Abstract
The TOR (target of rapamycin) kinase limits longevity by poorly understood mechanisms. Rapamycin suppresses the mammalian TORC1 complex, which regulates translation, and extends lifespan in diverse species, including mice. We show that rapamycin selectively blunts the pro-inflammatory phenotype of senescent cells. Cellular senescence suppresses cancer by preventing cell proliferation. However, as senescent cells accumulate with age, the senescence-associated secretory phenotype (SASP) can disrupt tissues and contribute to age-related pathologies, including cancer. MTOR inhibition suppressed the secretion of inflammatory cytokines by senescent cells. Rapamycin reduced IL6 and other cytokine mRNA levels, but selectively suppressed translation of the membrane-bound cytokine IL1A. Reduced IL1A diminished NF-κB transcriptional activity, which controls much of the SASP; exogenous IL1A restored IL6 secretion to rapamycin-treated cells. Importantly, rapamycin suppressed the ability of senescent fibroblasts to stimulate prostate tumour growth in mice. Thus, rapamycin might ameliorate age-related pathologies, including late-life cancer, by suppressing senescence-associated inflammation.
Several molecular pathways limit longevity in diverse species1, including that governed by the TOR (target of rapamycin) kinase. TOR senses nutrient and growth signals; high TOR activity favours somatic growth and limits lifespan, whereas dampened TOR activity favours longevity2,3. Rapamycin specifically suppresses activity of the mammalian TOR (MTOR) complex MTORC1, which regulates messenger RNA translation2, and was recently shown to extend lifespan in mice4. To understand how MTOR regulates longevity, we explored its role in regulating cellular senescence. Cellular senescence suppresses cancer by preventing the proliferation of cells at risk for malignant transformation5. Senescent cells accumulate with age, and express a complex senescence-associated secretory phenotype (SASP). SASPs can alter tissue microenvironments6–11, contributing to age-related pathologies, including, ironically, cancer8,12–16.
The incidence of cancer increases exponentially with age and therefore poses a major challenge to the longevity of many complex organisms. Unlike most age-related diseases, which generally cause cell and tissue degeneration and loss of function, cancer cells must acquire different, albeit aberrant, functions to progress to lethal disease. One link between age-related cancer and degeneration could be an inflammatory milieu driven by MTOR in senescent cells.
Persistent inflammation can cause or contribute to both degenerative diseases and cancer17–20. Further, a common feature of ageing tissues is low-level chronic inflammation, termed inflammaging21. The source of inflammaging is unclear. It may derive partly from a decline in immune homeostasis with age21,22. It may also derive partly from senescent cells that reside with increasing frequency within aged tissues23,24.
Many mitotically competent cells mount a senescence response following challenges that include DNA damage, disrupted chromatin and strong mitogenic signals (for example, those provided by activated oncogenes)5,25. In addition to a permanent cell-cycle arrest driven by the p53 (also known as TP53) and p16INK4a (also known as CDKN2A) tumour suppressors26, a major feature of senescent cells is the secretion of cytokines, growth factors and proteases6,7,9,10,14,27–33, termed the senescence-associated secretory phenotype8,9 (SASP).
The SASP is conserved between humans and mice, and includes inflammatory cytokines such as interleukin (IL) 6 and IL8 (otherwise known as CXCL8) (refs 6,8–10). The SASP can disrupt normal tissue structure and function and promote malignant phenotypes in nearby cells7,8,13,14,34. Further, senescent cells can promote tumour growth in mice8,13,14. As senescent cells increase with age35–37 and at sites of degenerative and hyperplastic pathology38–46, the SASP might contribute to inflammaging23,24,47. Further, DNA-damaging chemotherapies can induce senescence and a SASP in both normal and tumour cells, in culture and in vivo9,30,33,48–50, potentially fuelling the development of secondary cancers and some of the side effects of chemotherapy.
Our findings suggest that MTOR drives the SASP, and that rapamycin partly suppresses the SASP and its ability to promote tumour growth by targeting the upstream SASP regulator11, IL1A, for translational inhibition. Thus, tumours that are candidates for treatment with rapamycin derivatives (rapalogues) might less include those with a high proliferation index, but those with high DNA-damage signalling and inflammation, and rapalogues might ameliorate the inflammatory side effects of genotoxic chemotherapies.
RESULTS
Rapamycin decreases the SASP
To test the idea that MTOR activity and the SASP are linked, we exposed senescent human cells to the selective MTORC1 complex inhibitor rapamycin. We then evaluated secretion of the pro-inflammatory cytokine IL6, a major SASP component in mouse and human cells8,9, by enzyme-linked immunosorbent assay (ELISA).
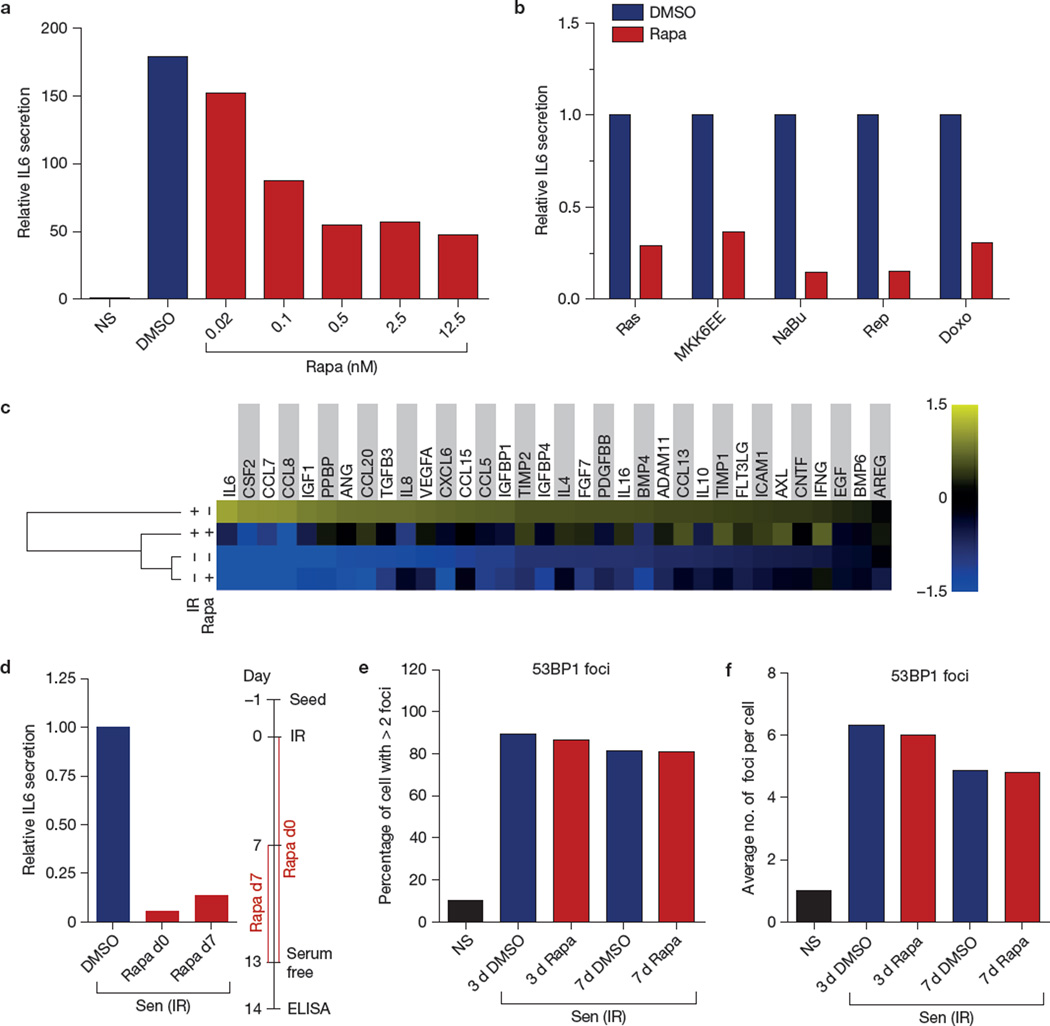
Rapamycin significantly decreased IL6 secretion by 3 strains of normal human fibroblasts (HCA2 neonatal foreskin, Fig. 1a; WI-38 fetal lung and PSC27 adult prostate, Supplementary Fig. 1A), and 2 strains of immortal, but non-tumorigenic human breast epithelial cell lines (MCF-10A and 184A1, Supplementary Fig. 1A)— all induced to senesce by ionizing radiation (10 Gy X-irradiation). This decrease was dose dependent, reaching maximal suppression of ~80% at <10 nM (Fig. 1a). We used 12.5 nM rapamycin for subsequent experiments.
