Abstract
JAK2V617F is the most common oncogenic lesion in patients with myeloproliferative neoplasms (MPN). Despite the ability of JAK2V617F to instigate DNA damage in vitro, MPN is nevertheless characterized by genomic stability. In this study, we address this paradox by identifying the DNA helicase RECQL5 as a suppressor of genomic instability in MPN. We report increased RECQL5 expression in JAK2V617F-expressing cells and demonstrate that RECQL5 is required to counteract JAK2V617F-induced replication stress. Moreover, RECQL5 depletion sensitizes JAK2V617F-mutant cells to hydroxyurea (HU), a pharmacological inducer of replication stress and the most common treatment for MPN. Using single-fiber chromosome combing we show that RECQL5 depletion in JAK2V617F-mutant cells impairs replication dynamics following HU treatment, resulting in increased double-stranded breaks and apoptosis. Cumulatively, these findings identify RECQL5 as a critical regulator of genome stability in MPN and demonstrate that replication stress-associated cytotoxicity can be amplified specifically in JAK2V617F-mutant cells through RECQL5-targeted synthetic lethality.
INTRODUCTION
The propensity of cancer cells to undergo clonal evolution is enabled by a heightened state of genomic instability wherein cancer cells are continuously accumulating and repairing DNA damage. This increase in genomic flux allows cancer cells to accumulate somatic mutations that can drive disease progression. However, heightened genomic instability can also activate DNA damage-associated checkpoints which can lead to apoptosis or cellular senescence. As such, cancer cells continuously thread a fine balance between cell death and survival in response to DNA damage(Negrini et al., 2010).
The chronic myeloproliferative neoplasms (MPN) encompass a spectrum of clonal hematological disorders with an inherent tendency to transform to more aggressive disease in the form of acute myeloid leukemia (AML). As such, MPN provide a window into cancer early during its ontogeny and give insights to the processes which regulate genome stability during malignant clonal evolution. The most common recurrent lesion in MPN patients is an activating V617F mutation in the JAK2 non-receptor tyrosine kinase (JAK2V617F), which causes hyperactive JAK-STAT signaling and confers a capacity for cytokine-independent growth(Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005). Recently, a growing body of work has suggested that JAK2V617F is associated with increased DNA damage: (i) increased numbers of γH2Ax-marked double-strand breaks (DSBs) have been detected in Ba/F3 pro-B cells over-expressing JAK2V617F(Marty et al., 2013) and in lineage-negative, Sca1-positive, c-Kit-positive (LSK) cells (enriched for hematopoietic stem cell (HSC) activity) from 6-month old JAK2V617F-heterozygous knock-in mouse(Li et al., 2010); (ii) JAK2V617F expression is associated with increased levels of DNA damaging reactive oxygen species(Marty et al., 2013); (iii) RAD51-positive foci indicative of increased DSB repair have been observed in CD34+ hematopoietic cells obtained from JAK2V617F-positive MPN patients(Plo et al., 2008); and (iv) JAK2V617F expression in both human diploid fibroblasts and in primary erythroblasts from MPN patients leads to higher rates of stalled replication forks, with improper processing of stalled replication intermediates representing a potential source of DSBs(Chen et al., 2014).
Given the genome destabilizing functionalities of JAK2V617F and the inherent tendency for leukemic transformation in patients with MPN, a reasonable supposition is that oncogenic JAK2 signaling imposes a mutator phenotype on MPN cells, accelerating the accumulation of mutations and promoting clonal evolution and disease progression. However, longitudinal studies of MPN patients indicate that JAK2V617F-positive polycythemia vera (PV) and essential thrombocythemia (ET) patients (i.e. chronic phase MPN) typically remain clinically and cytogenetically stable over decades(Tefferi et al., 2014). A recent copy number analysis of the genome of chronic phase MPN patients showed that cytogenetic abnormalities are rare(Klampfl et al., 2011), and an analysis of the mutational landscape of PV and ET patients revealed that each MPN patient harbors a modest number of mutations per exome (approximately 6.5)(Nangalia et al., 2013).
To reconcile the apparent paradox of JAK2V617F-induced DNA damage with the clinical and cytogenetic stability characteristic of chronic phase MPN, we hypothesized that JAK2V617F, in addition to instigating a state of increased DNA damage, could also in parallel activate protective pathways that counteract and prevent DNA damage-induced apoptosis. In this report, we identify increased expression of the DNA repair helicase RECQL5 in JAK2V617F-expressing cells and characterize its role in constraining JAK2V617F-induced replication stress and maintaining genomic integrity in MPN.
