SUMMARY
Fertilization is a conserved process in all sexually reproducing organisms whereby sperm bind and fuse with oocytes. Despite the importance of sperm-oocyte interactions in fertilization, the molecular underpinnings of this process are still not well understood. The only cognate ligand-receptor pair identified in the context of fertilization is sperm-surface Izumo and egg-surface Juno in mouse [1]. Here we describe a genetic screening strategy to isolate fertilization mutants in Caenorhabditis elegans in order to generate a more complete inventory of molecules required for gamete interactions. From this screening strategy, we identified, cloned, and characterized spe-45, a gene that encodes an Izumo-like immunoglobulin superfamily protein. Mammalian Izumo is required for male fertility and has the same basic mutant phenotype as spe-45. Worms lacking spe-45 function produce morphologically normal and motile sperm that cannot fuse with oocytes despite direct contact in the reproductive tract. The power of this screen to identify proteins with ancient sperm functions suggests that characterization of additional mutants from our screen may reveal other deeply conserved components in fertility pathways and complement studies in other organisms.
RESULTS AND DISCUSSION
A genetic screen for sterile mutants
We designed a forward genetic screen to isolate C. elegans mutants defective in fertilization. We used the strain sem-2(n1343);Is[Pelt-7::gfp; rol-6(su1006)] for our screening (Figure S1), and isolated twelve temperature-sensitive (ts) mutants and 23 non-conditional sterile mutants. Since this was a pilot screen, we did not attempt to pick every single candidate from our screen, and our screen did not reach saturation. Hence, the same screening strategy can be employed to isolate a wealth of mutants going forward that would help us understand the biology of fertilization. The feasibility of high throughput screening in C. elegans can complement the use of other model organisms in which other strategies are more conducive, such as biochemical analyses of sperm or eggs in sea urchin, and in vitro fertilization in mouse [2, 3]. To validate the utility of our screening strategy, we decided to characterize one of our new ts mutants, spe-45(as38). Adult spe-45 mutants are healthy and all pre-fertilization events [16] are normal in spe-45(as38) at all culture conditions (Table S1).
SPE-45 is required for fertility in both sexes
Hermaphrodites that are homozygous for spe-45(as38) mutation are completely self-sterile at 25°C. However, at 20°C and 16°C, the spe-45(as38) mutants produce modest numbers of progeny, suggesting that the as38 allele that we isolated is a temperature-sensitive mutant (Figures 1A). In contrast, spe-45(tm3715) hermaphrodites were profoundly sterile under all culture conditions (Figure 1A, see accompanying paper by Nishimura et al.). We conclude that SPE-45 is required for fertility in hermaphrodites. Further, spe-45 mutant hermaphrodites with either allele proved to be fertile upon crossing with wild-type males, indicating that the fertility defect in spe-45 mutants is restricted to their sperm and not their oocytes.
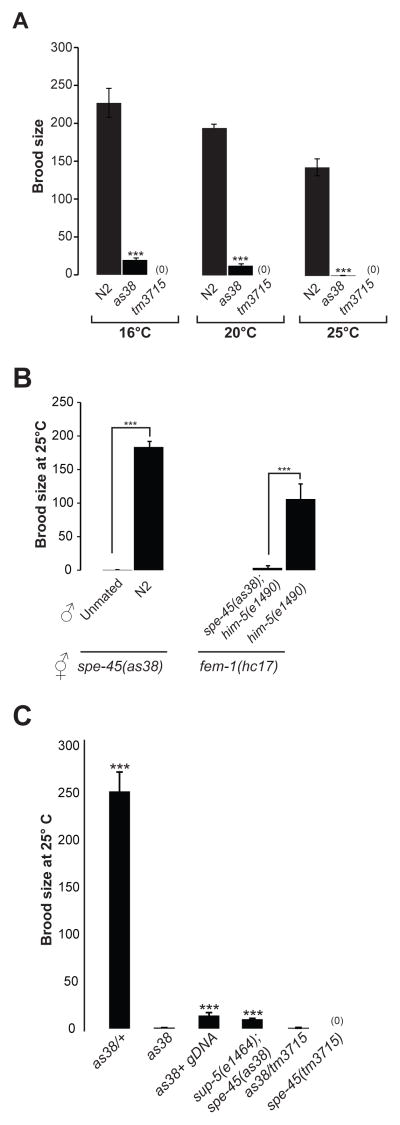
Figure 1. Worms with Mutations in spe-45 are Sterile.