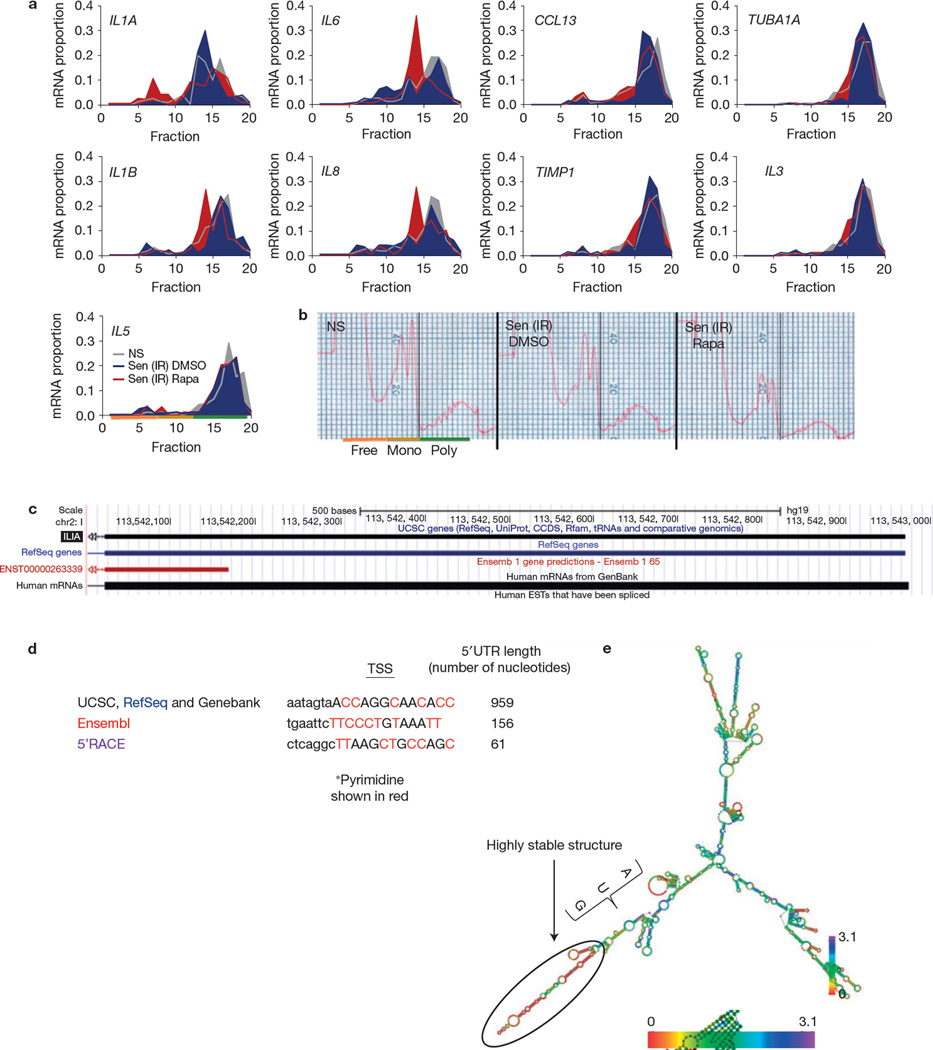
Figure 1.
Rapamycin decreases the SASP through MTORC1. (a) HCA2 fibroblasts, non-senescent (NS) or induced to senesce by ionizing radiation (IR; 10 Gy), were treated with the indicated concentrations of rapamycin (Rapa; or the highest concentration of DMSO as a control) immediately after ionizing radiation exposure. Conditioned media were collected 7 days later and analysed by ELISA for IL6. The level of IL6 secretion by non-senescent cells was arbitrarily set at 1. (b) HCA2 cells were induced to senesce by lentiviral-mediated overexpression of RAS or MKK6EE, 2 mM sodium butyrate (NaBu) for 3 days, replicative exhaustion (Rep), or 250 nM doxorubicin for 24 h (Doxo). The cells were given 12.5 nM rapamycin or DMSO (control) after treatment. Conditioned media were collected 7 days later and analysed by ELISA for IL6. (c) Conditioned media from non-senescent (NS) or senescent (ionizing radiation; Sen (IR)) cells treated with rapamycin or DMSO for 6 days were analysed by antibody arrays. The average signal from DMSO-treated cells was used as the baseline. Colour intensities represent log2-fold changes from the baseline. Signals higher than the baseline are shown in yellow; signals lower than the baseline are shown in blue. Shown is the average of three independent experiments. (d) Effects of two different rapamycin treatment regimens on IL6 secretion by senescent (ionizing radiation; Sen (IR)) HCA2 cells, one starting immediately after irradiation (d0) and continuing for 13 days, the other starting 7 days after irradiation and continuing for an additional 6 days. (e,f) Senescent (ionizing radiation; Sen (IR)) HCA2 cells were treated with DMSO or rapamycin and immunostained for the DNA-SCAR marker 53BP1. The number of 53BP1 foci was determined using CellProfiler. Shown is the percentage of cells with >2 53BP1 foci (e), and the average number of foci per cell (f). NS cells are shown for comparison. For all panels except c, shown is one representative of two independent experiments, each with triplicate samples. For raw data, see Supplementary Table 4.
Rapamycin also suppressed IL6 secretion by human fibroblasts induced to senesce by other stimuli (Fig. 1b): ectopic expression of oncogenic RASV12 (ref. 51), the stress-activated p38MAPK activator MKK6EE (ref. 15), sodium butyrate52, replicative exhaustion (see also Supplementary Fig. 1A for IMR-90 fetal lung fibroblasts) and doxorubicin. In non-senescent fibroblasts, rapamycin mildly suppressed the low basal secretion of IL6 (Supplementary Fig. 1B).