RESULTS
Activated JAK2 signaling regulates expression of the RECQL5 helicase in MPN cells
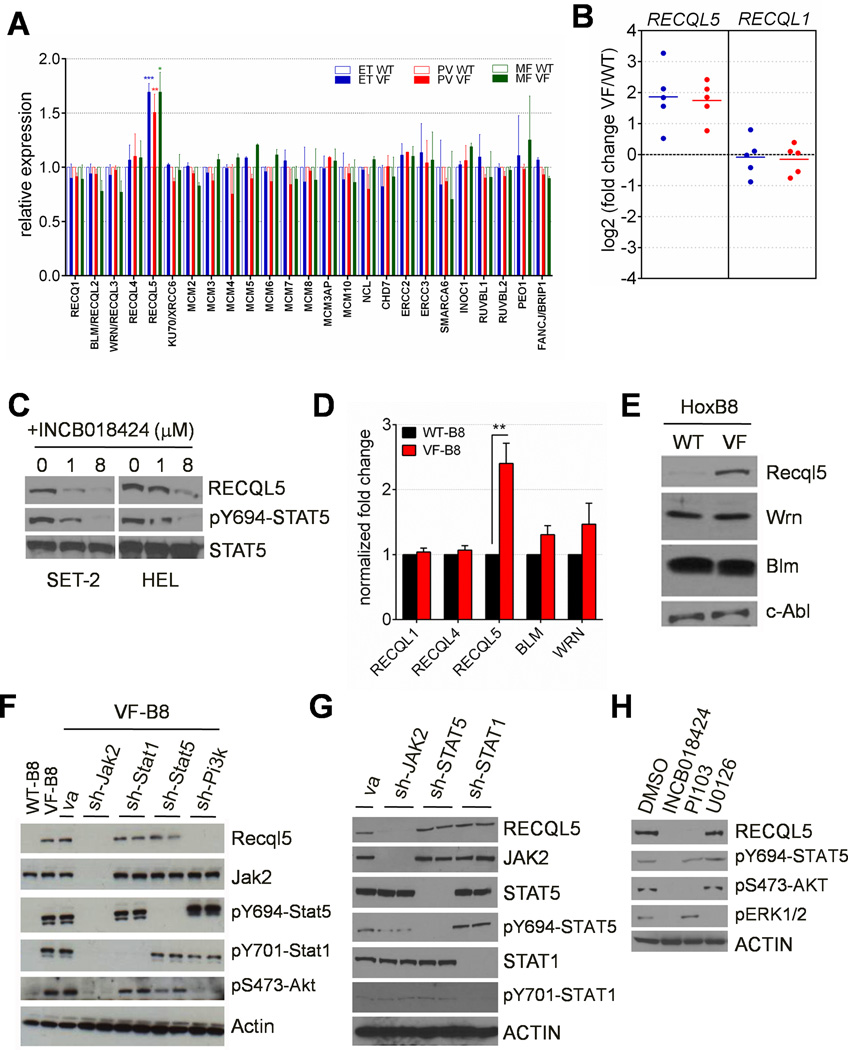
We analyzed gene expression profiles of autologous normal and JAK2V617F-heterozygous burst forming unit erythroid (BFU-E) colonies from 40 MPN patients (Chen et al., 2010). To explore the hypothesis that JAK2V617F may activate genes that counteract DNA damage, we evaluated the expression of DNA helicases in JAK2-mutant and autologous normal cells in this data set. Of 25 DNA helicases expressed, RECQL5 was the sole DNA helicase enzyme whose expression was significantly elevated in mutant-JAK2 BFU-Es relative to autologous BFU-Es in ET, PV and MF patients after multiple hypothesis testing (q<0.05) (Figure 1A). Real-time qPCR analysis of 10 MPN patients (5 PV, 5 ET) demonstrated increased expression of RECQL5 (but not the related RECQ family member, RECQL1) in mutant-JAK2 colonies relative to autologous wild-type colonies in both PV and ET (Figure 1B). We next assessed RECQL5 levels following JAK2 inhibition in two independent cell lines that harbor the JAK2V617F mutation - HEL and SET-2 cells. We found that treatment of the erythroleukemic cell line, HEL and the megakaryoblastic cell line, SET-2 with the JAK2 inhibitor, INCB018424 reduced RECQL5 expression concomitant with decreased STAT5 phosphorylation (Figure 1C). In aggregate, these data indicates that RECQL5 expression is regulated by activated JAK2 signaling in human disease-relevant contexts.
Figure 1. JAK2V617F increases expression of RECQL5.
(A) Gene expression profiles depicting expression of 25 DNA helicases in 40 MPN patients (20 ET, 16 PV and 4 MF). (B) qPCR validation of RECQL5 and RECQL1 expression in ET (blue) or PV (red) patients. (C) RECQL5 expression in SET-2 and HEL cells following treatment with 0–8 mM of the JAK2 inhibitor INCB18424. (D–E) Expression of Recq family members in WT-B8 and VF-B8 cells by qPCR (Panel D) and Western immunoblot (Panel E). (F) Recql5 expression in WT-B8 cells and in VF-B8 cells following knockdown of Jak2 (sh-Jak2), p85 subunit of Pi3-kinase (sh-Pi3k), Stat1 (sh-Stat1) or Stat5 (sh-Stat5). (G) RECQL5 expression in HEL cells following knockdown of JAK2 (sh-JAK2), STAT5 (sh-STAT5) or STAT1 (sh-STAT1). (H) RECQL5 expression in HEL cells following treatment for 16h with INCB018424 (1 µM), PI3K inhibitor PI103 (1 µM) or ERK1/2 inhibitor U0126 (5 µM). See also Figure S1.
Physiological levels of Jak2V617F lead to increased Recql5 expression
To investigate the role of Recql5 under conditions that more closely recapitulate chronic phase MPN in patients, we generated disease-relevant cell lines from Jak2V617F knock-in mice that we have previously developed and extensively characterized(Mullally et al., 2013; Mullally et al., 2010). This mouse model closely recapitulates the features of human MPN and Jak2V617F expression is physiological in the model, being driven from the endogenous Jak2 promoter. To generate cell lines, we engineered bone marrow progenitors from littermate wild-type or Jak2V617F-knockin mice to express a β-estradiol-regulated Hoxb8 homeodomain-containing protein. Following 3 weeks of serial passaging and antibiotic selection, immortalized myeloid progenitor cells were generated from both wild-type mice (WT-B8) and Jak2V617F-knockin mice (VF-B8)(Wang et al., 2006). Both WT-B8 and VF-B8 lines resemble cells at a granulocyte-macrophage progenitor (GMP) stage of myeloid maturation by cell surface immunophenotype analysis (Figure S1A). Immunoblotting showed equivalent levels of Jak2 expression in both cell lines but increased levels of phosphorylated Stat5 in the VF-B8 cells relative to WT-B8 cells (Figure S1B). Additionally, VF-B8 cells exhibited increased proliferative capacity and survival at reduced serum conditions (Figure S1C–D), indicating that VF-B8 cells recapitulate this key pathognomonic feature of MPN biology.
