Abstract
Whether aged hematopoietic stem and progenitor cells (HSPCs) have impaired DNA damage repair is controversial. Using a combination of DNA mutation indicator assays, we observe a 2-3 fold increase in the number of DNA mutations in the hematopoietic system upon aging. Young and aged HSCs and HPCs do not show an increase in mutation upon irradiation-induced DNA damage repair, and young and aged HSPCs respond very similarly to DNA damage with respect to cell cycle checkpoint activation and apoptosis. Both, young and aged HSPCs show impaired activation of the DNA-damage induced G1-S checkpoint. Induction of chronic DNA double strand breaks by zinc-finger nucleases suggest that HSPCs undergo apoptosis rather than faulty repair. These data reveal a protective mechanism in both the young and aged hematopoietic system against accumulation of mutations in response to DNA damage.
Keywords: HSC, DNA DAMAGE, DNA MUTATION, AGING, STEM CELL
INTRODUCTION
Hematopoietic stem cells (HSCs) are tissue-specific stem cells that reside in the bone marrow (BM) and ensure the lifelong production of blood cells. This is achieved by the ability of HSCs to differentiate into a variety of specialized cells and to self-renew to achieve tissue homeostasis. HSC function declines from adulthood to old age, which contributes hematopoiesis dysfunction in older adults. Aging of HSC and the hematopoietic system is characterized by senescence associated immune remodeling and anemia (Van Zant and Liang, 2012; Geiger et al., 2013). Aged HSCs exhibit reduced self-renewal activity, a deficiency in their ability to produce erythrocytes and show a bias towards the myeloid lineage (Linton and Dorshkind, 2004; Signer and Morrison, 2013). Furthermore, aged HSCs present with a distinct gene expression signature and apolar distribution of proteins compared to young HSCs (Chambers et al., 2007; Florian et al., 2012).
The paradigm of the DNA damage theory of stem cell aging states that aging-associated changes in the DNA repair system in HSCs, together with changes in cell cycle regulation due to increased DNA damage with age (Pietras et al., 2011; Rossi et al., 2007a) are thought to result in elevated DNA mutations, which then causally contribute to the decrease in HSCs function with age. The paradigm is in part based on the finding that mice lacking a distinct set of DNA damage repair proteins display reduced function of HSCs, including an impaired repopulating potential and an overall depletion of the HSC pool (Ito et al., 2004; Navarro et al., 2006; Nijnik et al., 2007; Parmar et al., 2010; Prasher et al., 2005; Reese et al., 2003; Rossi et al., 2007a; Ruzankina et al., 2007; Zhang et al., 2010; Geiger et al., 2013), although in naturally aged mice there is actually an expansion of the number of phenotypic stem cells instead of a depletion of the HSCs pool. HSC aging also correlates with an increase in DNA double strand breaks (DSBs). Both human and mouse HSCs present upon aging with a 2-3fold elevated number of γH2AX foci, a bona fide surrogate marker for unresolved DSBs (Rossi et al., 2007a; Rübe et al., 2011). Unresolved DSBs accumulated in quiescent but not cycling HSCs upon aging (Beerman et al., 2014). γH2AX foci though were very recently shown to co-localize in HSCs with proteins associated with replication and ribosomal biogenesis stress (Flach et al., 2014), rendering γH2AX foci as a general marker for persistent DNA DSBs in HSCs questionable.
Signaling cascades activated in HSCs in response to DNA damage will determine the ultimate outcome of the damage. The cascades might run in parallel, comprising initiation of DNA repair (intact or erroneous repair), apoptosis as well as senescence and differentiation signaling. DNA double strand breaks (DSBs) can also be very potent inducers of cellular senescence in murine and human fibroblasts and murine HSCs (Leonardo et al., 1994; Nakamura et al., 2008; Shao et al., 2014). Differentiation in response to DNA damage has been described for various tissues including HSCs, melanocytic stem cells and embryonic stem cells (Inomata et al., 2009; Li et al., 2012; Wang et al., 2012). DNA damage induces apoptosis by p53-dependent and p53-independent pathways, and in lymphocytes and germ cells for example apoptosis represents the primary response to DNA damage (Lee et al., 1998). In murine HSCs, DNA damage induced by telomere attrition or DNA DSBs leads to an induction of lymphoid differentiation (Wang et al., 2012). Quiescent murine HSPCs that are mostly residing in G0 phase of the cell cycle are thought to undergo an error-prone NHEJ pathway to repair DSBs (Mohrin et al., 2010). Upon irradiation though HSCs enter the cell cycle via elevation of p21 which implies that HSCs might use an error-free HR pathway to repair DNA damage (Insinga et al., 2013; Beerman et al., 2014). However, the outcome of DNA damage in HSCs with respect to mutational load upon damage resolution as well as stem cell function in correlation to aging has not been investigated in great detail.
In this study we therefore investigated DNA damage outcomes of aged and young HSPCs in more detail. Aging resulted in a 2-3 fold increase of the mutational load in the hematopoietic system. DNA damage response and outcomes in response to irradiation or chronic individual DSBs in young and aged HSCs though were similar. HSCs, both young and aged, were also very resilient towards further accumulation of DNA mutations in response to damage, which might be linked to unexpected differences in the DNA damage response between HSCs and differentiated cells regarding G1/S cell cycle checkpoint activation. Collectively, we show that differentiated cells and HSCs respond differently to DNA damage, while the response is very similar for young and aged HSCs with respect to mutation accumulation and fitness in response to DNA DSBs.
RESULTS
Distinct DNA damage checkpoint activation in HSCs upon irradiation
DNA damage induces cell cycle checkpoints as well as apoptosis, which are critical signaling cascades that directly impact DNA damage outcomes (Kastan and Bartek, 2004). Murine embryonic fibroblasts (MEFs), cell lines as well as differentiated primary cells activate a G1/S cell cycle checkpoint upon DNA damage through inhibition of the transcription factor E2F by the retinoblastoma (Rb) repressor complex. This results in a transcriptional arrest and a halt of progression into S-phase and is regarded to be a hallmark of the DNA damage response. For example though embryonic stem cells skip activation of the G1/S checkpoint upon DNA damage (Aladjem et al., 1998; Hirao et al., 2000; Hong and Stambrook, 2004). We thus first asked which DNA-damage checkpoints are active in HSCs and whether at stage of checkpoint activation HSCs undergo apoptosis in response to DNA damage. To this end, flow cytometry assays to measure cell cycle profiles and at the same time apoptotic status of young and aged HSPCs and differentiated hematopoietic cells were performed 16 hours post in vivo irradiation (Figure 1a). In response to 7 Gy (sublethal) of irradiation, the total number of young, HPCs (c-Kit+ cells), early hematopoietic progenitor cells (LSK cells) and long-term repopulating HSCs (LT-HSCs) was significantly reduced, but not yet that of terminally differentiated (Lin+) cells (Figure 1b). These data are consistent with a positive correlation between radiation sensitivity and primitiveness of hematopoietic cells.
