NF-κB [36]. It is interesting to note that the induction of STS was human-specific because LPS treatment had little effect on the expression of Sts in the mouse liver. The species specificity of the regulation may be due to the substantial divergence between the human and mouse STS genes. The genomic DNA of the mouse Sts gene spans approximately 9-kb, which is substantially smaller than its 146-kb human counterpart [37, 38]. Despite the much-appreciated biological significance of STS, the transcriptional regulation of this gene has been poorly understood. It has been reported that the human STS gene promoter resembled neither a tightly regulated gene that often contains a TATA box to position the RNA polymerase, nor a housekeeping gene that is usually GC rich and contains binding sites for the Sp1 transcriptional factor. The human STS promoter is GC poor and lacks the TATA box and Sp1 binding sites [39]. In the current study, we established the human STS gene as a transcriptional target of NF-κB. NF-κB transactivates STS gene expression through its binding sites in the STS gene promoter.
The biological and clinical relevance of the inflammatory regulation of STS is also intriguing. Our results clearly show that the inflammatory induction of STS enhances estrogen activity, which subsequently attenuates the NF-κB activation and inflammatory response, possibly through the inhibition of IKK activation. The NF-κB and STS mediated negative feedback loop to limit the NF-κB activity may however represent a double-edged sword. Although the consequent increase in estrogen activity can limit inflammation as suggested by our results, the induction of STS does have a potential to prevent the host’s ability to launch an efficient and pro-survival inflammatory response. It is possible that the significance of this negative feedback loop in liver diseases is disease stage-specific, because the NF-κB-mediated inflammation in hepatocytes plays a dual role in the progression of liver diseases. In the early stages of liver diseases, activation of NF-κB helps to fight infection and prevent hepatocyte death by inducing the anti-apoptotic genes. In the late stages, however, NF-κB also promotes the survival of hepatocytes harboring oncogenic mutations, which increase the risk of hepatocellular carcinoma. Future studies are necessary to pinpoint the stage when NF-κB activation becomes oncogenic, so that more precise treatment can be launched. On the other hand, the anti-inflammatory effect of estrogen may not be limited to the suppression of NF-κB. Activation of the estrogen signaling has been reported to inhibit the development of fatty liver, which is an important cause of hepatic inflammation [40]. Although estrogens may have beneficial effect in inhibiting inflammation, potential adverse effects, such as tumor promotion, risk of cardiovascular disease and feminization of males, limit the utility of systemic estrogen therapies. It will be ideal that selective ER modulators can be developed to protect against liver injury without causing adverse effect to extrahepatic tissues. More studies are needed in order to validate STS activation or other estrogen-related therapies in the management of chronic liver diseases.
Among the limitations, although the chemical inhibitors we used in this study, such as the STS inhibitor STX64, NF-κB inhibitor PDTC, and ER antagonist ICI 182,780, are well-established inhibitors to manipulate protein activities, we cannot exclude the possibilities that these chemicals may interfere with the off-target regulatory pathways [29]. It is encouraging to note that in the case of STS and NF-κB, the results from the use of pharmacological inhibitors were consistent with those obtained from the genetic siRNA knockdowns. In conclusion, our results provide a novel endocrine basis for the estrogen excess in chronic liver diseases, pointing to a critical and comprehensive role of the hepatic microenvironment in the regulation of estrogen homeostasis. We propose that the inflammatory regulation of STS represents a novel mechanism to control estrogen homeostasis and inflammation.
Supplementary Material
Acknowledgement
We thank Dr. Stephen J. Winters (University of Louisville) for his invaluable comments on the manuscript and Dr. Gutian Xiao (University of Pittsburgh) for the NF-κB reporter gene.
Financial support: This work was supported in part by National Institutes of Health grants DK083952, HD073070 (to W.X.), K23AA18399 (to M.C.C.), 1U24DK097154 (to N.W.G.), and by the German Federal Ministry of Education and Research Virtual Liver Network Grant 0315755 and the Robert Bosch Foundation, Stuttgart (to U.M.Z.). Normal human hepatocytes were obtained through the Liver Tissue Cell Distribution System, Pittsburgh, Pennsylvania, which was funded by NIH Contract # HHSN276201200017C.
Abbreviations
- ChIP
chromatin immunoprecipitation
- CRP
C-reactive protein
- CYP
cytochrome P450
- E2
estradiol
- E2S
estradiol sulfate
- EMSA
electrophoretic mobility shift assay
- ER
estrogen receptor
- EST
estrogen sulfotransferase
- FBS
fetal bovine serum
- HPH
human primary hepatocytes
- ICI
ICI 182,780
- IHC
immunohistochemistry
- IL-8
interleukin-8
- IKK
IκB kinase
- LPS
lipopolysaccharide
- MCP-1
monocyte chemotactic protein-1
- PDTC
pyrrolidine dithiocarbamate
- PMA
phorbol 12-myristate 13-acetate
- STS
steroid sulfatase
- STX
STX64
- TFF1
trefoil factor 1
- TNFα
tumor necrosis factor α
- VCAM
vascular cell adhesion molecule
- VEH
vehicle
- WT
wild-type
- XBP1
X-box binding protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author’s contributions: Study concept and design (MJ, MK, UMZ, WX); acquisition of data (MJ, MK, MKM, NWG, KWS, YG, JH, XZ, QS, JL); analysis and interpretation of data (MJ, MK, UMZ, MCC, WQ, SL, WX); drafting of the manuscript (MJ, MK, UMZ, WX); critical revision of the manuscript (MCC, KWS, WQ, SL); statistical analysis (MJ, MK); obtained funding (UMZ, MCC, NWG, WX).