We next determined whether the expression of Recql5 was modulated by mutant Jak2 in these Hoxb8-immortalized cell lines. In accord with the primary human data, VF-B8 cells exhibited elevated expression of Recql5 relative to WT-B8 cells by qPCR analysis (Figure 1D) and Western immunoblotting (Figure 1E). No significant differential expression of other Recq family members (Recql1, Recql4, Blm and Wrn) was observed. These data demonstrate that mutant JAK2 increases expression of the DNA repair helicase RECQL5 in both human and murine cells.
RECQL5 is a target of JAK2-PI3K signaling
To determine which signalling pathways were necessary for the regulation of Recql5 by Jak2V617F, we used shRNAs to knockdown Jak2 and downstream effector molecules of Jak2 in VF-B8 cells. Expression of shRNAs targeting Jak2 led to diminution of Recql5 levels relative to cells transduced with an empty vector control (Figure 1F), which is consistent with the finding that Recql5 is a downstream target of Jak2 signaling. Next, we assessed Recql5 levels in VF-B8 cells following transduction of shRNAs targeting Stat1, Stat5 or the p85 subunit of the Pi-3 kinase. Levels of Recql5 in VF-B8 cells were reduced following knockdown of Pi3k but not Stat1 or Stat5 (Figure 1F). Similarly, RECQL5 levels were also attenuated following knockdown of JAK2 in HEL cells but were not attenuated following knockdown of STAT5 or STAT1 (Figure 1G), and treatment of HEL cells with the PI-3 kinase inhibitor PI103 (but not the ERK1/2 inhibitor U0126) led to attenuated levels of RECQL5 accompanied by decreased AKT phosphorylation (Figure 1H). Taken together, these data indicate that RECQL5 expression is increased upon JAK2 activation in a PI-3 kinase-dependent manner.
Recql5 depletion sensitizes Jak2V617F-expressing cells to replication stress
We next sought to clarify the function of RECQL5 up-regulation in JAK2V617F-expressing cells. Previously, we had demonstrated that over-expression of JAK2V617F in human diploid fibroblasts resulted in increased replication fork stalling and replication stress(Chen et al., 2014). As RECQL5 has been functionally linked to regulating stalled replication forks in normal cells, we therefore hypothesized that the up-regulation of RECQL5 may function to mitigate the deleterious consequences of replication stress in JAK2-mutant cells.
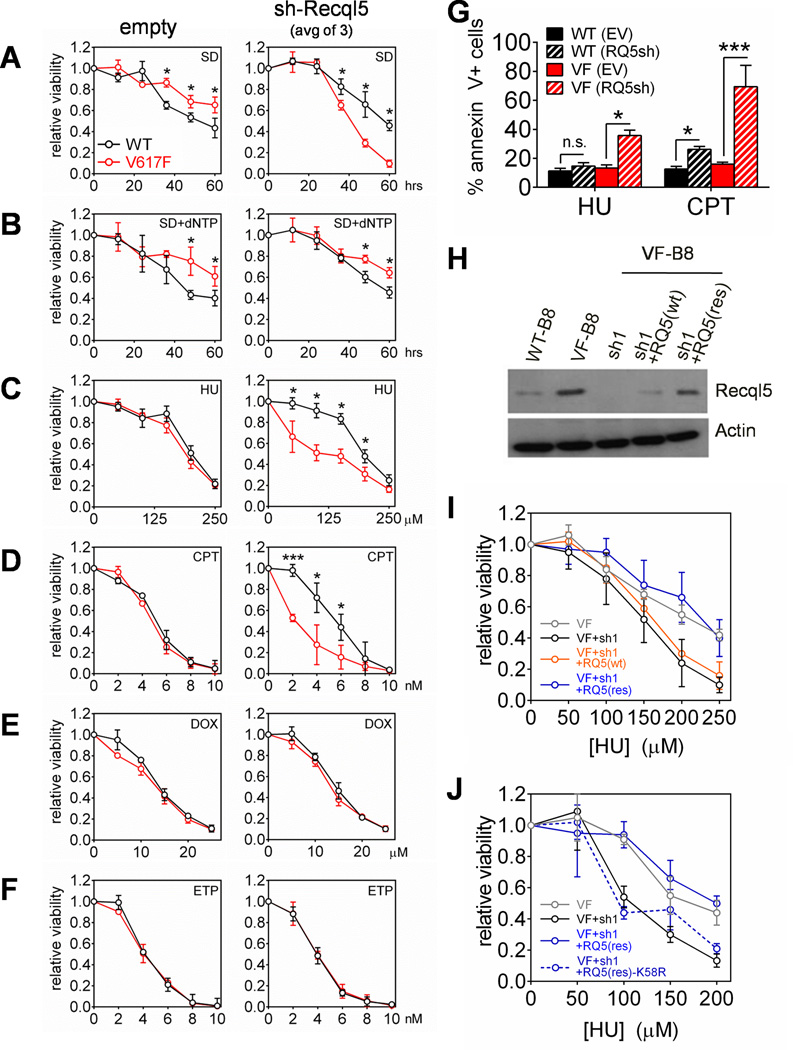
To test this, we knocked down Recql5 in VF-B8 cells using 3 independent shRNAs (Figure S2A). Densitometric analysis revealed a knockdown efficiency of 61–95% for the three hairpins. Under normal growth conditions, Recql5 knockdown in VF-B8 cells did not affect proliferation rate (Figure S2B) or apoptosis (Figure S2C) relative to either equivalently modified WT-B8 cells or to VF-B8 cells expressing empty vector alone (VA) controls. To simulate replication stress, we subjected WT-B8 and VF-B8 cells to low serum conditions to deprive cells of dNTPs(Bester et al., 2011). We observed that VA-expressing VF-B8 cells exhibited greater viability relative to WT-B8 cells in low serum conditions (Figure 2A, left), which accord with our previous data in non-genetically perturbed cells. In contrast, Recql5-depleted VF-B8 cells exhibited decreased viability relative to equivalently modified WT-B8 cells (Figure 2A, right). Hypersensitivity of Recql5-depleted VF-B8 cells to low serum conditions was completely abrogated upon repletion of deoxynucleotide (dNTPs) into the culture medium (Figure 2B). The results shown represent the average of 3 independent Recql5-targeting shRNAs, and are consistent with data obtained for each individual hairpin (Figure S2D). Collectively, these data demonstrate that Recql5 protects VF-B8 cells from endogenous replication stress instigated by low dNTP levels.