Figure 1. Distinct DNA damage checkpoint activation dynamics in young and aged HSCs upon irradiation.
a: Experimental setting and gating strategy. Young (2-3 months) and aged (18-24 months) C57Bl6 mice were irradiated with 3 or 7 Gy. After 16 h, 500 μg BrdU were injected intraperitoneally per mouse and 45 min later the mice were sacrificed. BM was isolated and Lineage− cells, c-Kit+ cells, LSK cells and LT-HSCs were analyzed by flow cytometry. Each population was scanned for cell cycle status (BrdU incorporation over DNA content) and apoptosis (AnnexinV+). b: Total number of Lin+, Lin−, c-Kit+, LSK and LT-HSC cells per tibiae and femur determined by flow cytometry 16 h after total body irradiation with 7 Gy. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001; columns are means +1 SEM; n=5. c and d: Percentage of cells in either G0/G1- or S-phase of the cells cycle. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 statistics relative to control group; columns are means +1 SEM; young control n=5, young 7 Gy n=3, old control n=5, old 7 Gy n=5. e and f: Percentage of AnnexinV+ cells either in G0/G1- or S-phase of the cell cycle. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 statistics relative to control; young control n=5, young 7 Gy n=3, old control n=5, old 7 Gy n=5. (see also Figure S1)
Differentiated hematopoietic cells (Lin+ as well as Lin− cells) presented with the same frequency of cells in G0/G1 upon irradiation, while more primitive young as well as aged c-Kit+, LSK or LT-HSCs showed a lower frequency of cells in G0/G1. This implies an impaired activation of the G1-S checkpoint in primitive hematopoietic cells (PHCs) (Figure 1c+d). All cell types accumulated to a similar extent in the G2/M phase upon irradiation, implying a functional G2/M or mitotic checkpoint (Figure S1a). The impaired activation of the G1-S checkpoint was not restricted to the C57BL/6 strain (HSCs from DBA/2 mice show a similar phenotype) and can already be observed at lower doses of irradiation (Figure S1c+d, S2c). An impaired activation of the G1-S restriction checkpoint upon DNA damage was further implied by the finding that the frequency of HSPCs (c-Kit+, LSK, LT-HSCs) in S-phase of the cell cycle increases upon irradiation (Figure 1d).
Loss of the retinoblastoma (Rb) protein in fibroblasts results in a loss of a G1-S checkpoint activation and genetic instability (van Harn et al., 2010). A lack of G1-S activation in HSPCs would thus render Rb in HSCs dispensable with respect to the DNA damage response. We therefore tested activation of DNA damage signaling in hematopoietic cells deficient for Rb (Rb HemKO (Daria et al., 2008)). Consistent with our prediction, HSPCs devoid of Rb did not show any difference in checkpoint activation upon irradiation nor in the number of γH2AX foci or tail length in a comet assay, another surrogate assay for DNA strand breaks (Figure S2a-c). However, we observed a small but significant increase of cells in S-phase of the cell cycle in steady state in Rb HemKO mice (Figure S2c), supporting a role for Rb in regulation of the G1/S checkpoint in HSPCs with respect to basic proliferation.
Activation of the G1-S checkpoint is supposed to stop a cell from entering S-phase of the cell division cycle to allow for DNA repair processes in G0/G1, which in turn inhibit apoptosis initiated by the DNA damaging event (Sancar et al., 2004). Our analyses revealed that differentiated hematopoietic cells showed low levels of apoptosis in G0/G1-phase in response to irradiation, and elevated levels in the S and G2-M stages of the cell division cycle (Figure 1e+f, Figure S1b). Surprisingly though, there was already a strong induction of apoptosis in G0/G1 in more primitive cells (c-Kit+, LSK and LT-HSCs) in response to irradiation, with levels of apoptosis remaining high in S and G2-M. This indicated that HSPCs undergo apoptosis in response to irradiation, independent of their cell cycle checkpoint position.
The lack of G1-S checkpoint activation correlated with very low levels of expression of the checkpoint kinase Chk2 (critical for G0/G1 checkpoint activation) in HSPCs upon irradiation (data not shown) as well as sequestration of Chk2 to the Pericentrin-2 positive centrosome in primitive hematopoietic cells, most likely rendering Chk2 ineffective (Figure S2d). Elevated expression of Chk2 in hematopoietic cells activated a G0/G1 arrest upon DNA damage while maintaining the relatively high level of apoptosis already seen in G0/G1. This resulted in impaired stem and progenitor cell function in transplantation assays (Figure S2e-g), which was dependent on the kinase function of Chk2. These data imply that induction of Chk2 expression activates a G0/G1 checkpoint in HSPCs at the expense of a high level of apoptosis upon DNA damage (see Figure 1e,f). In summary, young as well as aged HSPCs actively suppress activation of the G1-S checkpoint upon irradiation and present at the same time with high levels of apoptosis upon DNA damage, independent of their position within the cell division cycle.
Robust protection of genomic integrity in young and aged HSCs
Various reports indicate persistence of DNA damage after irradiation in HSCs and elevated steady-state damage in aged HSCs (Rossi et al., 2007b; Mohrin et al., 2010; Insinga et al., 2013) or elevated levels of stalled replication forks (Flach et al., 2014). We therefore determined the frequency of γH2AX foci in response to irradiation in young and aged LT-HSCs, which has been regarded as a surrogate marker for the frequency of DSBs in the genome. Aged LT-HSCs presented with a small, but significant increase in the number of foci per cell as also previously reported (Beerman et al. 2014; Flach et al., 2014; Rossi et al., 2007a) (Figure S3a). The comet assay quantifies changes in DNA migration caused by DSBs, alkaline labile sites, and transient repair sites. Aged LT-HSCs presented with a minor elevated tail moment under steady state conditions compared to young cells (Figure S3b) (see also Beerman et al. 2014) . Both young and aged LT-HSCs though presented with a similar absolute increase in the tail moment as well as γH2AX foci number after irradiation. Interestingly, theses changes were not linked to oxidative stress, since levels of 8-oxo-dG (a quantitative marker of oxidative damage of DNA) did not show significant differences between young and aged LT-HSCs in steady state and after irradiation (Figure S3c).
We also determined the loss of heterozygosity (LOH) at distinct microsatellite marker locations to further quantify the outcome of DNA damage. Monoclonal colonies (from a CFC assay) derived from individual young and aged hematopoietic progenitor (LK cells) as well as early hematopoietic progenitor cells (LSK cells) presented with similar LOH frequency. Upon irradiation at 5 Gy ex vivo, aged LSK presented with a 2-3 fold increase in the LOH frequency compared to young LSK cells (Table S1). The CFC assay is performed under supraphysiological cytokine conditions, which have been reported to suppress apoptosis in hematopoietic cells upon irradiation (Chute et al., 2004; Hérodin and Drouet, 2005). The extent in increase in LOH frequency is similar in magnitude to the increase for example in tail moment of aged HSCs in the comet assay (Figure S3b).