Conflict of interest: The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
References
- [1].Adlercreutz H. Oestrogen metabolism in liver disease. J Endocrinol. 1970;46:129–163. doi: 10.1677/joe.0.0460129. [DOI] [PubMed] [Google Scholar]
- [2].Gavaler JS. Alcohol effects on hormone levels in normal postmenopausal women and in postmenopausal women with alcohol-induced cirrhosis. Recent Dev Alcohol. 1995;12:199–208. doi: 10.1007/0-306-47138-8_11. [DOI] [PubMed] [Google Scholar]
- [3].Long RS, Simmons EE. The liver and estrogen metabolism; report of cases. AMA Arch Intern Med. 1951;88:762–769. doi: 10.1001/archinte.1951.03810120063006. [DOI] [PubMed] [Google Scholar]
- [4].Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- [5].Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- [6].Glass SJ, Edmondson HA, Soll SN. Sex hormone changes associated with liver disease. Endocrinology. 1940;27:749–752. [Google Scholar]
- [7].Becker U. The influence of ethanol and liver disease on sex hormones and hepatic oestrogen receptors in women. Dan Med Bull. 1993;40:447–459. [PubMed] [Google Scholar]
- [8].Hobkirk R. Steroid sulfation Current concepts. Trends Endocrinol Metab. 1993;4:69–74. doi: 10.1016/s1043-2760(05)80018-9. [DOI] [PubMed] [Google Scholar]
- [9].Reed MJ, Purohit A, Woo LW, Newman SP, Potter BV. Steroid sulfatase: molecular biology, regulation, and inhibition. Endocr Rev. 2005;26:171–202. doi: 10.1210/er.2004-0003. [DOI] [PubMed] [Google Scholar]
- [10].Traupe H, Happle R. Clinical spectrum of steroid sulfatase deficiency: X-linked recessive ichthyosis, birth complications and cryptorchidism. Eur J Pediatr. 1983;140:19–21. doi: 10.1007/BF00661898. [DOI] [PubMed] [Google Scholar]
- [11].Miyoshi Y, Ando A, Hasegawa S, Ishitobi M, Taguchi T, Tamaki Y, et al. High expression of steroid sulfatase mRNA predicts poor prognosis in patients with estrogen receptor-positive breast cancer. Clin Cancer Res. 2003;9:2288–2293. [PubMed] [Google Scholar]
- [12].Matsuoka R, Yanaihara A, Saito H, Furusawa Y, Toma Y, Shimizu Y, et al. Regulation of estrogen activity in human endometrium: effect of IL-1beta on steroid sulfatase activity in human endometrial stromal cells. Steroids. 2002;67:655–659. doi: 10.1016/s0039-128x(02)00016-8. [DOI] [PubMed] [Google Scholar]
- [13].Newman SP, Purohit A, Ghilchik MW, Potter BV, Reed MJ. Regulation of steroid sulphatase expression and activity in breast cancer. J Steroid Biochem Mol Biol. 2000;75:259–264. doi: 10.1016/s0960-0760(00)00177-1. [DOI] [PubMed] [Google Scholar]
- [14].El-Serag HB, Mason AC, Key C. Trends in survival of patients with hepatocellular carcinoma between 1977 and 1996 in the United States. Hepatology (Baltimore, Md) 2001;33:62–65. doi: 10.1053/jhep.2001.21041. [DOI] [PubMed] [Google Scholar]
- [15].Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28:521–574. doi: 10.1210/er.2007-0001. [DOI] [PubMed] [Google Scholar]
- [16].Schroder A, Klein K, Winter S, Schwab M, Bonin M, Zell A, et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2013;13:12–20. doi: 10.1038/tpj.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gaikwad NW. Ultra performance liquid chromatography-tandem mass spectrometry method for profiling of steroid metabolome in human tissue. Anal Chem. 2013;85:4951–4960. doi: 10.1021/ac400016e. [DOI] [PubMed] [Google Scholar]
- [18].Selcer KW, Difrancesca HM. Characterization of steroid sulfatase in the MC3T3-E1 mouse pre-osteoblastic cell line. Steroids. 2012;77:696–702. doi: 10.1016/j.steroids.2012.02.024. [DOI] [PubMed] [Google Scholar]
- [19].Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. BioTechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- [20].Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cervoni JP, Thevenot T, Weil D, Muel E, Barbot O, Sheppard F, et al. C-reactive protein predicts short-term mortality in patients with cirrhosis. J Hepatol. 2012;56:1299–1304. doi: 10.1016/j.jhep.2011.12.030. [DOI] [PubMed] [Google Scholar]
- [22].Devaraj S, Kumaresan PR, Jialal I. Effect of C-reactive protein on chemokine expression in human aortic endothelial cells. J Mol Cell Cardiol. 2004;36:405–410. doi: 10.1016/j.yjmcc.2003.12.005. [DOI] [PubMed] [Google Scholar]