Figure 2. Recql5 depletion sensitizes Jak2V617F-expressing cells to replication stress.
(A–F) Viability of WT-B8 (black) and VF-B8 (red) cells transduced with empty vector (VA) or Recql5-targeting shRNAs, following serum deprivation in 0.5% FCS (SD) (Panel A), SD with 100 µM dNTPs (SD+dNTPs) (Panel B), HU (0–250 µM) (Panel C), CPT (0–10 nM) (Panel D), DOX (0–25 µM) (Panel E) or ETP (0–10 nM) (Panel F). For VA-transduced cultures, each point represents the mean of 3 independent cultures. For Recql5-knockdown cultures, each point represents the mean of 3 separate Recql5-targeting shRNAs. (G) Quantitation of annexin V-positivity in cells treated with 100 µM HU or 4 nM CPT. (H–J) VF-B8 cells were transduced with Recql5-targeting shRNA simultaneously with either a wildtype Recql5 cDNA (RQ5wt), shRNA-resistent Recql5 cDNA (RQ5res), or RQ5res cDNA harboring a K58R mutation (RQ5res-K58R). Immunoblotting for Recql5 protein levels was performed (Panel H) and cell viability was assessed (Panel I–J). Testing for statistical significance was performed using a student’s t-test (*: p<0.05; **: p<0.01; ***: p<0.001). See also Figure S2.
We next tested whether Recql5 depletion would also sensitize VF-B8 cells to exogenous instigators of replication stress, such as pharmacological agents. We tested hydroxyurea (HU) and camptothecin (CPT), which impair DNA replication by limiting production of dNTPs and inhibiting topoisomerase I activity, respectively. Strikingly, contemporaneous knockdown of Recql5 and exposure to either HU or CPT led to significantly decreased viability of VF-B8 cells compared to like-treated WT-B8 cells after 24 hours of drug treatment (Figure 2C–D and 2G, Figure S2D). Decreased cell viability was noted as early as 12 hours post-drug treatment (Figure S2E–F). In contrast, Recql5-depleted VF-B8 cells were not hypersensitive to the double-stranded breaking agents, doxorubicin (DOX) and etoposide (ETP) (Figure 2E–F and S2D). Concordant with these data, shRNA depletion of human RECQL5 also enhanced cytotoxicity in HEL and SET2 cells only to pharmacological instigators of replication stress (HU, CPT, aphidicolin and irinotecan) and not to pro-oxidants or DSB–generating drugs (Figure S2G–H).
Replication stressors such as HU and CPT can also induce double-stranded breaks (DSBs) at sufficiently high doses. We therefore validated whether the HU and CPT doses associated with preferential cytotoxicity of Recql5-depleted VF-B8 cells were causing increased replication stress or excessive formation of DSBs. To differentiate between these two phenomena, we treated parental WT-B8 and VF-B8 cells with HU (6 µM) and CPT (4 nM) and performed immunocytochemical staining for foci containing the RPA protein (which marks stalled replication forks) or 53BP1 (which localizes to DSBs) (Figure S3A–B). At these HU and CPT dosages, we observed a marked increase in foci containing RPA (Figure S3C) with only a marginal increase in numbers of 53BP1-positive foci (Figure S3D). In contrast, etoposide (4 nM) generated both RPA-positive and 53BP1-positive foci (Figure S3C–D). In aggregate, these findings verify that the dosage of HU and CPT used to enhance cytotoxicity of Recql5-depleted VF-B8 cells causes enhanced replication stress with only a slight elevation of DSBs.
Given the potential off-target effects of shRNAs that may potentially influence the observed phenotypes, we next confirmed the specificity of the Recql5 shRNAs. We designed a Recql5 complementary DNA (cDNA; RQ5(res)) that has been mutated at every third nucleotide so as to disrupt each of the shRNA-binding sites while retaining the correct amino acid encoded at each triplet codon. Expression of a wildtype Recql5 (RQ5(wt)) in VF-B8 cells together with a Recql5-targeting shRNA did not rescue Recql5 expression (Figure 2H) and failed to abrogate the increased sensitivity of Recql5-depleted VF-B8 cells to HU (Figure 2I). In contrast, expression of RQ5(res) resulted in high levels of Recql5 expression that was maintained despite expression of a Recql5 shRNA (Figure 2H) and successfully abrogated the hypersensitivity to HU of the VF-B8 cells co-expressing the a Recql5 shRNA (Figure 2I). Moreover, a RQ5(res) cDNA harboring a K58R mutation within the helicase domain is incapable of abrogating HU hypersensitivity, revealing the essentiality of Recql5 helicase activity in protecting against replication stress (Figure 2J). Together, this data demonstrates that the effects of the shRNAs were on-target, and that Recql5 depletion was the critical factor for conferring increased sensitivity of VF-B8 cells to replication stress.