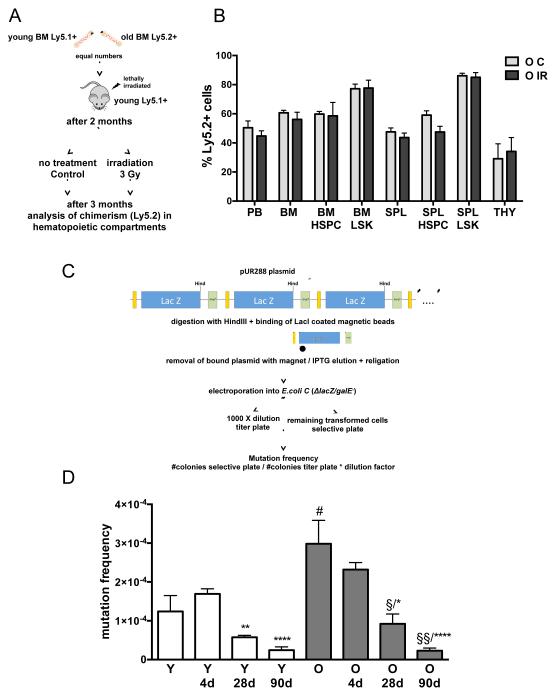
To test whether DNA damage will alter stem cell function in vivo distinctly in young and aged HSPCs, competitive transplantation - irradiation - recovery experiments were performed, in which young and aged HSCs directly functionally compete post DNA-damage in the same recipient animal. Functionality of young and aged HSCs is indicated by the level of chimerism of donor cells in peripheral blood 3 months post irradiation. We detected almost identical chimerism ratios of old (Ly5.2+) vs. young cells (remaining cells, Ly5.1+) in control vs. irradiated animals (Figure 2a+b), strongly implying that stem cell function upon irradiation is very similar in young and aged HSCs. This conclusion is further supported by our finding that young and aged stem and progenitor cells showed almost identical survival frequencies upon irradiation in various functional in vitro assays (cobblestone-area forming cell (CAFC) assay as well as colony-forming cell assays, data not shown).
Figure 2. Protection of genomic integrity in young and aged hematopoietic cells upon DNA damage in vivo.
a: Schema of experimental setup. b: Chimerism of Ly5.2+ cells within the different hematopoietic compartments. Control n=5, 3 Gy n=7; PB=peripheral blood; BM=bone marrow; SPL=spleen, THY=thymus, LSK=lineage negative/c-Kit+/Sca-1+ BM cells; HSPC=hematopoietic stem and progenitor cells. (see also Figure S2) c: Experimental setup of the assays to determine genomic mutations (see Material and Methods for details). d: Mutation frequency (number of mutated plasmids per 104 plasmids analyzed) of BM cells derived from young (2-3 months) and aged (18-24 months) mice before, after 4 d, 28 days and 3 months after irradiation with 3 Gy. n= 18 for young, n= 5 for aged, n=7 for young 90d, n=5 for aged 90d, n= 3 for young 4d, 28d and aged 28d and n=2 for aged 4d; *p < 0.05, **p < 0.01, ****p < 0.0001 (compared to values after 4d within group), §p < 0.05, §§p < 0.05 (compared to values before irradiation within group) #p = 0.0524 (Y compared to O); columns are means +1 SEM.
Using a validated and widely accepted transgenic plasmid based mutation detection assay (Figure 2c) (Boerrigter et al., 1995; Geiger et al., 2006, 2009), we next analyzed DNA repair outcomes in hematopoiesis with respect to fidelity of the DNA damage repair process in vivo by determining mutation frequencies in response to DNA damage. Mutation frequencies in BM cells from transgenic mice were determined in response to total body irradiation (3 Gy) at four days after irradiation (resembling activity of differentiated cells), 28 days after (indicative of progenitor cell activity) and three months after (hematopoiesis indicative of HSCs activity). BM cells in aged mice had a trend towards an increased mutational load in steady state hematopoiesis (around 2-fold) (Figure 2d). The majority of mutations detected were translocations and/or deletions (data not shown) and not point mutations, which is a unique pattern of mutations among all tissues analyzed so far with this indicator strain (Dollé et al., 2000). Surprisingly, the mutation frequency in BM cells did not increase in response to irradiation. To the contrary, the mutation frequency significantly decreased 28 days post irradiation in BM cells as well as at three months post irradiation in both age groups, with translocations still being the majority of the remaining mutations (Figure 2d and Figure S3d). This implies that the DNA damage response of the hematopoietic system avoids accumulation of genomic mutations upon irradiation and this mechanism remains fully intact in aged animals. The data also demonstrate that young and aged HSPCs show a similar in vivo response to DNA damage and a similar DNA damage repair outcome in response to irradiation, with an overall low mutational load.
In summary, both young and aged PHCs strongly avoid having DNA-damaged cells in the G0/G1 state. They present with a high level of apoptosis upon DNA damage induced by irradiation, and show for a selective type of DNA mutations (LOH) in response to in vitro damage an 2-3 fold increase, while DNA damage surviving cells in vivo do not accumulate DNA mutations as determined by the indicator strain. These data imply a mechanism of either repair with an overall low error rate or a strong selection for undamaged cells. Also clearing of damaged cells, which exceed a threshold for a still tolerated mutation number or induction of replicative senescence might contribute to this outcome. This suggests that the hematopoietic system is actively managing the DNA damage response outcome in vivo with respect to mutational load.
Generation of a defined DNA DSB in vivo in stem cells via a LacZ-specific zinc-finger nuclease
We further tested this “reduce the mutational load upon damage” hypothesis of PHCs in vivo. Specifically, a defined DSB was induced by homodimeric zinc-finger nucleases (ZFNs) (we generated two distinct ZNFs versions, 1.25 and 1.34, Figure S4a) specific to a palindromic target site within the lacZ gene (at bp 407-430, Figure 3a) (Maeder et al., 2009). ZFNs bind as dimers to their specific target site (in our case LacZ, transgenic mouse) and a DNA DSB is generated via the attached Fok1 nuclease within the spacer region (6 bp), separating the two binding domains (9 bp each, Figure 3a). A plasmid-based single strand annealing repair assay in which the activity of the ZFN is proportional to the expression of GFP (Porteus and Baltimore, 2003) demonstrated increased activity of the zinc-fingers relative to standard controls (113% for 1.25 and 192% for 1.34), Figures 3b+c). A cell survival assay, in which the non-toxic endonuclease I-SceI from yeast was used as a negative standard for reference and transfection with caspase-activated DNAse (CAD) served as positive control, revealed no toxicity of the zinc-fingers and thus no unspecific off-target activity (Figure 3d). Transfection with ZNFs did not result in a significantly elevated number of γH2AX foci in cells containing no lacZ target site (data not shown), further confirming specificity of the ZNFs for their target site. For stable expression of the ZNFs in cell lines and HSCs, a bicistronic retroviral vector SF91/ZFN-IRES-eGFP was used (Figure 3e). Expression of ZFN proteins in hematopoietic cells was confirmed by western blotting of cells transduced with the ZFNs (Figure 3f). To further determine activity in vivo, we next investigated the activity of nuclear extracts from GFP+ fibroblast cells transduced with the ZFNs on purified pUR288 plasmid containing the lacZ target sequence. Incubation of the plasmid for one hour with the nuclear extract resulted in linearization of the plasmid (5.3kb band), which intensified in response to a 3 hour incubation, confirming specific ZFN activity in nuclear extracts (Figure 3g). In order to verify activity on genomic lacZ DNA in vivo, DNA from lacZ murine (small blue mouse) cells, which were transfected with the ZNF 8 hours prior to analysis by southern blot, was used. Since the pUR288 plasmid is integrated as a concatamer of 20 copies within the genome, the creation of a DNA DSB at the ZFN target site in vivo will result in DNA fragments the size of the plasmid (5.3 kb) (Figure 3h). ZFN 1.25 displays a distinct band at 5.3 kb, which is a unique result as usually “free” degradation products of ZNFs in vivo are very difficult to track due to their short half-life in vivo. In summary the two ZFNs generated are active on lacZ-DNA in primary cells in vivo and specific and thus are a unique tool to determine the response of stem cells to defined DSBs.