(A) Brood sizes (self-progeny) of wild-type, spe-45(as38) and spe-45(tm3715) mutants at indicated temperature; “***” indicates where the P value is <0.001 between the wild type and spe-45(as38). See Figure S1 for screening method that identified spe-45(as38) and Table S1 for summary of pre-fertilization events.
(B) Brood sizes of spe-45(as38) hermaphrodites left unmated or mated with N2 males at 25°C. Brood sizes of fem-1(hc17) females mated with spe-45(as35);him-5(e1490) or him-5(e1490) males. “***” indicates where the P value is <0.001.
(C) Brood sizes of indicated genotypes at 25°C. spe-45 gDNA refers to extrachromosomal array comprising the admixture of the fosmids WRM061bG12, WRM0641cH06, WRM066cF06, WRM0631aA01 and WRM0624aC09.
To test the fertility of males carrying spe-45 mutations, we crossed them to fem-1 mutant hermaphrodites. fem-1 mutants do not produce any sperm, and hence do not produce any self-progeny [4]. When crossed with him-5 males, the fem-1 animals produce cross-progeny. However, the spe-45(as38);him-5 males fail to sire progeny when crossed with fem-1 at 25°C, indicating that SPE-45 is required for fertility in males as well. (Figure 1B).
Spermatogenesis and sperm activation is normal in both sexes of spe-45(as38)
The spermathecae of spe-45 hermaphrodites contain and retain sperm, just like wild-type control animals (Figure 2A), indicating that the sperm production is uncompromised and the spermatozoa are likely to be motile in spe-45(as38) hermaphrodites. To assess male-derived sperm morphology, sperm were isolated from spe-45;him-5 males, either in sperm media alone or in the presence of the in vitro activator, pronase (Figure 2B). As viewed under DIC optics, both the spermatids and spermatozoa were indistinguishable in both quality and apparent quantity from that of control him-5 males. SPE-45 is dispensable for in vitro activation since pseudopods were present in over 90% of sperm from both wild-type and spe-45 mutant sperm. We also examined hermaphrodite-derived sperm from dissected spe-45 mutants. These sperm were activated in vivo with fully formed pseudopods and were indistinguishable from wild-type sperm (Figure 2B). Taken together, we conclude that spermatogenesis and sperm activation is normal in both sexes of spe-45 mutants.
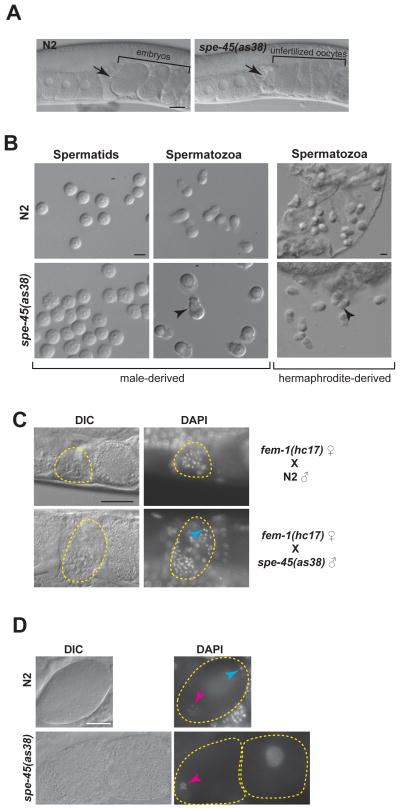
Figure 2. Despite Normal Sperm Morphology and Migratory Behavior, spe-45 Mutant Sperm Cannot Fertilize Oocytes.
(A) DIC images of wild-type and spe-45(as38) reproductive tract. Black arrow indicates spermatheca containing sperm. Uterus is located at the right side of spermatheca; ovary is located at the left side of spermatheca. Scale bar: 20μm.