Recql5 depletion increases severity of replication fork stalling in Jak2V617F–expressing cells exposed to exogenous replication stress
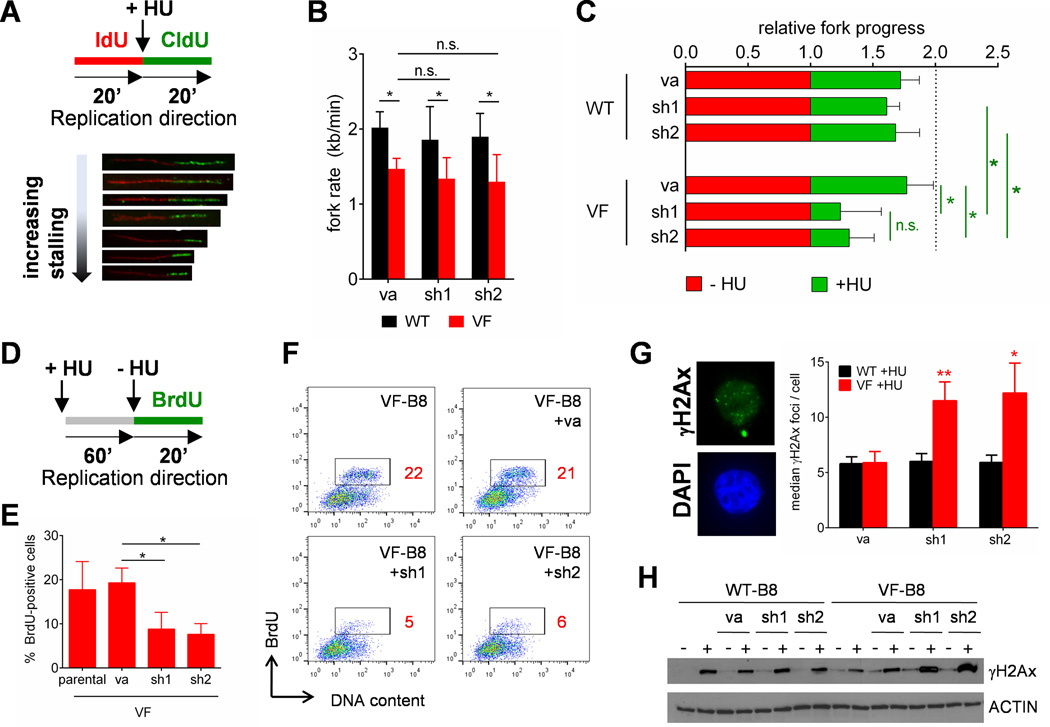
To understand the molecular mechanisms by which Recql5 depletion in VF-B8 cells leads to increased cytotoxicity by exogenous replication stressors (such as HU), we performed chromosome combing, which allows the direct visualization and analysis of replication tracts on individual, BrdU-labelled DNA fibers. We subjected WT-B8 or VF-B8 cells transduced with Recql5-targeting shRNAs or VA controls to this procedure. All cultures underwent a first labelling step with the BrdU analogue iodo-deoxyuridine (IdU) under normal growth conditions, followed by a second labelling step with another BrdU analogue chlorodeoyuridine (CldU) in culture medium supplemented with HU (Figure 3A). In this way, the extent of fork progression can be measured both in the absence and in the presence of HU to determine any differential effects of HU on the cultures.
Figure 3. Recql5 protects against collapse of stalled replication forks in Jak2V617F-expressing cells exposed to replication stress.
(A) Schematic of time course for chromosome combing experiments (top). Representative replication structures of a single combed DNA molecule labelled with IdU (red) and CldU (green) (bottom). (B) Fork rate in WT-B8 and VF-B8 cells transduced with empty vector alone (va) or with two different Recql5-targeting shRNAs (sh1 and sh2). Results represent the mean±S.D. for at least 50 fibres. (C) Quantification of HU-induced effects on fork dynamics. Relative fork progress is depicted as normalized ratio of the second (HU-treated) labelling step relative to the first (HU-free) labelling step. (D–F) Schematic of timeline for fork restart experiments (Panel D). Quantification of BrdU-positive cells (Panel E). Representative flow cytometric plots of BrdU staining (Panel F). (G–H) Quantification of γH2Ax-marked double-stranded breaks as assessed by immunofluorescent detection (Panel G) and Western immunoblot (Panel H). (D–H) Testing for statistical significance was performed using a student’s t-test (*: p<0.05; **: p<0.01). See also Figure S2.
We focused our initial analysis on fibers generated solely from the first labelling step in order to ascertain whether Recql5 depletion in the absence of HU altered DNA replication kinetics. For this analysis, we measured the length of these fibers and calculated the average fork rate, as decreased fork processivity is a robust indicator of fork stalling. In control cells, we observed a significant decrease in the mean replication rate in VF-B8 cells as compared to WT-B8 cells (1.47 ± 0.18 Kb/min (n=88) in VF-B8 versus 1.97 ± 0.21 Kb/min (n=94) in WT-B8 (p < 0.01)) (Figure 3B), consistent with previously published reports indicating a replication processivity impairment in JAK2V617F-expressing cells (Chen et al., 2014). However, the mean replication rate was not significantly altered by Recql5 knockdown in either WT-B8 or VF-B8 cells (WT-B8+sh1: 1.88 ± 0.44 Kb/min (n=80); WT-B8+sh2: 1.91 ± 0.28 Kb/min (n=55); VF-B8+sh1: 1.33 ± 0.31 Kb/min (n=101); VF-B8+sh2: 1.32 ± 0.39 (n=50)). Critically, no difference was observed in fork rate between VF-B8 cells depleted for Recql5 relative to those transduced with an empty vector (Figure 3B). These data indicate that physiological levels of Jak2V617F expression in myeloid cells gives rise to a replication stress phenotype as evidenced by decreased fork processivity, but that Recql5 depletion alone has no additional affect.
We next explored the possibility that depletion of Recql5 in WT-B8 and VF-B8 cells could lead to altered replication dynamics in the presence of HU. As replication fork progress in VF-B8 cells is globally impaired relative to WT-B8 cells, in order to facilitate comparison, the extent of fork progress during the first (HU-free) labelling step was normalized and arbitrarily designated at 1, and the second (HU-treated) labelling step has been depicted relative to the normalized first label. Using this analysis, we observed that the presence of HU impaired fork progression in all cultures tested (Figure 3C). However, the impairment in fork progression caused by exposure to HU was significantly greater in Recql5-depleted VF-B8 cells relative to control VF-B8 cells and to Recql5-depleted WT-B8 cells (Figure 3C). This indicates that Recql5 depletion differentially impairs replication fork progression in VF-B8 cells following HU exposure as compared to WT-B8 cells.