Figure 3. Confirmation of in vivo activity of a LacZ-specific zinc-finger nuclease.
a: Each three-finger zinc finger (F1, F2, F3) linked to the FokI nuclease domain (zinc-finger nuclease (ZNF) binds to a 9-bp half of a palindromic pUR288 plasmid target site. The amino acid sequences of the zinc-finger domains (F1, F2, F3) of the two ZFNs (1.25 and 1.34) are depicted. b: Schematic representation of the SSA-assay. In the assay the activity of the ZFN is proportional to the expression of GFP. c: Activity of the ZFNs relative to a positive standard. n=3. Columns: means +1 SEM. d: ZNF toxicity assay: Survival of fibroblasts co-transfected with a GFP plasmid and either with the ZFN or with the non-toxic endonuclease I-Sce-I (negative control) or caspase-activated Dnase (CAD, positive control), n=3. Columns: means +1 SEM. e: Gammaretroviral bicistronic SF91/ZFN-IRES-eGFP vector used for stable transduction of cells (LTR: long terminal repeat with strong enhancer element, wPRE: woodchuck hepatitis virus posttranscriptional regulatory element). f: Expression of the ZFNs 1.25 and 1.34 and actin in fibroblasts transduced with the SF91/ZFN-IRES-eGFP vector (representative Western blot) g: Schematic representation of experimental setup. Agarose gel electrophoreses of pUR288 plasmid incubated with nuclear extracts for 1 or 3 hours from cells transduced with the SF91/ZFN-IRES-eGFP virus containing the ZFNs, pUR288 alone (negative control) and pUR288 digested with the restriction enzyme HindIII (positive control). h: Schematic representation of experimental setup. Representative Southern blot against pUR288. CTL= genomic DNA digested with HindIII (positive control). The linear plasmid has a size of 5.3 kb.
Young and aged HSCs sense single DSBs to protect their genome
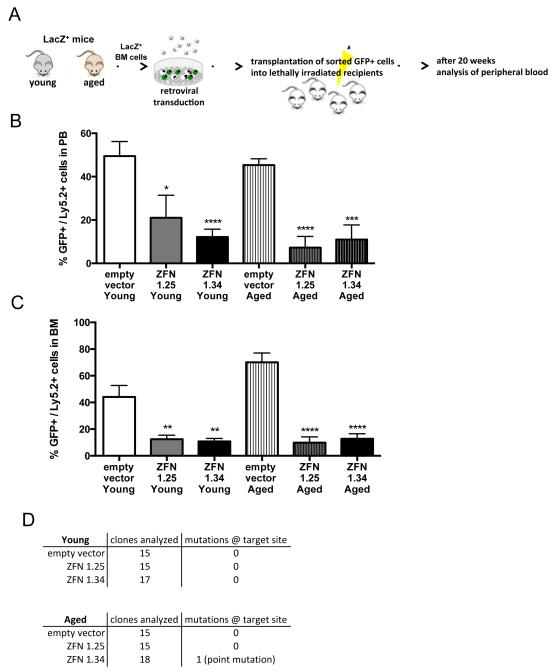
ZFN transduced and subsequently sorted (eGFP+) Lin− BM cells from the LacZ transgenic mouse were expanded in vitro for three days prior to analysis to obtain the cell numbers and thus the amount of DNA necessary for the mutation assay (Figure S4b). Lin− BM cells transduced with the lacZ-specific ZNFs showed a slight non-significant increase in the mutation frequency (Figure S4b). This finding implies a resilience of these cells to acquire DNA mutations and it correlates with the fact that PHCs do not activate a G1/S checkpoint and might thus avoid repair in G0/G1 and the DNA repair program associated with G0/G1. Mutation frequencies in PHCs are very difficult to determine due to the amount of DNA necessary for the assay. We instead focused on functional assays for lacZ positive HSCs transduced with ZNFs. Transduced Lin− cells (eGFP+) from young and aged mice were transplanted into lethally irradiated recipient mice (Figure 4a). 18-21 weeks after transplantation, when hematopoiesis in the periphery is driven by transplanted HSCs, ZFN positive young as well as aged HSCs presented with as significant decrease in eGFP+ cells among donor cells in PB and BM (Figure 4b+c). The fitness/status though of the transplanted HSPCs was not altered right after transduction, as there was no difference in the frequency of colony forming activity between control and ZFN transduced cells (Figure S4c). Because the active ZNF will constantly target the lacZ locus in stably transduced cells, a likely outcome is that almost all “surviving” eGFP+ clones show mutations in lacZ. Surprisingly, but consistent with the data presented so far, the remaining small number of eGFP+ transduced cells that could be recovered from the BM after transplantation did present with only a very low number of DNA mutations within the transgene, (one single clone with a point mutation at the ZFN target site in one mouse transplanted with aged Lin−cells (Figure 4d)). Our data therefore demonstrate that HSCs are resilient against acquiring mutations upon DNA damage in vivo, implying a ridged quality control mechanism to preserve genomic integrity of the stem cell pool. Interestingly, this approach and its underlying mechanisms are not altered upon aging of the hematopoietic system.
Figure 4. Young and aged HSCs sense single DSBs and protect their genome.
a: Experimental setup b+c: Contribution of transduced cells (GFP+/Ly5.2+ cells) to peripheral blood (PB) and bone marrow (BM) 18-21 weeks after transplantation. *p < 0.05, **p < 0.01, ****p < 0.0001; columns represent means +1 SEM. n=3 with a cohort of three to five recipient mice per group. d: Number of mutations in the lacZ ZNF target site sequence in GFP+BM cells (clone) 18-21 weeks post transplant.
DISCUSSION
The loss of maintenance of genomic integrity is central to most theories on somatic and stem cell aging. Analysis of HSCs with respect to the frequency of DNA damage (Comet assay, γH2AX foci, DNA mutation frequency, LOH assay) revealed an about 2-3 fold increase in these parameters upon aging. The difference in mutation frequency in steady state between young and aged BM cells was approximately twofold, which is in the range of changes in mutation frequencies recently reported for aged human BM cells via deep-sequencing approaches (Cancer Genome Atlas Research Network, 2013; Genovese et al., 2014; Welch et al., 2012). In a diploid genome that harbors around 6 × 109 nucleotides the total estimated mutational load per diploid BM cell is then around 300 mutations in young and 600 mutations in an aged animals. Whether such an increase upon aging is causatively linked to aging-associated diseases though still needs to be determined. A 22-fold increase in the mutational load (Geiger et al., 2006) is able to initiate leukemia in a mutator gene type setting (Kushner et al., 2008; Noronha et al., 2006; Su et al., 2005), while a 2-3 fold mutation load increase in BM cells does not induce leukemia (Krejci et al., 2008). The concept of a robust response of aged stem cells to DNA damage is further supported by the finding that muscle stem cells do not present with a significant accumulation of DNA damage upon aging (Cousin et al., 2013).