To determine whether rapamycin decreased the secretion of other SASP factors, we measured the levels of 120 secreted proteins using antibody arrays. Of the factors detected by the arrays and secreted at significantly higher levels by senescent, relative to proliferating or quiescent, cells, rapamycin reduced the secretion of 12/34 (35%), including several pro-inflammatory cytokines, chemokines and growth factors (Fig. 1c). Several SASP proteins were unaffected, indicating that rapamycin is a selective SASP modulator. Notably, all of the rapamycin-sensitive SASP factors were previously identified as targets of the NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) transcription factor15.
In culture, the SASP requires 4–5 days to develop once cells experience a senescence-inducing stimulus9,53. Rapamycin decreased IL6 secretion not only when added immediately after a synchronous senescence stimulus such as ionizing radiation (Fig. 1a), but also after the SASP fully developed (for example, 7 d after ionizing radiation-induced senescence; Fig. 1d). Thus, rapamycin suppressed both establishment and maintenance of the SASP.
Maintenance of the SASP requires persistent DNA-damage response (DDR) signalling, emanating from stable nuclear foci termed DNA-SCARS (DNA segments with chromatin alterations reinforcing senescence)53,54. To test the possibility that rapamycin inhibits the SASP by interfering with DDR signalling, we assessed its effect on persistent 53BP1 foci, a feature of DNA-SCARS (Fig. 1e,f). Rapamycin did not significantly alter the number of 53BP1 foci in senescent HCA2 (Fig. 1e,f) or PSC27 cells (Supplementary Fig. 1C,D), suggesting that it acts downstream of DDR signalling.
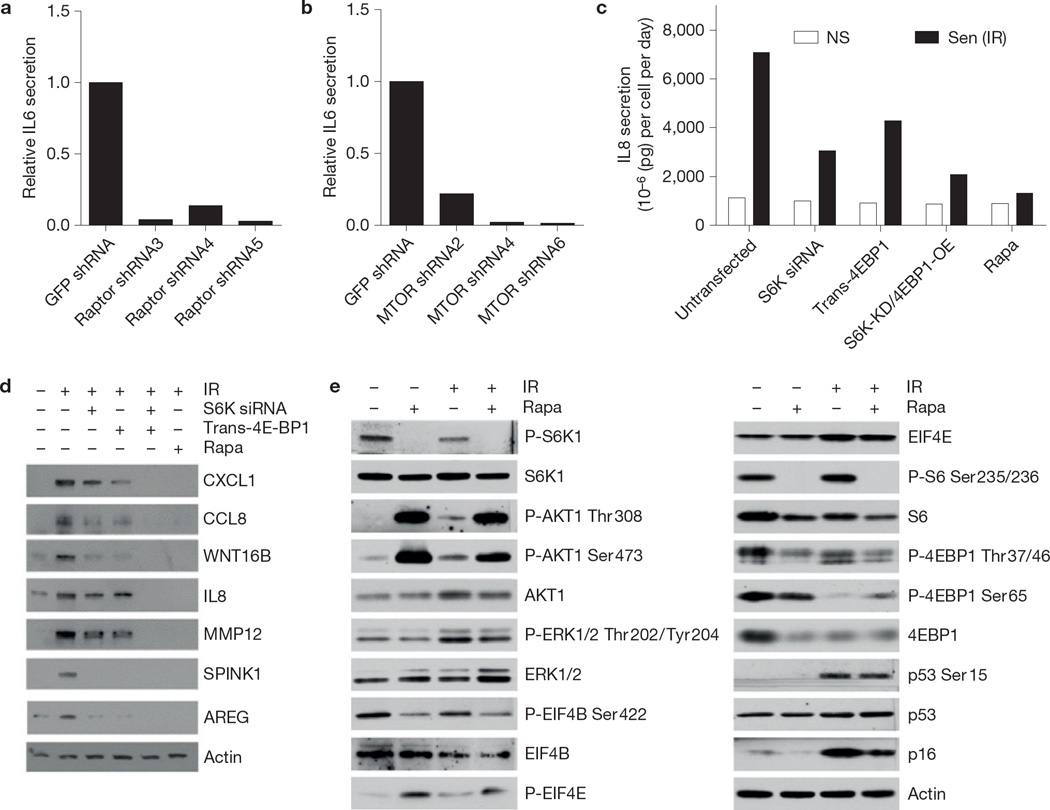
To confirm that the effects of rapamycin on the SASP are MTOR dependent, we used short hairpin RNA (shRNA) to deplete cells of the MTORC1 component raptor (otherwise known as RPTOR). shRNA against raptor, but not GFP (control; Supplementary Fig. 2A), severely blunted IL6 secretion by senescent cells (Fig. 2a). Likewise, shRNA-mediated depletion of MTOR reduced senescence-associated IL6 secretion by >90% (Fig. 2b and Supplementary Fig. 2B).
Figure 2.
SASP production is MTOR dependent. (a) HCA2 cells were infected with lentiviruses expressing GFP shRNA (control) or one of three different shRNAs targeting raptor. Cells were then irradiated; 7 days later, conditioned media were collected and analysed by ELISA for IL6. (b) HCA2 cells were infected with lentiviruses expressing GFP shRNA (control) or one of three different shRNAs targeting MTOR. Cells were then irradiated; 7 days later, conditioned media were collected and analysed by ELISA for IL6. (c) IL8 was quantified by ELISA in conditioned media from PSC27 cells in which S6K was depleted by siRNA or 4EBP1 overexpressed by transfection with a 4EBP1-encoding plasmid. Cells treated with rapamycin (Rapa) are shown for comparison. (d) After inducing senescence by ionizing radiation, protein extracts from PSC27 cells transfected with S6K siRNA or trans-4EBP1, or treated with rapamycin were analysed by western blotting for the indicated SASP factors. Unprocessed original scans of blots are shown in Supplementary Fig. 9. (e) Extracts from non-senescent (NS) or senescent (ionizing radiation; Sen (IR)) HCA2 cells, treated or not with rapamycin, were assayed for the indicated proteins by western blotting. β-actin served as the loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 9. For a–c, shown is one representative of two independent experiments, each with triplicate samples. For d and e shown is one representative of two independent biological replicates; each replicate required multiple blots, which were probed at the same time. For raw data, see Supplementary Table 4.
ELISAs confirmed that rapamycin suppressed the secretion of IL8, another prominent SASP component (Fig. 1c), by senescent cells (Fig. 2c). Likewise, shRNA-mediated depletion of the MTORC1 target S6K (also known as RPS6KB), phosphorylation of which stimulates translation, and overexpression of the MTORC1 target 4EBP1 (otherwise known as EIF4EBP1), phosphorylation of which alleviates translation inhibition, suppressed IL8 secretion (Fig. 2c). Additionally, western blot analysis showed that depletion of S6K1 or overexpression of 4EBP1 significantly decreased the expression of several SASP proteins (Fig. 2d). Rapamycin more effectively inhibits the S6K arm of the MTOR pathway compared with the 4EBP1 arm55; however, an additive effect of combining S6K depletion and 4EBP1 overexpression suggests that the 4EBP1 arm of MTOR could also be exploited to reduce the SASP (Fig. 2c). In agreement, the ATP-competitive MTOR inhibitor PP242 (ref. 56) suppressed the SASP more robustly than rapamycin (Supplementary Fig. 2C).