The exacerbation in impairment of fork progression may be due to increased frequency of fork collapse. To test for this, we measured the restart efficiency of a stalled replication fork, a process which is highly impaired following fork collapse. VF-B8 cells transduced with a control shRNA or shRNAs targeting Recql5 were initially exposed to HU for 60 minutes, followed by removal of HU and addition of BrdU (Figure 3D). Flow cytometric detection of BrdU-positive cells reflect cells which had efficiently restarted stalled forks. We observed significantly lower BrdU-positive cells in HU-treated Recql5-depleted VF-B8 cells compared to cells expressing control hairpins (Figure 3E–F). Consistent with a role for Recql5 in protecting VF-B8 cells against HU-induced fork collapse, we noted that following HU exposure Recql5-depleted VF-B8 cells exhibited an increase in DSBs, as indicated by higher numbers of γH2Ax-positive foci (Figure 3G) and higher levels of γH2Ax by immunoblot analysis (Figure 3H). Altogether, these data demonstrate that Recql5 is essential to maintain fork stability in mutant JAK2-expressing cells following exposure to HU.
RECQL5 depletion increases sensitivity of JAK2V617F-positive cells from MPN patients to HU
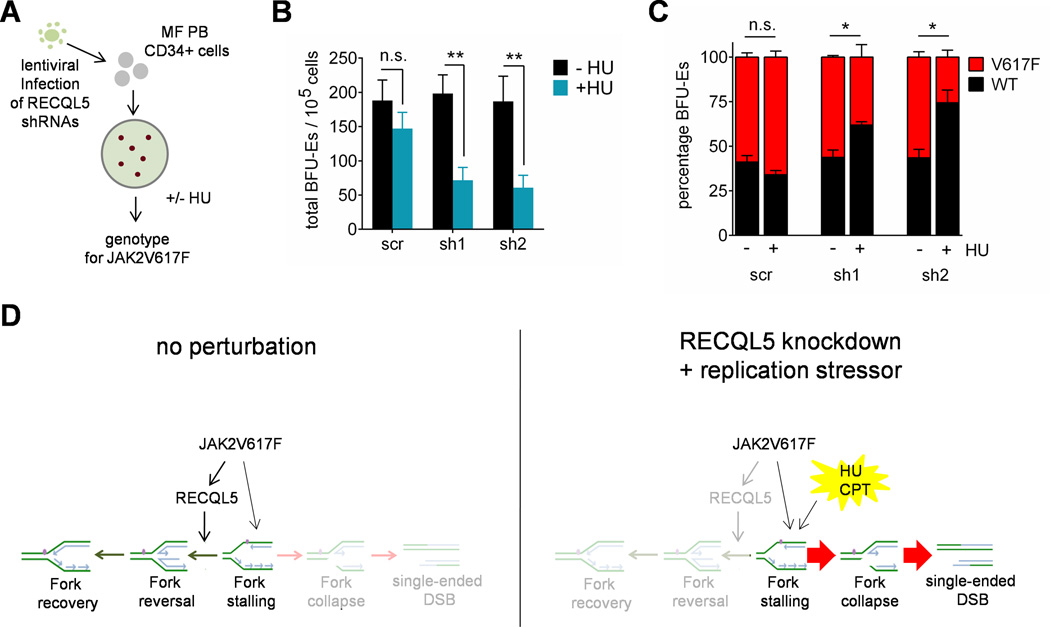
Finally, we tested whether modulation of RECQL5 could also increase sensitivity of JAK2V617F-positive cells from primary MPN patients to HU. Following depletion with RECQL5, CD34-positive peripheral blood mononuclear cells from JAK2V617F-positive myelofibrosis patients were grown in semi-solid medium supplemented with HU for 14 days (Figure 4A). Strikingly, following RECQL5 depletion, there was a preferential eradication of total BFU-E colonies relative to a control shRNA (Figure 4B). Moreover, genotyping of the colonies for JAK2V617F-positivity revealed a preferential elimination of JAK2V617F-positive BFU-E colonies compared to autologous wild-type colonies following exposure to HU (Figure 4C; Figure S4). Collectively, this indicates that RECQL5 knockdown preferentially sensitizes JAK2V617F-positive BFU-E colonies from MPN patients to pharmacological induction of replication stress with HU.
Figure 4. RECQL5 depletion sensitizes JAK2V617F-positive cells from MPN patients to replication stressors.
(A) Work flow for determining effect of RECQL5 knockdown on HU sensitivity of primary MPN samples. (B–C) CD34+ peripheral blood mononuclear cells from 3 myelofibrosis patients were transduced with a non-targeting shRNA (scr) or RECQL5-targeting shRNA (sh1 and sh2) and cultured on methylcellulose in the presence and absence of 2 µM HU. After 14 days, total BFU-Es were counted (Panel B) and genotyped for JAK2V617F (Panel C). (D) Model depicting interactions between JAK2V617F, RECQL5 depletion and exogenous pharmacological replication stress. See also Figure S4.
DISCUSSION
The chronic myeloproliferative neoplasms (MPN) represent a model of early stages of leukemogenesis and can provide insights into the balance between oncogene-associated DNA damage and the mechanisms that act to constrain it. JAK2V617F is the most common molecular driver of MPN, and in PV and ET, it is frequently the sole genetic driver identified. Although there is experimental evidence demonstrating that JAK2V617F induces DNA damage, the clinical evidence indicates that chronic phase MPN follows a relatively indolent course over decades and MPN genomes remain generally stable over time. In this study we help resolve this apparent conundrum by demonstrating a role for the DNA repair helicase RECQL5 in regulating the balance between JAK2V617F-induced DNA replication stress and genomic integrity in patients with MPN.