(B) Spermatids and pronase-activated spermatozoa of N2 and spe-45(as38) males (Left panels). Spermatozoa from N2 and spe-45(as38) hermaphrodites (Right panels). Arrow head indicates pseudopod of sperm. Scale bar: 5μm.
(C) DAPI staining of the fem-1(hc17) females mated with either N2 males or spe-45(as38) males. Yellow, dotted region shows spermatheca regions are indicated by. Blue arrow head indicates an example of sperm DNA. Scale bar: 20μm.
(D) DAPI staining of N2 and spe-45(as38). Blue arrowhead indicates the compacted sperm chromatin mass in a newly fertilized oocyte; the pink arrowheads indicate the meiotic oocyte chromosomes in a newly fertilized oocyte. The white arrowhead indicates the chromosomes of an unfertilized oocyte that have undergone endomitotic replication. See related Figure S2. Scale bar: 10μm
Sperm migration is normal in spe-45(as38) mutants
Upon mating, the sperm ejaculated from males should get activated and directionally migrate from the site of ejaculation (vulva) to the site of fertilization (spermatheca). Furthermore, sperm are dislodged from the spermatheca into the uterus during the passage of fertilized oocytes into the hermaphrodite’s uterus. These sperm then crawl back into the spermatheca. Several factors orchestrate the directional migration of sperm into spermatheca [5]. As a defect in sperm migration into the spermatheca can compromise fertility, we asked if this is the case in spe-45(as38) mutants. fem-1(lf) worms do not produce any sperm, and hence the spermatheca of fem-1 animals are devoid of sperm (Figure 2C). Just like wild-type, spe-45 male derived sperm migrate from just inside the vulva to the spermatheca (Figure 2C). This also indicated that male-derived spe-45 sperm activated normally in vivo. Further, spe-45 sperm were able to maintain their position in the spermatheca despite passing oocytes. We conclude that spe-45 mutant sperm are fully motile and display normal migratory behavior in the reproductive tract.
The spe-45 gene belongs to the spe-9 class of genes
Spermatogenesis-abnormal (spe) genes are classified based on the phase of the sperm development or sperm differentiation in which their roles are critically important [6]. The “spe-classes” are named after the founding member of the class. For example, the “spe-9 class” refers to genes whose loss of function phenocopies spe-9 mutants where sperm development is normal but sperm are unable to fertilize oocytes despite direct contact with passing oocytes in the spermatheca [7].
Unlike in wild-type worms, whose uteri are filled with developing embryos, the spe-45 uteri are filled with unfertilized oocytes (Figure 2A), suggesting that despite being motile and in the correct position in the reproductive tract, spe-45 spermatoza are incapable of fertilizing oocytes. Staining wild-type worms with DAPI showed the entry of sperm into the oocytes as evidenced from the localization of a small, compact nuclear material, characteristic of the sperm DNA, inside the oocyte (Figure 2D, blue arrow). In contrast, the spe-45 mutant worms showed no sign of sperm entry into the oocytes, indicating that SPE-45 is required for fertilization in C. elegans. The phenotypes of spe-45 mutants are consistent with spe-45 belonging to the spe-9 class of spe genes – all phases of sperm development and differentiation remain unaffected; however since the mutant sperm are incapable of fertilizing oocytes, the unfertilized oocytes undergo endomitotic replication (EMO phenotype) (Figure 2D, white arrow) [20, 21].
A further indictor of lack of sperm entry can be found in the nature of the EMO phenotype seen in oocytes from unmated spe-45 hermaphrodites. In addition to paternal DNA, sperm also deliver centrosomes to the zygote. When sperm do not enter the oocyte, they do not deliver centrosomes and the EMO DNA forms a single mass as seen in spe-45 mutants (Figure 2D white arrow, Figure S2) [8].