Might the elevated γH2AX foci and increased tail moment in comet assays upon aging also be linked to other aging-associated changes in stem cells than just DNA damage? In murine hair follicle stem cells, 53BP1 foci, a mechanistic marker for DNA DSBs, did not overlap with γH2AX foci, and instead γH2AX foci were identified as a sign for chromatin alterations upon aging (Schuler and Rube, 2013). Very recently it could be shown that elevated levels of γH2AX foci in aged HSCs can be also associated with replication and ribosomal biogenesis stress and might therefore not be an optimal marker for persistent DNA damage (Flach et al., 2014). Instead, elevated levels of γH2AX foci in aged HSCs might be linked to broader chromatin changes, which can, but need not to be, linked to DNA damage (Florian et al., 2012; Liu et al., 2013) Such changes might be linked to migration velocity of DNA in assays like the comet assay. Furthermore, it was shown by Beerman et al. (Beerman et al., 2014) that quiescent HSCs acquire DNA damage upon aging but when these cells are pushed into the cell cycle the damage is repaired. Our data suggest a mechanistic explanation for this finding, demonstrating that aged cells with DNA damage are pushed into the cell cycle and either repair properly without mutations or simply just die.
In summary, we demonstrate that both young and aged HSCs that survive irradiation are repaired properly with a low mutation rate regardless of their age. Our data strongly imply that both the young and the aged hematopoietic system are very resilient towards acquiring mutations upon DNA damage induced by irradiation, trying to maintain a pristine pool of HSCs contributing to hematopoiesis and to strongly suppressing leukemia. How is this resilience achieved? Multiple cellular and molecular mechanisms seem to contribute to that. Firstly, LT-HSCs are more sensitive to irradiation than their progeny as 3 Gy resulted in the loss of LT-HSCs whereas Lin−, c-Kit+ and LSK cells did not show a significant decrease in cell number (our own data and (Mohrin et al., 2010)). Secondly, and probably more importantly, our results show an unexpected loss of activation of the G1-S cell cycle checkpoint in HSPCs already at lower doses of irradiation (3 Gy), which was even more pronounced after high dose irradiation. Such an observation is similar to what has been described for embryonic stem cells, which also show absence of G1-S checkpoint upon damage (Aladjem et al., 1998; Hong and Stambrook, 2004). A lack of checkpoint activation in HSPCs was further supported by our finding that deletion of Rb in hematopoietic cells did not alter DNA damage response parameters. One alternative hypothesis, based on these observation is thus that genetic translocations and deletions that are found in leukemia cells are actually not a consequence of faulty repair by NHEJ in G0/G1, and rather imply that translocations and deletions are generated via faulty repair mechanisms at later stages of the cell division cycle, like in S, G2 or M phase.
In fibroblasts irradiation leads to a prolonged cell cycle arrest in G1-phase to halt the cell for repairing DNA damage repair before entering the cell cycle (Deckbar et al., 2011; Leonardo et al., 1994) whereas the same challenge could also lead to apoptosis, senescence or differentiation. In hematopoietic cells we observed that differentiated cells that do arrest in G0/G1-phase did not undergo apoptosis whereas more primitive cells did not arrest and showed high levels of apoptosis. Also human CD34+ HSPCs show increased apoptosis in response to DNA damaging agents when compared to differentiated cells (Buschfort-Papewalis et al., 2002). An elevated level of apoptosis in S-phase is supported by the finding that HSPCs, when proliferating, present with a decreased expression of prosurvival genes compared to quiescent cells (Mohrin et al., 2010).
Murine ES cells also lack G1-S checkpoint activation, which is due to intracellular mislocalization of the checkpoint kinase Chk2 (Hong and Stambrook, 2004). In murine LT-HSCs the Chk2 protein is not expressed throughout the nucleus, like differentiated cells, but sequestered at the centrosome. Ectopic expression of Chk2 in HSCs reduced function of HSCs, which is consistent with our finding that primitive hematopoietic cells, when in G0/G1, respond primarily with apoptosis to DNA damage. Taken together, the most likely outcome of DNA damage in a primitive hematopoietic cell might be dual in nature: apoptosis or repair without mutations. Such a model might not only hold true for DNA damage found at multiple locations within the cell as in response to irradiation. Our results further demonstrate that PHCs transduced with a ZNF that will create one site of DSBs per cells do not show an increase in mutation frequency. When transplanted, we further demonstrate that these cells are unable to contribute to PB chimerism as they most likely undergo apoptosis or senescence.
In aggregation, our results demonstrate an 2-3 fold increase in the steady-stated mutational load in the hematopoietic system, but almost equal DNA damage repair outcomes in young and aged HSCs. The role for DNA damage outcomes with respect to aging of HSPCs will need to be further investigated. Most importantly, these data reveal a heretofore unrecognized resilience of the hematopoietic system in general to acquire DNA mutations in response to DNA damage in vivo.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice (8-12 week-old and 18-26 month old) as well as C57BL/6.SJL-Ptprca/Boy (BoyJ) mice were obtained from Janvier or the divisional stock (derived from The Jackson Laboratories) L30 LacZ+ mice (small blue mouse, backcrossed on C57BL/6 background) were described previously (Boerrigter et al., 1995). Mice were housed under specific pathogen-free conditions at the University of Ulm or at CCHMC. Experiments were performed in compliance with the German Law for Welfare of Laboratory Animals and were approved by the Regierungspräsidium Tübingen or the IACUC at CCHMC.
Generation of a L30 lacZ+ adult fibroblasts cell line
Fibroblasts of L30 lacZ+ and lacZ− mice were generated as described before (Bosco and Knudsen, 2005). L30 lacZ+ adult fibroblasts started to become immortalized between passage 11 and 15 and were then used for experiments.
Mutation Assay
The mutation frequency analysis using the L30/small blue mouse model was performed as published (Boerrigter et al., 1995; Dollé et al., 1997; Geiger et al., 2009; Vijg et al., 1997)
Determination of loss of heterozygousity upon DNA damage via analysis of loss of inbred strain specific microsatellites in B6D2F1 mice
Clonal colonies (CFCs in complete methylcellulose medium, Stem Cell Technologies) from Lin−, c-Kit+ cells or Lin−, c-Kit+ Sac-1+ cells from young (2-3 months) or aged (22 month old) B6D2F1 mice were picked between day 7 and 9, washed in PBS and subsequently lysed (0.91 mg/ml Proteinase K, 0.5% Tween20, 0.5% Nonidet P40). DNA was subjected to multiplex-cocktails of fluorescently labeled primers that flank small tandem nucleotide repeats (microsatellites) polymorphic in length between DBA2 and B6. PCR a mplified DNA (95°C 15min; then 38 cycles of 94°C 30sec, 57°C 1:30min and 72°C 1min; 60°C 30min; and 4°C forever) was analyzed by capillary electrophoreses and peak calling relative to B6 and DBA/2 controls was performed with Gene Mapper software. (primers for LOH assay, picked randomly among the microsatellite markers that are distinct in length between C57BL/6 and DBA/2 and readable in multiplex setup while covering most chromosomes: D1Mit380, D9Mit123, DXMit64, D8Mit45, D12Mit143, D4Mit17, D16Mit60, D14Mit39, D3Mit57, D18Mit177, D10Mit230, D5Mit309, D2Mit66, D13Mit256, D19Mit96, D1Mit102, D6Mit284, D7Mit350, D15Mit67).