RECQ helicases are a family of highly-conserved genome surveillance enzymes. Humans possess five RECQ helicases - RECQL1, BLM, WRN, RECQL4 and RECQL5 - which have both unique and over-lapping roles in the regulation of DNA replication, repair and transcription(Larsen and Hickson, 2013). Strikingly, we observed that the increased expression of RECQL5 in JAK2V617F-mutant cells was specific and not seen with other RECQ helicases. This finding precludes the possibility that increased RECQL5 levels are due to an excess of S-phase cells in cultures of JAK2V617F-expressing cells or that they represent an epi-phenomenon of increased DNA damage load. Rather, using shRNA knockdown and various pharmacological agents to inhibit individual signalling pathways, we demonstrate that JAK2V617F increases RECQL5 expression through the PI3K-AKT signaling axis. In terms of the effectors downstream of PI3K-AKT signalling responsible for mediating RECQL5 activation, it is possible that a direct transcriptional mechanism, such as by modulating activity of the FOXO family of transcription factors, is involved. Indeed, a similar pathway has previously been shown to be active in JAK2-mutant cells to modulate expression of the antioxidant protein catalase(Marty et al., 2013).
The mechanisms by which RECQL5 maintains genomic integrity in normal and cancer cells are on-going areas of investigation. Germline loss-of-function mutations in RECQ family members have been associated with a predisposition to developing cancer and a key role was recently described for RECQL4 in hematopoiesis(Smeets et al., 2014). However, RECQL5 dysregulation has not been previously implicated in human disease. Our data identify increased expression of RECQL5 as a means by which oncogene-induced replication stress is counteracted in JAK2-mutated MPN. Concomitant induction of replication stress and RECQL5 expression downstream of JAK2-PI3K signalling potentially forms a feedback loop that regulates DNA damage accumulation. Depleting RECQL5 in JAK2-mutant cells disrupts this homeostasis and exposes a synthetic lethal vulnerability of JAK2-mutant cells with pharmacological inducers of replication stress. Following depletion of RECQL5, JAK2V617F-expressing cells exposed to replication stressors (such as HU or CPT) exhibit more severe fork stalling and impaired fork restart in comparison to isogenic wild-type cells. Moreover, RECQL5-depleted JAK2-mutant cells exhibited higher numbers of DSBs and underwent apoptosis at lower doses of HU when compared to RECQL5-depleted wild-type cells (Figure 4D).
We envision two mutually non-exclusive scenarios to explain why JAK2 mutant cells are more sensitive than isogenic wild-type cells to contemporaneous RECQL5 depletion and exogenous replication stress. Firstly, JAK2V617F-expressing cells are known to have more replication fork stalling and replication stress(Chen et al., 2014). As such, increased RECQL5 expression in these cells may be required to mitigate replication-associated genomic instability and maintain an appropriate balance of DNA damage/repair to ensure cell viability. RECQL5 depletion in JAK2V617F-expressing cells may therefore be more deleterious than to equivalently-modified wild-type cells, as JAK2-mutant cells may have less leeway to cope with the additional replication stress in the form of exogenous administration of HU. Secondly, JAK2-mutant cells may exhibit specific replication fork structures whose resolution is especially dependent on RECQL5 and thus would be particularly sensitive to RECQL5 depletion. The mechanism of JAK2V617F-induced replication stress remains unresolved, but is likely to involve unscheduled euchromatinization during S phase by JAK2 directly(Dawson et al., 2009) or following activation of downstream STATs(Shi et al., 2006) leading to physical collisions between replication machinery and transcriptional apparatus on the same competing DNA template. The resulting DNA:RNA hybrid structures (called R-loops) are potentially mutagenic and genome destabilizing. RECQL5 may play an important role in resolving R-loops in JAK2-mutant cells. Future studies to distinguish between these possibilities will be interesting.
Finally, our data may have broader pharmacological implications for cancer therapy. While HU remains the front-line treatment for patients with ET and PV(Harrison et al., 2005), most studies have demonstrated that HU does not preferentially target the V617F-positive cell fraction in ET and PV patients(Antonioli et al., 2010) and as a result HU does not alter the natural history of MPN. Our findings highlight a potential approach to amplify replication stress-associated cytotoxicity through helicase-targeted synthetic lethality. Indeed, there is precedent for this approach. Inhibition of the Werner helicase using small molecules has been shown to sensitize HeLa and U2OS cells to the replication stressor topotecan(Aggarwal et al., 2011). Nonetheless, the development of small molecule inhibitors of RECQL5 in MPN would need to be approached with caution. In particular, the safety of RECQL5 inhibition with respect to its potential to provoke more genomic insults would need to be carefully examined. However, exploiting the differential dependencies of cancer cells on DNA repair pathways has already demonstrated clinical efficacy, through the use of PARP1 inhibitors in BRCA-mutant cancers(Bryant et al., 2005). Identifying and leveraging synthetic lethal relationships with DNA helicases may represent another similar approach for improved anti-cancer treatment strategies more broadly.
MATERIALS AND METHODS
Generation of myeloid progenitor cell lines from Jak2V617F-knockin mice
Myeloid progenitor cell lines were generated from a C57Bl/6 Jak2V617F-expressing mouse(Mullally et al., 2010) and from a Jak2 wild-type litter-mate control, by retroviral over-expression of an estrogen-dependent Hoxb8 transcription factor as previously described(Wang et al., 2006). Conditionally immortalized myeloid progenitor cell lines from wild-type mice (WT-B8) and Jak2V617F-knockin mice (VF-B8) were were maintained in Myeloid Medium (RPMI+10% FBS supplemented with 50 ng/ml mSCF and 1 µM β-estradiol (Sigma).
Cell viability assays
WT-B8 and VF-B8 cells were counted and 5×104 cells were seeded in triplicate in each well of a 96-well plate in 100 µL of Myeloid Medium. For drug treatments, agents were added fresh. For serum deprivation studies, cells were rinsed with 1× PBS twice and 1×105 cells were seeded per well of a 96-well plate in RPMI+0.5% FBS supplemented with 1 uM β-estradiol. Cell growth was measured using a standard Alamar blue assay (Life Technologies).