A positive role for S6K and 4EBP1 in driving the SASP suggested that the SASP might be under translational regulation at the level of mRNA structure. The EIF4A1 helicase is important for resolving stable secondary mRNA structures and is regulated by EIF4B and EIF4E, which in turn are regulated by phosphorylation57,58. Western blot analyses showed that rapamycin reduced EIF4B phosphorylation and increased EIF4E phosphorylation (Fig. 2e). Further, although ERK (otherwise known as MAPK1) and S6K are both upstream of EIF4B, rapamycin reduced only S6K phosphorylation. Rapamycin also partly reduced 4EBP1 phosphorylation, potentially sequestering EIF4E. Together, the results suggest that rapamycin impairs the helicase machinery in senescent cells, leading to reduced translation of mRNAs with stable secondary structures.
Rapamycin decreases NF-κB activity by decreasing IL1A production
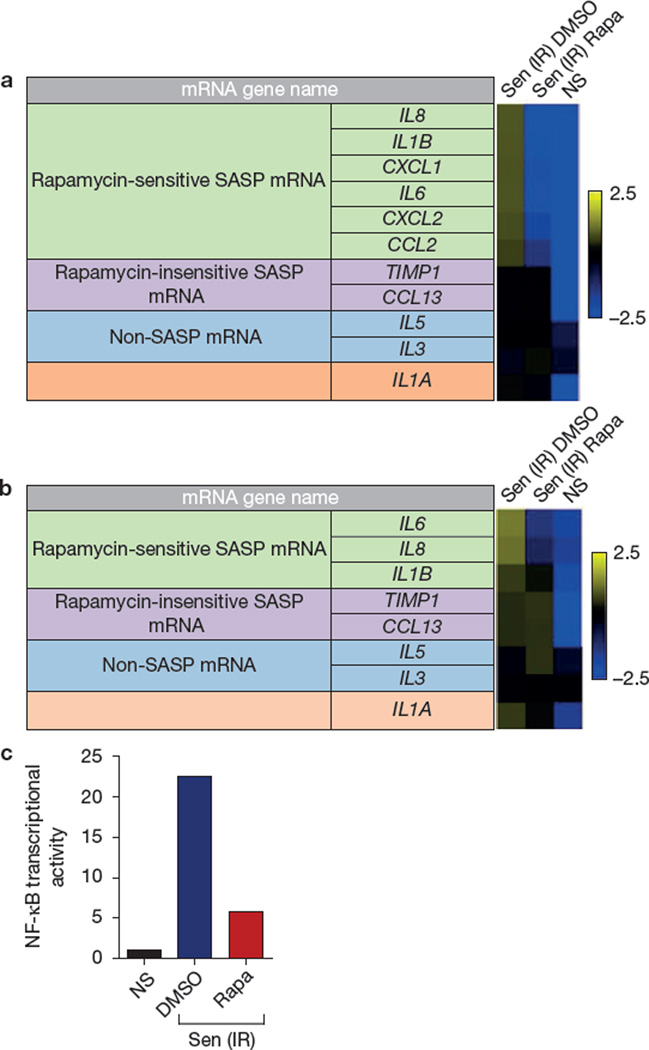
Although MTORC1 primarily regulates translation, most SASP proteins are upregulated at the level of mRNA abundance6–10. Surprisingly, quantitative PCR (qPCR) showed that rapamycin reduced the transcript levels of six rapamycin-sensitive SASP proteins in senescent HCA2 cells (Fig. 3a) and ten such transcripts in senescent PSC27 cells (Supplementary Fig. 3A). mRNAs encoding two rapamycin-insensitive and two non-SASP secreted proteins were unaffected, indicating that rapamycin does not indiscriminately reduce transcripts encoding secreted proteins. Importantly, rapamycin reduced mRNA levels as early as 2 d after ionizing radiation exposure (Fig. 3b), suggesting that it acts early in the development of the SASP.
Figure 3.
Rapamycin suppresses NF-κB transcriptional activity. (a) Transcripts for SASP factors secreted by DMSO- or rapamycin (Rapa)-treated senescent (ionizing radiation; Sen (IR)) HCA2 cells were quantified by qPCR 8 days after ionizing radiation exposure. Cells were incubated in serum-free media without rapamycin for the last 24 h. For the heatmap, the average signal from non-senescent (NS), senescent (ionizing radiation; Sen (IR)) DMSO-and rapamycin-treated HCA2 cells was used as the baseline (for numbers, see Supplementary Table 1). Colour intensities represent log2-fold changes from the baseline. Signals higher than the baseline are shown in yellow; signals lower than the baseline are shown in blue. (b) Transcripts for SASP factors from DMSO- or rapamycin-treated senescent (ionizing radiation; Sen (IR)) HCA2 cells were quantified by qPCR 2 days after ionizing radiation exposure. For the heatmap, the average signal from non-senescent, senescent DMSO-and rapamycin-treated cells was used as the baseline (for numbers, see Supplementary Table 2). Colour intensities represent log2-fold change from the baseline. Signals higher than the baseline are shown in yellow; signals lower than the baseline are shown in blue. (c) Cell extracts were prepared from non-senescent (NS) and senescent (ionizing radiation; Sen (IR)) HCA2 cells expressing an NF-κB–luciferase reporter construct. Cells were treated with DMSO (control) or rapamycin for 7 days, and analysed for luciferase activity as described previously11,15. Non-senescent luciferase activity was set at 1. For a and b, shown are the average of three independent experiments. For c, shown is one representative of two independent experiments, each with triplicate samples. For raw data, see Supplementary Table 4.
As NF-κB stimulates the transcription of many SASP genes6,11,15, we tested the possibility that rapamycin decreased NF-κB activity in senescent cells. We infected HCA2 cells with a lentivirus carrying an NF-κB–luciferase reporter, and measured the effect of rapamycin on reporter activity. Rapamycin reduced NF-κB activity by ~80% (Fig. 3c and Supplementary Fig. 3B). As rapamycin acts primarily to suppress translation through MTORC1 inhibition, these results suggest that rapamycin suppresses NF-κB activity and SASP mRNAs by an indirect mechanism.
Clues to how rapamycin might regulate SASP gene transcription came from noting a prominent exception to its effects on mRNA levels: the mRNA encoding the pro-inflammatory cytokine IL1A was only slightly affected by rapamycin (Fig. 3a and Supplementary Fig. 3A).