Oocytes function normally in spe-45(as38)
The observed sterility in spe-45(as38) could be due to the defect in sperm or oocyte or both. At least two observations suggest that SPE-45 is not required in oocytes. First, the brood size of spe-45(as38) hermaphrodites could be restored to that of N2 upon crossing with N2 males (Figure 1B), indicating that SPE-45 is dispensable in oocytes. Second, the spe-45(as38) hermaphrodites lay large numbers of oocytes (mean = 91± 9, s.e.) suggesting that oogenesis and ovulation are not compromised in spe-45(as38) [8].
The genetic nature of spe-45(as38) allele
Compared to spe-45(as38) homozygous mutants, which do not produce any progeny at 25°C, the spe-45(as38) heterozygous mutant produce a significant number of progeny (mean=252±22 s.e., n=25, p <0.001), indicating that spe-45(as38) is recessive (Figure 1C). The phenotype of spe-45(as38)/sDf27 is Spe, which suggests that the null mutant phenotype is sperm sterile (Spe). The tm3715 mutation from the Japanese Knockout Consortium harbors a large deletion within the protein coding region of F28D1.8 (see below). As expected, the tm3715 allele exhibited a Spe phenotype (Figure 1C). Next, we expected that the as38 allele should fail to complement the tm3715 allele, if both are alleles of the same gene. As shown in Figure 1C, there is no significant difference between the brood sizes of as38/as38 homozygotes and as38/tm3715 trans-heterozygotes (p=0.36), indicating that same gene is affected in both the as38 and tm3715 alleles. The molecular lesions for both alleles also map to the same transcript (Figure 3).
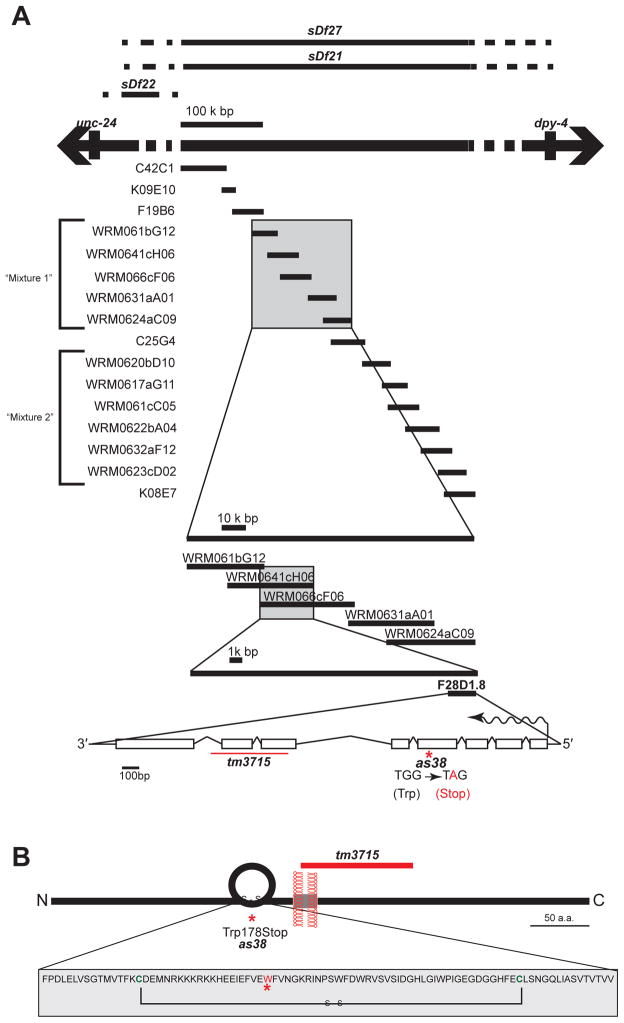
Figure 3. Molecular Analysis Reveals that SPE-45 is an Immunoglobulin Superfamily Protein.
(A) Genomic locus of spe-45(as38). See related Table S2. Complementing and non-complementing deficiencies are indicated at the top of the figure. List of cosmids and fosmids tested for the genetic rescue of spe-45(as38) is indicated. Single extrachromosomal array carrying a mixture of all fosmids listed in “mixture 1” -- but not “mixture 2” -- rescued spe-45(as38) phenotype. Introns and exons of spe-45 gene are indicated as lines and boxes, respectively; the direction of the transcription is shown as wavy arrow.