Chromosome combing
Chromsome combing was performed as previously described(Chen et al., 2014). Briefly, 1×105 WT-B8 or VF-B8 cells were seeded in a well of a 96-well plate in a volume of 100 µL. Replicating DNA was first labelled with 25 uM 5-iodo-2’-deoxyuridine (IdU; Sigma), followed by 250 uM 5-chloro-2’-deoxyuridine (CldU; Sigma) for 20 min each. Cells were then harvested, genomic DNA was extracted and individual DNA molecules were stretched on glass slides, stained with fluorescence detection were anti-IdU (1:300) and anti-CldU (1:150) and analyzed under oil immersion on a Zeiss Axioscop 2 fluorescent miscroscope.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
We identify the DNA repair helicase, RECQL5 as a suppressor of genomic instability in JAK2V617F-driven myeloproliferative neoplasms (MPN) and demonstrate that RECQL5 is required to counteract replication stress in JAK2V617F-expressing cells. We find that RECQL5 depletion sensitizes JAK2V617F-mutant cells to hydroxyurea (HU), a pharmacological inducer of replication stress and the most common treatment for MPN.
ACKNOWLEDGEMENTS
AM is supported by the NIH (K08 HL109734), the MPN Research Foundation, the Jeanne D. Housman Fund for Research on Myeloproliferative Disorders and is a recipient of a Damon Runyon clinical investigator award. EC is a recipient of a Lady Tata Memorial Trust Award. ARG is supported by Bloodwise (13003), the Wellcome Trust (104710/Z/14/Z), the Medical Research Council, the Kay Kendall Leukaemia Fund, the Cambridge NIHR Biomedical Research Center, the Cambridge Experimental Cancer Medicine Centre, the Leukemia and Lymphoma Society of America (07037), and a core support grant from the Wellcome Trust and MRC to the Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute. We thank Drs. Benjamin Ebert and Steven Lane for critically reviewing the manuscript and Dr. Ross Levine for providing helpful insights during manuscript revision.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing interests
REFERENCES
- Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioli E, Carobbio A, Pieri L, Pancrazzi A, Guglielmelli P, Delaini F, Ponziani V, Bartalucci N, Tozzi L, Bosi A, et al. Hydroxyurea does not appreciably reduce JAK2 V617F allele burden in patients with polycythemia vera or essential thrombocythemia. Haematologica. 2010;95:1435–1438. doi: 10.3324/haematol.2009.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Chen E, Ahn JS, Massie CE, Clynes D, Godfrey AL, Li J, Park HJ, Nangalia J, Silber Y, Mullally A, et al. JAK2V617F promotes replication fork stalling with disease-restricted impairment of the intra-S checkpoint response. Proc Natl Acad Sci U S A. 2014;111:15190–15195. doi: 10.1073/pnas.1401873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, Ingle CE, Dermitzakis ET, Campbell PJ, Green AR. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell. 2010;18:524–535. doi: 10.1016/j.ccr.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CN, Campbell PJ, Buck G, Wheatley K, East CL, Bareford D, Wilkins BS, van der Walt JD, Reilly JT, Grigg AP, et al. Hydroxyurea compared with anagrelide in high-risk essential thrombocythemia. N Engl J Med. 2005;353:33–45. doi: 10.1056/NEJMoa043800. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Klampfl T, Harutyunyan A, Berg T, Gisslinger B, Schalling M, Bagienski K, Olcaydu D, Passamonti F, Rumi E, Pietra D, et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood. 2011;118:167–176. doi: 10.1182/blood-2011-01-331678. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Larsen NB, Hickson ID. RecQ Helicases: Conserved Guardians of Genomic Integrity. Adv Exp Med Biol. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Li J, Spensberger D, Ahn JS, Anand S, Beer PA, Ghevaert C, Chen E, Forrai A, Scott LM, Ferreira R, et al. JAK2 V617F impairs hematopoietic stem cell function in a conditional knock-in mouse model of JAK2 V617F-positive essential thrombocythemia. Blood. 2010;116:1528–1538. doi: 10.1182/blood-2009-12-259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty C, Lacout C, Droin N, Le Couedic JP, Ribrag V, Solary E, Vainchenker W, Villeval JL, Plo I. A role for reactive oxygen species in JAK2 V617F myeloproliferative neoplasm progression. Leukemia. 2013;27:2187–2195. doi: 10.1038/leu.2013.102. [DOI] [PubMed] [Google Scholar]
- Mullally A, Bruedigam C, Poveromo L, Heidel FH, Purdon A, Vu T, Austin R, Heckl D, Breyfogle LJ, Kuhn CP, et al. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-alpha in a murine model of polycythemia vera. Blood. 2013;121:3692–3702. doi: 10.1182/blood-2012-05-432989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 2010;17:584–596. doi: 10.1016/j.ccr.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al. Somatic CALR Mutations in Myeloproliferative Neoplasms with Nonmutated JAK2. N Engl J Med. 2013 doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Plo I, Nakatake M, Malivert L, de Villartay JP, Giraudier S, Villeval JL, Wiesmuller L, Vainchenker W. JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood. 2008;112:1402–1412. doi: 10.1182/blood-2008-01-134114. [DOI] [PubMed] [Google Scholar]
- Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets MF, DeLuca E, Wall M, Quach JM, Chalk AM, Deans AJ, Heierhorst J, Purton LE, Izon DJ, Walkley CR. The Rothmund-Thomson syndrome helicase RECQL4 is essential for hematopoiesis. J Clin Invest. 2014;124:3551–3565. doi: 10.1172/JCI75334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Guglielmelli P, Larson DR, Finke C, Wassie EA, Pieri L, Gangat N, Fjerza R, Belachew AA, Lasho TL, et al. Long-term survival and blast transformation in molecularly annotated essential thrombocythemia, polycythemia vera, and myelofibrosis. Blood. 2014;124:2507–2513. doi: 10.1182/blood-2014-05-579136. quiz 2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.