- [23].Affo S, Dominguez M, Lozano JJ, Sancho-Bru P, Rodrigo-Torres D, Morales-Ibanez O, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fan S, Meng Q, Gao B, Grossman J, Yadegari M, Goldberg ID, et al. Alcohol stimulates estrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000;60:5635–5639. [PubMed] [Google Scholar]
- [25].Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- [26].Ding WX, Yin XM. Dissection of the multiple mechanisms of TNF-alpha-induced apoptosis in liver injury. J Cell Mol Med. 2004;8:445–454. doi: 10.1111/j.1582-4934.2004.tb00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Holden NS, Squires PE, Kaur M, Bland R, Jones CE, Newton R. Phorbol ester-stimulated NF-kappaB-dependent transcription: roles for isoforms of novel protein kinase C. Cell Signal. 2008;20:1338–1348. doi: 10.1016/j.cellsig.2008.03.001. [DOI] [PubMed] [Google Scholar]
- [28].Schreck R, Meier B, Mannel DN, Droge W, Baeuerle PA. Dithiocarbamates as potent inhibitors of nuclear factor kappa B activation in intact cells. J Exp Med. 1992;175:1181–1194. doi: 10.1084/jem.175.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Atanasov AG, Tam S, Rocken JM, Baker ME, Odermatt A. Inhibition of 11 beta-hydroxysteroid dehydrogenase type 2 by dithiocarbamates. Biochem Biophys Res Commun. 2003;308:257–262. doi: 10.1016/s0006-291x(03)01359-7. [DOI] [PubMed] [Google Scholar]
- [30].Pessara U, Koch N. Tumor necrosis factor alpha regulates expression of the major histocompatibility complex class II-associated invariant chain by binding of an NF-kappa B-like factor to a promoter element. Mol Cell Biol. 1990;10:4146–4154. doi: 10.1128/mcb.10.8.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Purohit A, Woo LW, Potter BV, Reed MJ. In vivo inhibition of estrone sulfatase activity and growth of nitrosomethylurea-induced mammary tumors by 667 COUMATE. Cancer Res. 2000;60:3394–3396. [PubMed] [Google Scholar]
- [32].Barkhem T, Haldosen LA, Gustafsson JA, Nilsson S. pS2 Gene expression in HepG2 cells: complex regulation through crosstalk between the estrogen receptor alpha, an estrogen-responsive element, and the activator protein 1 response element. Mol Pharmacol. 2002;61:1273–1283. doi: 10.1124/mol.61.6.1273. [DOI] [PubMed] [Google Scholar]
- [33].Sengupta S, Sharma CG, Jordan VC. Estrogen regulation of X-box binding protein-1 and its role in estrogen induced growth of breast and endometrial cancer cells. Horm Mol Biol Clin Investig. 2010;2:235–243. doi: 10.1515/HMBCI.2010.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Van Thiel DH, Lester R. Alcoholism: its effect on hypothalamic pituitary gonadal function. Gastroenterology. 1976;71:318–327. [PubMed] [Google Scholar]
- [36].Klein M, Thomas M, Hofmann U, Seehofer D, Damm G, Zanger UM. A systematic comparison of the impact of inflammatory signaling on absorption, distribution, metabolism, and excretion gene expression and activity in primary human hepatocytes and HepaRG Cells. Drug Metab and Dispos. 2015;43:273–283. doi: 10.1124/dmd.114.060962. [DOI] [PubMed] [Google Scholar]
- [37].Salido EC, Li XM, Yen PH, Martin N, Mohandas TK, Shapiro LJ. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts) Nat Genet. 1996;13:83–86. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- [38].Yen PH, Marsh B, Allen E, Tsai SP, Ellison J, Connolly L, et al. The human X-linked steroid sulfatase gene and a Y-encoded pseudogene: evidence for an inversion of the Y chromosome during primate evolution. Cell. 1988;55:1123–1135. doi: 10.1016/0092-8674(88)90257-7. [DOI] [PubMed] [Google Scholar]
- [39].Li XM, Alperin ES, Salido E, Gong Y, Yen P, Shapiro LJ. Characterization of the promoter region of human steroid sulfatase: a gene which escapes X inactivation. Somat Cell Mol Genet. 1996;22:105–117. doi: 10.1007/BF02369901. [DOI] [PubMed] [Google Scholar]
- [40].Monteiro R, Teixeira D, Calhau C. Estrogen signaling in metabolic inflammation. Mediators Inflamm. 2014;2014:615917. doi: 10.1155/2014/615917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.