(B) Schematic diagram of the SPE-45 protein. Complete sequence of the Ig-domain of the SPE-45 is indicated in the box. See related Figure S3 for full amino acid sequence of SPE-45.
Molecular identification of spe-45
We did standard two-factor, three-factor and SNP mapping (Figure 3, Table S2), and found that the spe-45 locus maps to the fourth chromosome between the genomic intervals defined by the cosmids C42C1 and K08E7 (Figure 3A).
We used strains deficient in a defined regions of the genome as a tool to further map the locus of spe-45(as38). In complementation tests, the deficiency sDf22 complemented spe-45(as38); the deficiencies, sDf27 and sDf21 failed to complement spe-45(as38) (Figure 3A). These results suggest that spe-45 lies within the region that is deleted by sDf27 and sDf21 and lies outside the region of sDf22. Further, spe-45 over a non-complementing deficiency has no new phenotypes and still has only a Spe mutant phenotype.
Results from three-factor mapping, deficiency mapping and complementation testing suggested that the as38 mutation resides within the interval of an approximately 400 kb region. We introduced the extrachromosomal arrays representing this genomic interval into the as38 mutant to test, which, if any, could rescue the as38 Spe phenotype. Among the tested arrays, the fosmids WRM0641cH06 and WRM066cF06 rescued the Spe phenotype of as38 (Figure 1C and 3A), indicating that the molecular lesion responsible for as38 phenotype should lie somewhere in the overlapping region of these two fosmids.
We performed whole-genome sequencing of the spe-45(as38) and our laboratory N2 strains to identify variants unique to spe-45(as38). We discovered 23 candidate mutations on chromosome IV, defined as homozygous, non-synonymous variants. Of those, only one mapped to the interval delimited by the rescuing fosmids RM0641cH06 and WRM066cF06. The presence of that variant in spe-45(as38) was confirmed independently by PCR amplification and Sanger sequencing. The variant is a G->A transition in the gene F28D1.8, and encodes a nonsense mutation predicted to truncate the protein at Trp185. The gene name oig-7 (for one immunoglobulin domain) has been proposed on the basis of structural prediction [9], but no functional studies had been reported. Additional analyses (described below) confirm the identity of F28D1.8 as spe-45, and we use that designation hereafter.
Suppression of spe-45(as38)
We observed a point mutation altering the tryptophan codon (TGG) to a stop codon (TAG) in the as38 allele of the spe-45 gene (Figure 3B). If this mutation is indeed responsible for the observed Spe phenotype, we hypothesized that sup-5(e1464) should be able to at least partially rescue the spe-45(as38) mutant phenotype [10]. The sup-5(e1464) allele harbors a mutation in a gene encoding tRNA, such that the anti-codon of tRNA carrying tryptophan is TAG. Therefore, sup-5(e1464) is expected to insert Trp in lieu of a stop codon TAG, allowing the protein synthetic machinery to continue translating past the premature stop codon TAG in our as38 allele. We tested our hypothesis by constructing a sup-5(e1464);spe-45(as38) double mutant. The spe-45(as38) essentially no progeny at 25°C. In contrast, the sup-5(e1464);spe-45(as38) double mutant showed a statistically significant increase in the number of progeny produced (Figure 1C), reaffirming that we have correctly identified the molecular lesion responsible for the as38 phenotype.
spe-45 encodes a single-pass transmembrane protein with a single immunoglobulin domain
The gene F28D1.8 is originally predicted to have seven exons and six introns in Wormbase. However, close examination of the sequence upstream of this gene revealed a predicted start codon. Therefore, we analyzed RT-PCR products of F28D1.8 and found the existence of additional sequences upstream of the predicted transcript. We conclude that F28D1.8 is composed of eight exons and seven introns, and is predicted to encode 492 amino acids (Figure S2).