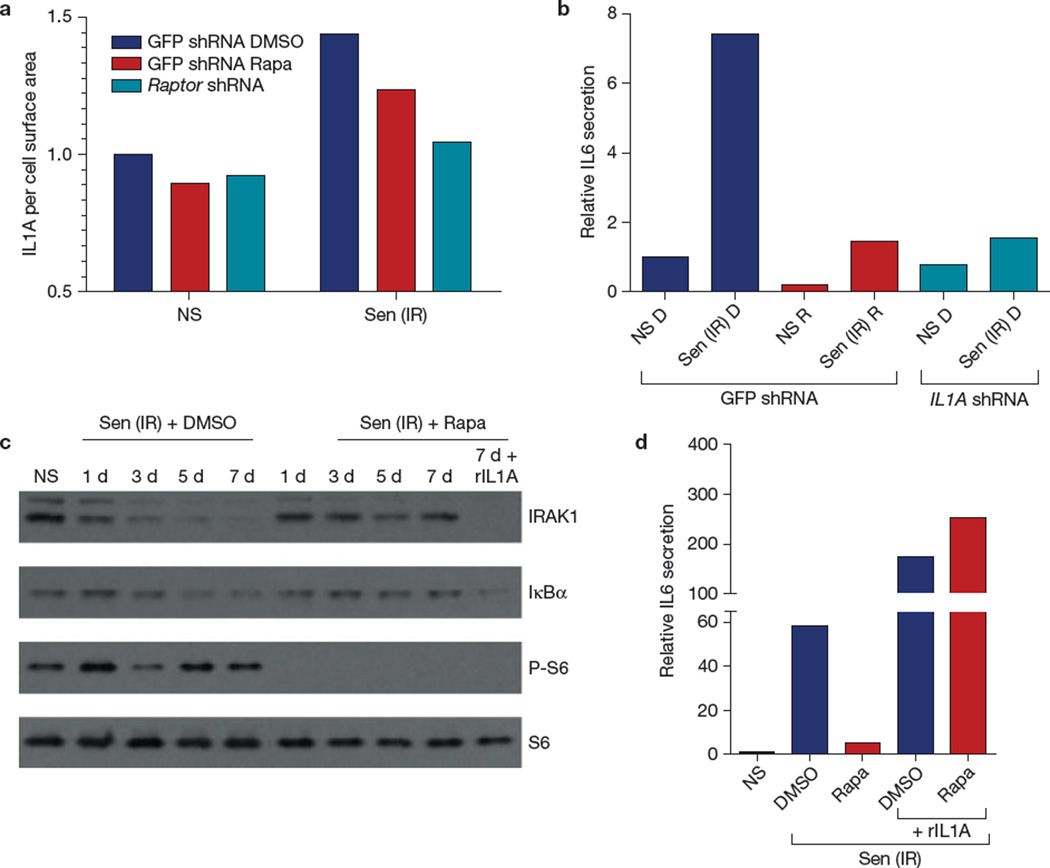
NF-κB and IL1A comprise a positive feedback loop that ultimately stimulates the transcription of several genes encoding inflammatory cytokines24. IL1A is cell surface bound and not secreted by senescent cells; however, its abundance on the senescent cell surface increases significantly, where it plays a key role in establishing and maintaining the SASP (ref. 11). Rapamycin, despite eliciting little change in IL1A transcript levels, significantly reduced IL1A protein levels on the surface of senescent cells (Fig. 4a and Supplementary Fig. 4A). Finally, shRNA-mediated depletion of IL1A in senescent cells suppressed IL6 secretion—similar to the suppression caused by rapamycin (Fig. 4b and Supplementary Fig. 4B). Thus, MTORC1 inhibition seemed to suppress the secretion of selected SASP components by interfering with the IL1A-NF-κB feedback loop.
Figure 4.
Rapamycin suppresses IL1A signalling. (a) HCA2 cells were infected with lentiviruses expressing shRNAs against GFP (control) or raptor. Senescent (ionizing radiation; Sen (IR)) cells, treated with rapamycin (Rapa) or DMSO for 10 days after ionizing radiation exposure, were analysed by flow cytometry for cell-surface IL1A using a FITC-tagged antibody. The fluorescence signal was divided by the forward scatter signals to account for cell size variations; 10,000 flow cytometry events were recorded. Shown is the result of one of two independent experiments. (b) HCA2 cells infected with lentiviruses expressing GFP shRNA or IL1A shRNA were irradiated and treated with DMSO (D) or rapamycin (R); 7 days later conditioned media were collected and analysed by ELISA for IL6. (c) Proteins were extracted from DMSO- and rapamycin-treated senescent cells and analysed by western blotting for IRAK1, IκBα, phospho-S6 and S6 at the indicated intervals after ionizing radiation exposure. Recombinant (r) IL1A protein was added to one senescent (ionizing radiation) sample treated with rapamycin (right lane). Unprocessed original scans of blots are shown in Supplementary Fig. 9. Shown is one of two independent biological replicates; each replicate required multiple blots, which were probed at the same time. (d) Non-senescent (NS) and senescent (ionizing radiation; Sen (IR)) HCA2 cells were treated with DMSO or rapamycin for 6 d, after which rIL1A in serum-free medium was added for 24 h. Conditioned media were collected and analysed by ELISA for IL6. For b and d, shown is one representative of two independent experiments, each with triplicate samples. For raw data, see Supplementary Table 4.
Consistent with this idea, rapamycin reduced IL1A signalling in senescent cells. IL1A binds its cell surface receptor (IL1R1) in a juxtacrine fashion, initiating a signalling cascade that ultimately degrades IRAK1 (interleukin-1 receptor-associated kinase 1) and IκBα (otherwise known as NFKBIA, nuclear factor of kappa light polypeptide gene enhancer of B-cells inhibitor alpha) to allow NF-κB nuclear translocation24. We analysed IRAK1 and IκBα protein levels in cells made senescent by ionizing radiation in the absence or presence of rapamycin. In the absence of rapamycin, IRAK1 and IκBα were reduced by ionizing radiation, indicating active IL1R1 signalling (Fig. 4c), as expected11. In the presence of rapamycin, IRAK1 and IκBα protein levels remained elevated (Fig. 4c), indicating a blockage of IL1R1 signalling. Addition of recombinant IL1A (rIL1A) rescued both IL1R1 signalling and IL6 secretion in the rapamycin-treated cells (Fig. 4c,d). Thus, rapamycin acts upstream of the IL1R1, and the signalling pathway downstream of IL1R1 remains intact. Further, phosphorylation of ribosomal protein S6, a substrate of S6K, remained low under rIL1A treatment, indicating that restoration of IL6 secretion by rIL1A was not due to reactivation of MTORC1 activity (Fig. 4c).
Rapamycin modulates the SASP by suppressing IL1A translation
As rapamycin decreased cell surface-bound IL1A levels and subsequent IL1R1 signalling, leading to decreased SASP gene transcription by NF-κB, IL1A might be a critical target for translational inhibition by rapamycin. To test this possibility, we determined mRNA translational efficiencies using polysome fractionation, which measures the fraction of an mRNA that is associated with polyribosomes. Rapamycin significantly decreased the translational efficiency of IL1A mRNA (Fig. 5a), and to a lesser extent IL1B, IL6 and IL8 mRNAs (Fig. 5a,b and Supplementary Fig. 5A,B), in senescent cells. In contrast, it had little effect on the translational efficiency of mRNAs encoding MCP-4 (monocyte chemotactic protein-4/CCL13), the secretion of which is not rapamycin sensitive (Fig. 1c), and α-tubulin (TUBA1A), a non-SASP gene (Fig. 5a). Thus, rapamycin decreased translational efficiency specifically.
Figure 5.
Rapamycin inhibits IL1A translation. (a) Senescent (ionizing radiation; Sen (IR)) HCA2 cells were treated for 7 days with rapamycin (Rapa) or DMSO followed by 1 day in serum-free media, after which cells were collected and mRNA was collected for polysome profiling as described in the Methods. qPCR was performed on each fraction for IL1A, IL6, CCL13, TUBA1A, IL1B, IL8, TIMP1, IL3 and IL5 mRNA (one representative experiment is shown). Fractions 1–7: free RNA; 8–12: 40–60S; 13–20: polysome. (b) Polysome profiles used to determine the translated fractions; representative polysome traces are shown. NS, non-senescent. (c) Using the UCSC genome browser sequence, analysis of the IL1A gene was performed to determine potential start sites of transcription. (d) Potential transcription start sites (TSSs) and their 5’UTRs are shown from various sources, including the determination based on 5’RACE (see c). Transcription start site sequences start at the capital letter and pyrimidines are shown in red. (e) IL1A mRNA minimum free energy structure was determined using the default parameter obtained from the University of Vienna RNAfold web service (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Scale represents the organizational entropy. The presence of a highly stable structure upstream of the start codon is detected. 5’RACE was performed once. For a and b, shown is one representative of two independent experiments, each with triplicate cell culture samples. For raw data, see Supplementary Table 4.