Generation of LacZ-specific zinc-finger nuclease
The lacZ specific zinc-finger nucleases (ZFN) 1.25 and 1.34 were generated using the OPEN method (oligomerized pool engineering) (Maeder et al., 2009). The homodimeric ZFN target site within the lacZ (bp 407-430, 5′-TCC GGC ACC AGA AGC GGT GCC GGA-3′) was identified using the web based software provided by the ZFN consortium. Then bacterial two-hybrid (B2H) selection strains were constructed harboring the ZFN target half-sites upstream of a B2H promoter. The zinc-finger array libraries were constructed by using DNA sequences encoding fingers from pre-selected ‘pools’ for each targeted triplicate (F1: GGA, F2: GCC, F3: GGT) that were fused together by overlap-PCR (Porteus, 2008). This resulted in a library of DNA sequences encoding random combinations of fingers. These DNA sequences were then cloned into low-copy expression phagemids and converted into infectious phage particles that were used to infect B2H selection cells (Kanamycin/Tetracyclin/Sucrose selection). Phagemids encoding the zinc-finger arrays that bind to the target site were isolated from colonies on the selection plates, the zinc-finger array DNA sequence amplified by PCR reaction, fused to a 5 amino acid linker sequence and ligated to the wildtype FokI nuclease domain. For sequences of LacZ-specific ZFNs see Figure S4.
For expression of the ZFN in hematopoietic cells the bicistronic retroviral vector SF91/IRES-eGFP was used. Cell-free supernatants containing retroviral particles were generated by transient transfection of Phoenix-gp packaging cells (ATCC number: CRL-3215) using Calcium Phosphate Transfection kit (Invitrogen).
Activity of ZFNs on target site (SSA assay)
The full ZFN target site was inserted into repeated sequences within the GFP gene. The reporter constructs also included the GFP1/2 full ZFN target site (5′-ACCATCTTC-ttcaag-GACGACGGC-3′) as a positive control and internal standard, previously described in (Pruett-Miller et al., 2008) as GFP1.4-B2H and GFP2-B2H. These SSA reporter plasmids were used to investigate the activity of the ZFNs on their target site. 100 ng of each ZFN-expression plasmid and 20 ng of reporter plasmid were co-transfected into HEK293 or 293T cells using the calcium phosphate transfection kit (Invitrogen). Percentage of GFP+ cells (DSB of ZFN at target site and subsequent SSA repair, restoring GFP expression) was determined at day 2 via flow cytometry. The activities of the ZFNs 1.25 and 1.34 were normalized to the activity of the internal standard.
Preparation of nuclear extract harboring ZFNs
Nuclear extract was prepared from stably transfected L30LacZ- fibroblasts with the SF91/ZFN-IRES-eGFP virus based on (Dignam et al., 1983). Protein concentrations obtained were between 2-6 μg/μL.
Activity of ZFNs on pUR288 plasmid (Plasmid Assay)
2 μg of supercoiled pUR288 plasmid were incubated at 37 °C for 1 to 3 hours in 1 x NEB buffer 2 with 50 μg of nuclear extracts from cells transduced with the SF91/ZFN- IRES-eGFP virus and applied to a 1 % agarose gel. Plasmid digested with HindIII served as positive control and undigested plasmid served as negative control.
Activity of ZFNs on genomic DNA (Southern Blot)
L30LacZ+ fibroblasts were transfected with the ZFN plasmids using the Attractene Transfection Reagent (QIAGEN). After 8 or 18 hours genomic DNA was isolated using the total DNA purification Kit (Epicenter). As positive control 10 μg of L30lacZ+ genomic DNA was digested with HindIII for 2 h at 37 °C. 15 μg of genomic DNA was loaded on a 1 % agarose gel. The Gel was incubated with 0.2 N HCl, denatured, neutralized and transferred overnight on a Hybond nylon membrane (GE Healthcare) using 10 x SSC buffer (pH7). Membrane was auto-crosslinked with 2400 mJoule UV irradiation, pre-hybridized for 1 h at 45 °C with salmon sperm DNA and Roti-Hybriquick solution (Roth) and finally hybridized with the radioactive labeled (32P-dCTP probe against bp 849-2442) over night at 48 °C. After stringent washing the membrane was exposed to a PhosphoImager screen and analyzed using AIDA Image Analyzer and ImageJ software. Radioactive labeled probe was generated using the random primed DNA labeling Kit (NEB) according to manufacturers instructions
Toxicity assay
Toxicity assays were performed as previously described (Pruett-Miller et al., 2008). To calculate the percent survival relative to I-SceI, the ratio after nuclease transfection was normalized to the ratio after I-SceI transfection and this determined the percent survival compared to I-SceI.
Western blot analysis of ZFN and Chk2-Protein
Transduced cells were re-suspended in Mg2+ lysis/wash buffer (Upstate cell signaling solutions) containing 10 % glycerol, 25 mM sodium fluoride, 1 mM sodium orthovanadate and a protease inhibitor cocktail (Roche Diagnostics), incubated for 15 minutes on ice and centrifuged. Equal amounts of protein were used for western blot analysis. ZFN was visualized using anti-FLAG-Tag antibody (OctA-Probe (D8): sc-807, Santa Cruz). Chk2 and Chk2 KD anti-Chk2 antibody (sc-17747 (a-11), Santa Cruz), β-actin (Sigma).
Isolation of BM and retroviral transduction
Lineage negative cells from BM were pre-stimulated for 2 days in IMDM medium (Lonza) supplemented with 10 % FBS, 1 % Penicillin/Streptomycin, 2 mM L-glutamine, 50 ng/mL ratSCF, 10 ng/mL mIL-3, 100 ng/mL mFlt3-Ligand and 100 ng/mL mIL-11 (Prospec) at a density of 6 - 8 × 106 cells/well. Viral transduction was performed on day 3 in RetroNectin-coated (TaKaRa) non-tissue culture plates that were pre-loaded with viral supernatant by centrifugation. Pre-stimulated BM cells were seeded on top (9 - 9.5 × 105 cells/well) and another layer of virus with cytokines was added. Media was changed the next morning and another round of transduction was carried out over night. The next day cells were harvested using cell dissociation buffer (Gibco) and GFP+ cells were sorted by FACS on a BD FACS Aria II or III (BD Bioscience).
CFC assay
CFC assays were performed as described elsewhere (Geiger et al., 2001; Xing et al., 2006).
Transplantation
For competitive transplantation, equal numbers (1 – 2 × 106) of young (2 - 4 months) Ly5.1+ total BM cells and aged (18 - 24 months) Ly5.2+ total BM cells were mixed and transplanted into lethally irradiated (11 Gy) Ly5.1+ recipient mice. After three months one cohort of transplanted mice (n=7) was irradiated with 3 Gy. Ly5.2-chimerism in all hematopoietic compartments was investigated after 3 months (PB, BM, BM-HSPCs, spleen, spleen-HSPCs and thymus) using flow cytometry. To investigate in vivo behavior of cells transduced with the ZFNs, 1 - 4 × 105 sorted GFP+ Lin- cells in 200 μL PBS/mouse were transplanted into lethally irradiated (11 Gy) 3 - 6 months old BoyJ (Ly5.1+) mice via retro-orbital injection. To determine effects of Chk2 overexpression and expression of Chk2 KD, Ly 5.2+ BM cells were tranduced with the appropriate virus and equal numbers of cells were mixed with (1-2 × 106) Ly5.1+ BM cells and transplanted into lethally irradiated Ly5.1+ recipient mice. PB chimerism was analyzed every 4 weeks by flow cytometry.