We find that spe-45 encodes a one immunoglobulin (OIG) transmembrane protein. This is a large family of proteins that is found in a broad range of species and likely functions in many different tissues. In C. elegans there are at least eight members of this gene family, oig-1 through oig-8 [9]. The spe-45 gene was originally annotated as oig-7. Where functional and cell biological data exists, single immunoglobulin proteins from various species are involved in cell-cell interactions [9]. Further, recent biochemical analysis suggest that OIG proteins can function in direct ligand-receptor interactions [1]. The OIG protein Izumo1 has been shown to bind to an oocyte specific GPI-anchored protein Juno [1]. Izumo1 is a sperm specific protein and knockout mice have male specific infertility [11]. Izumo1 was originally identified by biochemical characterization of an antigen that upon inhibition by monoclonal antibody prevented sperm-egg fusion in vitro [11]. Izumo1 mutant sperm are morphologically normal, with normal motility, the ability to find the egg, and transit through the egg coat (zona pellucida). However, these sperm cannot fuse with the egg plasma membrane. As presented in this paper, spe-45 mutants have the equivalent mutant phenotype in C. elegans. SPE-45 has a sperm specific function. spe-45 mutant sperm are morphologically normal but cannot fuse with the eggs despite direct contact at the site of fertilization in the reproductive tract. Based on this structural (Accompanying Manuscript Nishimura et al.) and functional similarity we propose the spe-45 encodes an Izumo-like function. A common ancestor for C. elegans and mammals existed about 700 million years ago [12] and our discovery of spe-45 represents the most deeply conserved and ancient fertility function discovered to date.
Use of many model organisms continues to shape our current understanding of fertilization. Large quantities of sperm and egg can be obtained from sea urchin, which makes it feasible to do a variety of biochemical assays. However, the current technology allows us to perform very limited genetic manipulation in this organism. In contrast, a variety of sophisticated genetic analyses are feasible in C. elegans. Since leveraging the strengths of all model organisms advances the field of fertilization, we have demonstrated that conducting an unbiased, forward genetic screen, a procedure particularly well suited in C. elegans, should aid in the discovery of new key components of fertilization pathways.
Supplementary Material
Acknowledgments
We thank Dr. Steven L’Hernault and Dr. Hitoshi Nishimura for sharing their unpublished results and for the critical reading of the manuscript. We would also like to thank members of the Singson Lab for assistance with experiments and manuscript preparation. We thank Dr. Joel Rothman and Dr. Keith Strohmaier for providing the transgenic worms carrying Pelt-7:gfp. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the Mitani Lab and National Bioresource Project for the Experimental Animal (Japan) for providing the tm3715 mutant strain. This work was supported by a grant from NIH (R01 HD054681) to Andrew Singson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlendieck K, Dhume ST, Partin JS, Lennarz WJ. The sea urchin egg receptor for sperm: isolation and characterization of the intact, biologically active receptor. The Journal of cell biology. 1993;122:887–895. doi: 10.1083/jcb.122.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidder BL. In vitro maturation and in vitro fertilization of mouse oocytes and preimplantation embryo culture. Methods in molecular biology. 2014;1150:191–199. doi: 10.1007/978-1-4939-0512-6_12. [DOI] [PubMed] [Google Scholar]
- 4.Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Developmental biology. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- 5.Hoang HD, Prasain JK, Dorand D, Miller MA. A heterogeneous mixture of F-series prostaglandins promotes sperm guidance in the Caenorhabditis elegans reproductive tract. PLoS Genet. 9:e1003271. doi: 10.1371/journal.pgen.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura H, L’Hernault SW. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:1502–1514. doi: 10.1002/dvdy.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singson A, Mercer KB, L’Hernault SW. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- 8.Geldziler BD, Marcello MR, Shakes DC, Singson A. The genetics and cell biology of fertilization. Methods in cell biology. 2011;106:343–375. doi: 10.1016/B978-0-12-544172-8.00013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook : the online review of C. elegans biology. 2013:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wills N, Gesteland RF, Karn J, Barnett L, Bolten S, Waterston RH. The genes sup-7 X and sup-5 III of C. elegans suppress amber nonsense mutations via altered transfer RNA. Cell. 1983;33:575–583. doi: 10.1016/0092-8674(83)90438-5. [DOI] [PubMed] [Google Scholar]
- 11.Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 12.Wood WB. The Nematode. Caenorhabditis elegans. 1988:16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.