MTORC1 can regulate translation through terminal oligopyrimidine tracts in the 5’UTR. Bioinformatics, plus experimental 5’ rapid amplification of cloned/cDNA ends (RACE), identified the main transcription start sites—but no terminal oligopyrimidine sequence (Fig. 5c–e)—in the IL1A mRNA. However, structure predictions revealed a highly stable secondary structure downstream of the start codon, consistent with a strong requirement for helicase activity to efficiently translate this mRNA (Fig. 5e). This finding is surprising because translational regulation by helicase activity is generally specific for the 5’UTR (ref 57).
Rapamycin does not reverse cellular senescence
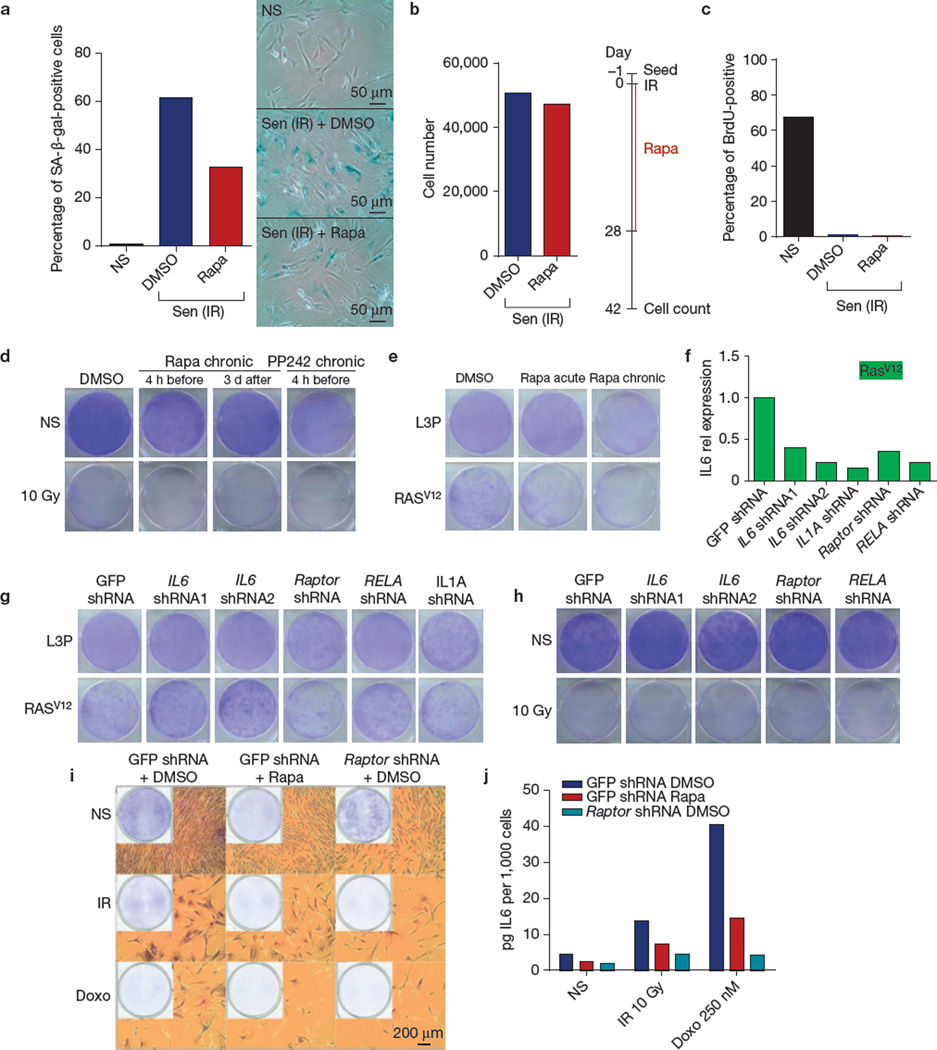
The rapamycin-sensitive SASP components IL6 and IL8 can reinforce the growth arrest of oncogene-induced senescent cells6,10, and rapamycin can suppress the senescence arrest in certain immortal cells59. In the primary human cells used here, rapamycin did not reverse the arrest caused by ionizing radiation, nor did it induce a senescence arrest in proliferating cells. It did decrease the percentage of cells that robustly expressed the senescence-associated β-galactosidase9,35 (SA-β-gal; Fig. 6a and Supplementary Fig. 6A), but cell numbers were unchanged when senescent cells were given rapamycin for 28 days and counted 2 weeks later (42 days after ionizing radiation exposure; Fig. 6b). Likewise, whether added before or after ionizing radiation exposure, rapamycin did not alter the low level of DNA synthesis or colony formation by senescent HCA2 or PSC27 cells (Fig. 6c,d and Supplementary Fig. 6B,C). Further, rapamycin did not prevent the arrest caused by oncogenic RAS (Fig. 6e).
Figure 6.
Rapamycin does not reverse cellular senescence. (a) Senescent (ionizing radiation; Sen (IR)) HCA2 cells were treated with DMSO or rapamycin (Rapa) for 6 days, and the percentage of cells expressing SA-β-gal was determined by light microscopy (right panel) and counting (left panel). (b) Cells were treated with rapamycin or DMSO for 28 days following ionizing radiation exposure, washed, and incubated in drug-free media for 14 days, at which point cell numbers were determined. (c) HCA2 cells, non-senescent (NS) or made senescent by ionizing radiation exposure and treated with DMSO or rapamycin for 6 days, were pulsed with BrdU for 24 h and the fraction that incorporated BrdU was determined by fluorescence microscopy. (d) Clonogenic assays comparing the effects of chronic treatment with rapamycin or PP242 (500 nM) of non-senescent (NS) and senescent (ionizing radiation; Sen (IR)) HCA2 cells. Cells were plated and drugs were added at the indicated times before or after ionizing radiation exposure and cells were cultured for 10–14 days (one representative experiment shown). (e) HCA2 cells were infected with a control lentivirus (L3P) or lentivirus carrying oncogenic RAS. Cell were grown in drug for three days (acute) and released or continuously treated (chronic) for 10–14 days after which clonogenic staining was performed. (f) HCA2 cells were co-infected with RAS and lentiviruses carrying shRNAs to deplete the indicated proteins. Transcripts for IL6 were quantified by qPCR. (g,h) HCA2 cells were infected with lentiviruses carrying shRNAs to deplete the indicated proteins. Clonogenic assays were performed on the infected cells induced to senesce by oncogenic RAS (g) or ionizing radiation (h) (one representative experiment is shown). (i) HCA2 cells were infected with a lentivirus carrying shRNAs against GFP (control) or raptor, induced or not (NS) to senesce by 10 Gy ionizing radiation (IR) or 250 nM doxorubicin (Doxo) for 24 h and treated with DMSO or rapamycin. Cells were cultured for 10–14 days and clonogenic staining was performed using crystal violet (one representative experiment is shown). (j) Cells were treated as in i. Seven days after senescence induction, conditioned medium was collected and analysed for IL6 by ELISA. For a–c, f and j shown is one representative of two independent experiments, each with triplicate cell culture samples. For d, e, g–i, shown is one representative clonogenic assay experiment replicated once. For raw data, see Supplementary Table 4.