Flow cytometry and cell sorting
For apoptosis and cell cycle analyses, young (2 - 3 months) and aged (18 - 24 month) C57BL/6 mice were irradiated with 3 or 7 Gy (n=4). After 16 h, 500 μg BrdU (BD, Cell Cycle Kit) in 200 μL PBS/mouse were injected i.p. After 45 min the mice were sacrificed, BM flushed and mononuclear cells isolated by low-density centrifugation. 2 × 106 cells were stained with a cocktail of biotinylated lineage antibodies after Fc block for 15 min: anti-Sca-1 (clone D7) (eBioscience), anti-c-Kit (clone ACK2) (eBioscience), anti-CD34 (clone RAM34) (eBioscience), and Streptavidin (eBioscience) for 1 h on ice. Then cells were washed and incubated in 1 x Binding Buffer (BD, Cell Cycle Kit) with anti-AnnexinV antibody (BD, Apoptosis Detection Kit) for 20 min at RT. After washing cells were fixed and permeabilized using BD Cytofix/Cytoperm buffer (BD, Cell Cycle Kit). Cells were kept overnight in 1 x Perm/Wash buffer (BD, Cell Cycle Kit) and again permeabilized the next morning. Cells were then treated with 30 μg DNAse (BD, Cell Cycle Kit) in PBS with Ca2+/Mg2+ for 1.5 h at 37 °C and after washing incubated with anti-BrdU antibody (BD, Cell Cycle Kit) for 20 min at RT. Finally, after washing 7AAD was added and cells were immediately analyzed with a LSRII flow cytometer (BD Biosciences). LT-HSCs were defined as Lin−/c-Kit+/Sca-1+/CD34−/low, LSK represent Lin−/c-Kit+/Sca-1+ cells and hematopoietic progenitor cells were gated Lin−/c-Kit+. Apoptosis staining on Chk2 and Chk2 KD transduced c-Kit+ BM cells was performed using the AnnexinV antibody according to manufacturer’s instruction (BD, Apoptosis Detection Kit). LT-HSCs were isolated as previously described (Florian et al., 2012).
8-oxo-dG staining
Lineage negative cells were harvested and incubated over night at 37 °C (5 % CO2, 3 % O2) in HBSS supplemented with 10 % FBS. After 16 h cells were irradiated with 2 Gy and incubated for 1 h at 37 °C (5 % CO2, 3 % O2) in HBSS supplemented with 10 % FBS. Then surface marker staining and permeabilization was completed as described in flow cytometry and cell sorting. Stained, fixed and permeabilizsed cells were incubated with 30 μg DNAseI (BD, Cell Cycle Kit) in PBS with Ca2+/Mg2+ for 1 h at 37 °C., washed and stained over night with the anti-8-oxo-dG antibody (clone 2E2, Trevigen). After 16 h cells were washed and stained with secondary antibody (Alexa Fluor® 488 Goat Anti-Mouse IgG, Invitrogen).
IF staining
IF staining was performed exactly as described previously (Florian et al., 2012).
Alkaline Comet Assay
LT-HSCs were incubated in IMDM medium (Lonza), 10 % FBS, 1 % Penicillin/Streptomycin, 2 mM L-glutamine at 37 °C (5 % CO2, 3 % O2) and irradiated the next day with 2 Gy (2 min 20 sec at RT), put on ice and centrifuged for 5 min at 1500 rpm at RT. Cell were resuspended (zero time point) and incubated at 37 °C (5 % CO2, 3 % O2) for two hours. Alkaline comet assay was performed with CometSlides™ and Comet Assay® Reagent Kit (Trevigen). 1000-1500 LT-HSCs were encapsulated in 49 μL of low-melting-point on a pre-warmed CometSlide™ and incubated at 4 °C in the dark for 30 min. Slides were immersed in 44 ml pre-chilled lysis solution (4 ml DMSO (Sigma) in 40 ml Lysis Solution®) on ice for 40 min, drained of excess lysis solution, immersed into the alkaline solution for 30 min and then placed into the electrophoresis chamber with alkaline buffer (pH=13) at 4 °C (30 V and 300 mA for 30 min). Excess buffer was drained, slides immersed twice in cold ddH2O for 10 min, fixed by immersing in cold 70 % ethanol for 5 min and dried at 37 °C for 30 min. For DNA staining 100 μl of diluted SYBR® Green 1 (1 μl of 10000 x concentrated SYBR® green 1 in 10 ml TE buffer; TE buffer: 10 mM Tris-HCL pH 7.5, 1 mM EDTA) were added onto each sample. Cells were stained 5 min at 4 °C before excess SYBR solution was removed. Dried slides were analyzed by fluorescence microscopy (AxioObserver Z1 microscope (Zeiss)). Images of 50 cells of randomly chosen fields with equal exposure time were captured. Comet tail length and tail moment (%DNA in tail multiplied by tail length) were analyzed with the image analysis software CometScore™ (TriTek Corporation).
Statistical analysis
Normal distribution of data was implied and the variance between the groups was similar. Data is displayed as mean +1 standard error of the mean (SEM). All statistical analyses were performed using Student’s t-test with GraphPad prism 6 software. In transplantation experiments only healthy engrafted mice were included in analysis. For in vitro experiments, samples were excluded due to technical problems (procedure or reagents). Number of biological repeats (n) is indicated in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Van Zant and Jose A. Cancelas for advice and critical reading of the manuscript. We thank the cores at Ulm University and CCHMC for cell sorting support and the Comprehensive Mouse and Cancer Core at CCHMC for support with animal experiments. This work was supported by grants (H.G) from the Deutsche Forschungsgemeinschaft SFB 1079, KFO 142, The German Scholar Organization, the BMBF-funded Program SyStaR and from the Edward P. Evans Foundation and the National Institute of Health, HL076604, DK077762 and AG040118 to H.G. and a “Bausteinprogramm” of the Department of Medicine Ulm to M.C.F.