Although shRNA-mediated depletion of IL6, RELA (a subunit of NF-κB), IL1A or raptor all reduced IL6 secretion (Fig. 6f), only IL6 depletion partially prevented the RAS- (but not ionizing radiation-) induced senescence growth arrest (Fig. 6g,h). Similarly, doxorubicin-induced senescence was not reversed by rapamycin or raptor depletion (Fig. 6i,j and Supplementary Fig. 6D). Thus, MTORC inhibition suppressed the expression and secretion of selected SASP factors, including IL6 and IL8, but did not interfere with the senescence growth arrest. Although IL6 and IL8 expression reinforce the oncogene-induced senescence arrest6,10, only IL6 depletion, not RELA, IL1A or raptor depletion, prevented the RAS-induced senescence arrest. The cause(s) for this phenomenon are unknown. We speculate that RELA, IL1A or raptor depletion not only suppresses IL6 expression, but also the expression of growth-stimulating factors, such as amphiregulin (AREG) and CXCL1, which are expressed and secreted by senescent cells7,9. This hypothesis has yet to be tested. Finally, rapamycin did not prevent the senescence growth arrest induced by 6 or 8 Gy ionizing radiation in HCA2 cells (Supplementary Fig. 6E). However, in BJ, another primary neonatal foreskin human fibroblast strain, a small proportion (<1%) of cells bypassed the senescence arrest at 5 Gy, indicating minor cell strain variation in the ability of rapamycin to prevent the senescence growth arrest (Supplementary Fig. 6F).
Sustained effect of rapamycin on the secretory phenotype
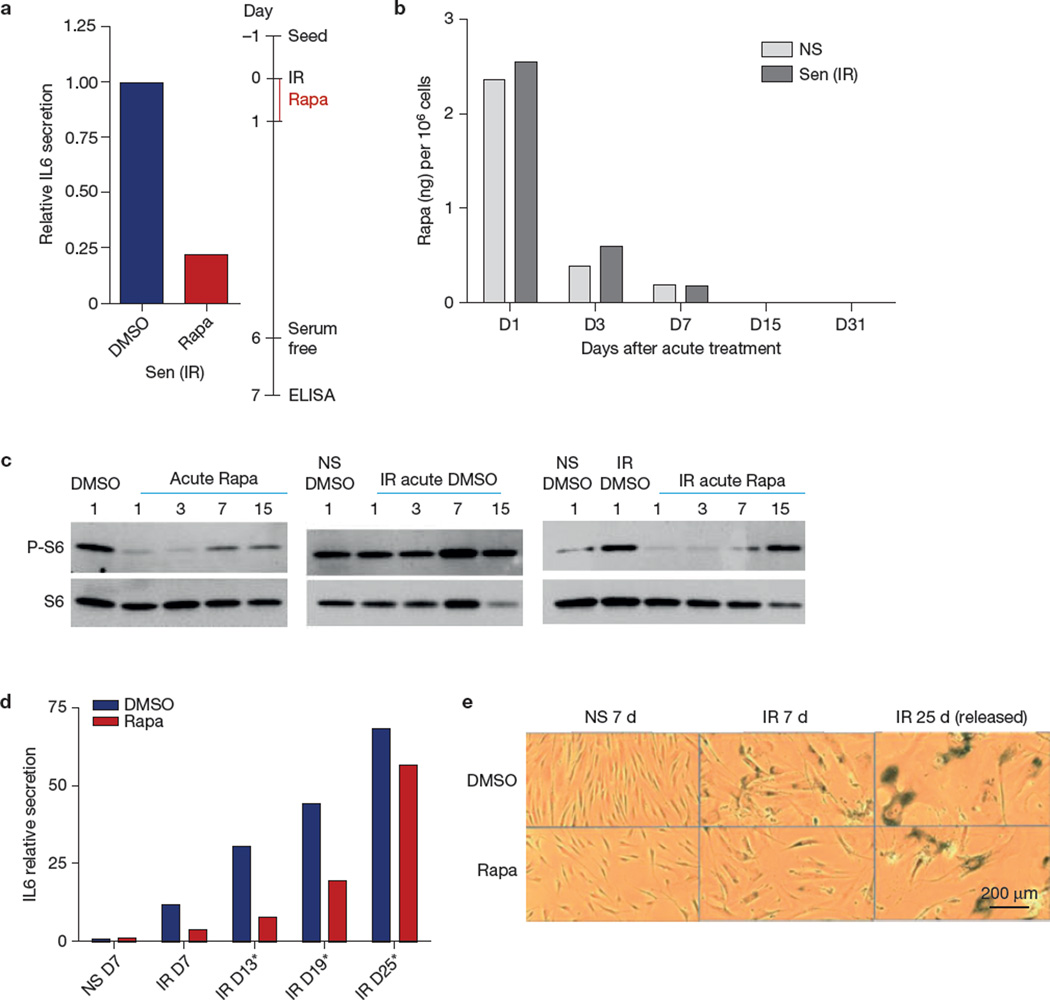
If rapamycin suppresses the SASP by preventing establishment of the IL1A–NF-κB feedback loop, transient treatment with rapamycin might suppress the SASP for an interval after removal of the drug. In culture, a single exposure of HCA2 cells to rapamycin for only 24 h immediately after ionizing radiation suppressed IL6 secretion by ~80% for 7 days thereafter (Fig. 7a), similar to the suppression seen after continuous treatment for 7 days (Fig. 1a). Although it is possible that the lingering effect of rapamycin after removal from the medium was due to intracellular retention, cell-associated rapamycin declined by ~80% and >90% 2 and 6 days, respectively, after removal (Fig. 7b). Interestingly, the MTOR pathway remained partly suppressed on the basis of S6 phosphorylation 2 and to a lesser extent 6 days after rapamycin removal (Fig. 7c). Eventually, however, the loop was re-established. Suppression of IL6 secretion and SA-β-gal activity slowly recovered after rapamycin withdrawal, reaching the level of untreated cells ~3 weeks after withdrawal (Fig. 7d,e).
Figure 7.
Sustained effect of rapamycin on the SASP. (a) HCA2 cells were treated with rapamycin (Rapa) for 1 day immediately after ionizing radiation exposure, and conditioned medium was collected 6 days later and analysed for IL6 secretion by ELISA; shown is one representative of two independent experiments, each with triplicate cell culture samples. (b) The amount of rapamycin was measured in non-senescent (NS) and senescent (ionizing radiation; Sen (IR)) HCA2 cells 1, 3, 7, 15 and 31 days after acute treatment (single 1-day dose immediately after ionizing radiation exposure). Rapamycin was measured by high-performance liquid chromatography at the Biological Psychiatry Laboratories Services of the University of Texas Health Sciences Center San Antonio. Six samples were collected and pooled in groups of two for each condition. (c) Western blot analysis of phospho-S6 and S6 using proteins extracted 1, 3, 7 and 15 days after acute treatment (as in a) of HCA2 cells with rapamycin (one representative or two representative experiments is shown). Unprocessed original scans of blots are shown in Supplementary Fig. 9. (d) IL6 secretion by X-irradiated (IR) senescent HCA2 cells at day 7, 13, 19 and 25 after rapamycin treatment, relative to non-senescent (NS) secretion; shown is one representative of two independent experiments, each with triplicate cell culture samples. (e) SA-β-gal activity in non-senescent and senescent cells treated with DMSO and rapamycin was determined by light microscopy 7 or 25 days after treatment (one representative of two representative independent experiments is shown). For raw data, see Supplementary Table 4.