Footnotes
AUTHOR CONTRIBUTIONS
B.M.M. and H.G. designed and interpreted experiments and wrote the manuscript. A.B., M.C.F., K.N., K.S., M.R., M.V., C.B., D.S. and D.W. performed and analysed experiments. A.A., D.P., M.D.M, P.S. and M.P. assisted in designing and interpreting experiments.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent Hematopoietic Stem Cells Accumulate DNA Damage during Aging that Is Repaired upon Entry into Cell Cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerrigter ME, Dollé ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- Bosco EE, Knudsen ES. Differential role of RB in response to UV and IR damage. Nucleic Acids Res. 2005;33:1581–1592. doi: 10.1093/nar/gki283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschfort-Papewalis C, Moritz T, Liedert B, Thomale J. Down-regulation of DNA repair in human CD34(+) progenitor cells corresponds to increased drug sensitivity and apoptotic response. Blood. 2002;100:845–853. doi: 10.1182/blood-2002-01-0022. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chute JP, Fung J, Muramoto G, Erwin R. Ex vivo culture rescues hematopoietic stem cells with long-term repopulating capacity following harvest from lethally irradiated mice. Exp. Hematol. 2004;32:308–317. doi: 10.1016/j.exphem.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Cousin W, Ho ML, Desai R, Tham A, Chen RY, Kung S, Elabd C, Conboy IM. Regenerative Capacity of Old Muscle Stem Cells Declines without Significant Accumulation of DNA Damage. PLoS ONE. 2013;8:e63528. doi: 10.1371/journal.pone.0063528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daria D, Filippi M-D, Knudsen ES, Faccio R, Li Z, Kalfa T, Geiger H. The retinoblastoma tumor suppressor is a critical intrinsic regulator for hematopoietic stem and progenitor cells under stress. Blood. 2008;111:1894–1902. doi: 10.1182/blood-2007-02-071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Jeggo PA, Lobrich M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit. Rev. Biochem. Mol. Biol. 2011;46:271–283. doi: 10.3109/10409238.2011.575764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat. Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014 doi: 10.1038/nature13619. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian MC, Dörr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, Filippi M-D, Hasenberg A, Gunzer M, Scharffetter-Kochanek K, et al. Cdc42 Activity Regulates Hematopoietic Stem Cell Aging and Rejuvenation. Cell Stem Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H, True JM, Haan G. de, Zant GV. Age- and stage-specific regulation patterns in the hematopoietic stem cell hierarchy. Blood. 2001;98:2966–2972. doi: 10.1182/blood.v98.10.2966. [DOI] [PubMed] [Google Scholar]
- Geiger H, Schleimer D, Nattamai KJ, Dannenmann SR, Davies SM, Weiss BD. Mutagenic potential of temozolomide in bone marrow cells in vivo. Blood. 2006;107:3010–3011. doi: 10.1182/blood-2005-09-3649. [DOI] [PubMed] [Google Scholar]
- Geiger H, David S, Nattamai KJ, Jan V. Quantification of genomic mutations in murine hematopoietic cells. Methods Mol. Biol. Clifton NJ. 2009;506:423–436. doi: 10.1007/978-1-59745-409-4_28. [DOI] [PubMed] [Google Scholar]
- Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013 doi: 10.1038/nri3433. advance online publication. [DOI] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérodin F, Drouet M. Cytokine-based treatment of accidentally irradiated victims and new approaches. Exp. Hematol. 2005;33:1071–1080. doi: 10.1016/j.exphem.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Hong Y, Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, et al. Genotoxic Stress Abrogates Renewal of Melanocyte Stem Cells by Triggering Their Differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Insinga A, Cicalese A, Faretta M, Gallo B, Albano L, Ronzoni S, Furia L, Viale A, Pelicci PG. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3931–3936. doi: 10.1073/pnas.1213394110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Krejci O, Wunderlich M, Geiger H, Chou F-S, Schleimer D, Jansen M, Andreassen PR, Mulloy JC. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111:2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner BH, Laquaglia MP, Kramer K, Modak S, Cheung N-KV. Recurrent metastatic neuroblastoma followed by myelodysplastic syndrome: Possible leukemogenic role of temozolomide. Pediatr. Blood Cancer. 2008;51:552–554. doi: 10.1002/pbc.21658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Leonardo AD, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct Regulatory Mechanisms and Functions for p53-Activated and p53-Repressed DNA Damage Response Genes in Embryonic Stem Cells. Mol. Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat. Immunol. 2004;5:133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BMC, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin Modifications as Determinants of Muscle Stem Cell Quiescence and Chronological Aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an “open-source” protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, Passegué E. Hematopoietic Stem Cell Quiescence Promotes Error-Prone DNA Repair and Mutagenesis. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura AJ, Chiang YJ, Hathcock KS, Horikawa I, Sedelnikova OA, Hodes RJ, Bonner WM. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin. 2008;1:6. doi: 10.1186/1756-8935-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, Puerto S, Surrallés J, Segovia JC, Bueren JA. Hematopoietic Dysfunction in a Mouse Model for Fanconi Anemia Group D1. Mol. Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Noronha V, Berliner N, Ballen KK, Lacy J, Kracher J, Baehring J, Henson JW. Treatment-related myelodysplasia/AML in a patient with a history of breast cancer and an oligodendroglioma treated with temozolomide: case study and review of the literature. Neuro-Oncol. 2006;8:280–283. doi: 10.1215/15228517-2006-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K, Kim J, Sykes SM, Shimamura A, Stuckert P, Zhu K, Hamilton A, Deloach MK, Kutok JL, Akashi K, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells Dayt. Ohio. 2010;28:1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. Design and testing of zinc finger nucleases for use in mammalian cells. Methods Mol Biol. 2008;435:47–61. doi: 10.1007/978-1-59745-232-8_4. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Prasher JM, Lalai AS, Heijmans-Antonissen C, Ploemacher RE, Hoeijmakers JH, Touw IP, Niedernhofer LJ. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1∣[minus]∣/∣[minus]∣ mice. EMBO J. 2005;24:861–871. doi: 10.1038/sj.emboj.7600542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair–deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007a;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Seita J, Czechowicz A, Bhattacharya D, Bryder D, Weissman IL. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle Georget. Tex. 2007b;6:2371–2376. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- Rübe CE, Fricke A, Widmann TA, Fürst T, Madry H, Pfreundschuh M, Rübe C. Accumulation of DNA Damage in Hematopoietic Stem and Progenitor Cells during Human Aging. PloS One. 2011;6:e17487. doi: 10.1371/journal.pone.0017487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the Developmentally Essential Gene ATR in Adult Mice Leads to Age-Related Phenotypes and Stem Cell Loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schuler N, Rube CE. Accumulation of DNA Damage-Induced Chromatin Alterations in Tissue-Specific Stem Cells: The Driving Force of Aging? PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0063932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Feng W, Li H, Gardner D, Luo Y, Wang Y, Liu L, Meng A, Sharpless NE, Zhou D. Total body irradiation causes long-term mouse BM injury via induction of HSC premature senescence in an Ink4a- and Arf-independent manner. Blood. 2014;123:3105–3115. doi: 10.1182/blood-2013-07-515619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Morrison SJ. Mechanisms that Regulate Stem Cell Aging and Life Span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-W, Chang M-C, Chiang M-F, Hsieh R-K. Treatment-related myelodysplastic syndrome after temozolomide for recurrent high-grade glioma. J. Neurooncol. 2005;71:315–318. doi: 10.1007/s11060-004-2028-0. [DOI] [PubMed] [Google Scholar]
- Vijg J, Dollé ME, Martus HJ, Boerrigter ME. Transgenic mouse models for studying mutations in vivo: applications in aging research. Mech. Ageing Dev. 1997;99:257–271. doi: 10.1016/s0047-6374(97)00155-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Groß A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, et al. A Differentiation Checkpoint Limits Hematopoietic Stem Cell Self-Renewal in Response to DNA Damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Ryan MA, Daria D, Nattamai KJ, Van Zant G, Wang L, Zheng Y, Geiger H. Increased hematopoietic stem cell mobilization in aged mice. Blood. 2006;108:2190–2197. doi: 10.1182/blood-2005-12-010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zant G, Liang Y. Concise review: hematopoietic stem cell aging, life span, and transplantation. Stem Cells Transl. Med. 2012;1:651–657. doi: 10.5966/sctm.2012-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-S, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, Watanabe-Smith K, Rathbun RK, Fleming WH, Bagby GC, et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. 2010;116:5140–5148. doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.