We propose that rapamycin selectively targets the SASP-initiating cytokine IL1A through translational inhibition, thereby suppressing establishment of the signalling cascade that activates NF-κB and the transcription of many genes encoding SASP factors (Supplementary Fig. 7).
Rapamycin represses the ability of senescent cells to stimulate cell proliferation and tumorigenesis in vivo
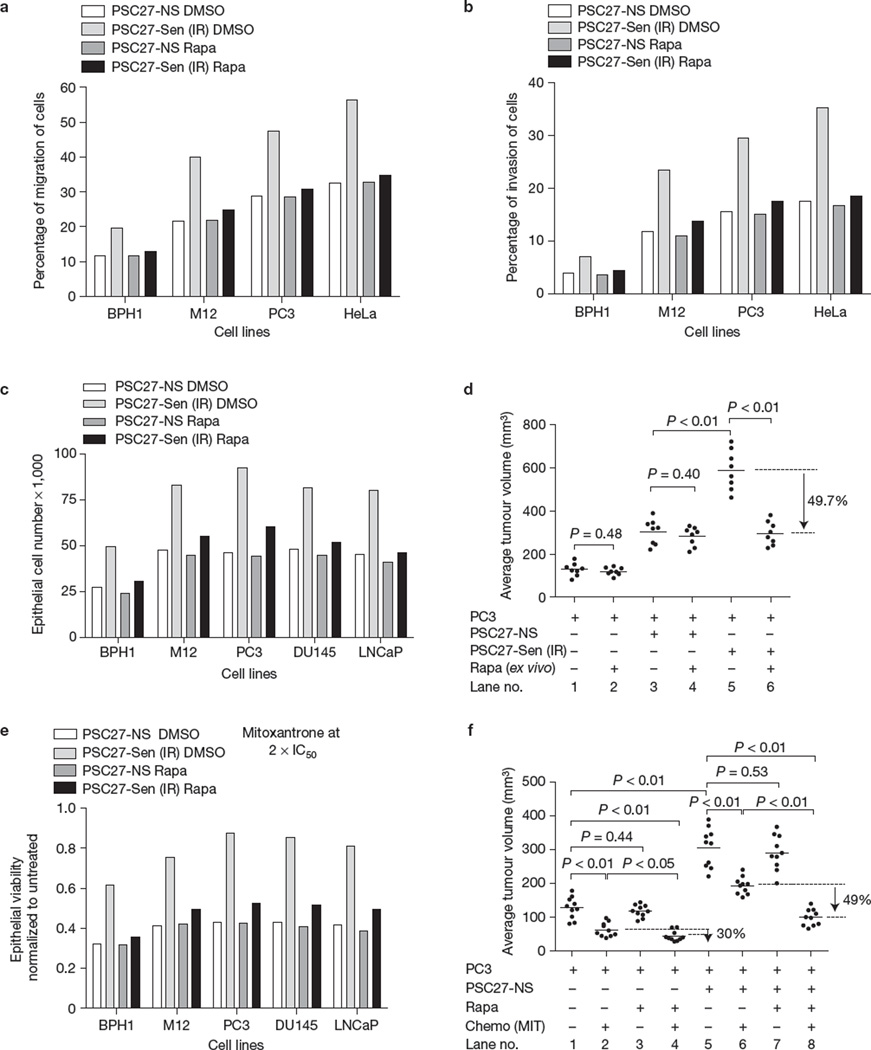
SASP factors can promote cancer cell proliferation and other malignant phenotypes in culture7–9,13. Notably, rapamycin substantially reduced the stimulation of proliferation, migration and invasion effects of conditioned media from senescent PSC27 cells on several immortalized, transformed or metastatic prostate cancer cells (Fig. 8a – c and Supplementary Fig. 8A). Further, senescent fibroblasts can promote tumour formation by premalignant epithelial cells and can stimulate tumour growth by malignant epithelial cells in mouse xenografts8,13,14. To determine the contribution of MTOR towards the tumour-promoting effects of SASPs in vivo, we grafted PC3 prostate tumour cells with or without PSC27 fibroblasts that were induced to senesce by ionizing radiation in culture. The tumour cells and fibroblasts were also treated separately, in culture, with rapamycin or vehicle for 8 days before grafting. Eight weeks after subcutaneous implantation, grafts comprised of PC3 cells alone averaged 129 mm3 and tumour growth was not significantly diminished by rapamycin exposure (Fig. 8d; compare lanes 1 and 2). Grafts comprised of tumour cells and irradiated PSC27 fibroblasts were substantially larger and averaged 589 mm3 (Fig. 8d, lanes 1 versus 5), whereas grafts comprised of tumour cells and irradiated fibroblasts co-treated with rapamycin ex vivo before implantation averaged 296 mm3 (49.7% smaller; P <0.01; Fig. 8d, lanes 5 versus 6; Supplementary Fig. 8B,C).
Figure 8.
Rapamycin suppresses the tumour-promoting activity of senescent cells. (a,b) Percentage of BPH1, M12 and PC3 prostate cancer cell migration (a) and invasion (b) was determined on co-culture with conditioned media from irradiated PSC27 fibroblasts that were treated with rapamycin (Rapa) or vehicle (control). The human cervical cancer line HeLa was used as a positive control in both experiments. (c) The indicated prostate epithelial cells were incubated with conditioned media from PSC27 human prostate fibroblasts, non-senescent (NS) or induced to senesce by irradiation (Sen (IR)), untreated or treated with rapamycin. Epithelial cell numbers were determined 3 days later. (d) PC3 prostate cancer cells were implanted subcutaneously with non-senescent (PSC27-NS) or senescent (ionizing radiation; PSC27-Sen (IR)) prostate fibroblasts that had been pretreated for 8 days in culture with DMSO or rapamycin. Tumour volumes were determined as described in the Methods (mean ± s.e.m., n = 8 animals). (e) Prostate cancer cell viability was determined by MTT assays following exposure to mitoxantrone for 72 h at twice the IC50 dose for each cell line in the presence of conditioned media from non-senescent (PSC27-NS) or senescent (PSC27-Sen (IR)) PSC27 fibroblasts that were co-treated with rapamycin or vehicle. (f) In vivo effect of rapamycin on chemotherapy resistance was determined by injecting PC3 cancer cells with or without PSC27 prostate fibroblasts into SCID mice followed by treatment with mitoxantrone or vehicle and co-administration of rapamycin or vehicle. Tumour volumes were determined after an 8-week treatment period (mean ± s.e.m., n = 10 animals). For a–c and e shown is one representative of three independent experiments, each with triplicate cell culture samples. For d and f, a standard t-test served to determine P values. For raw data, see Supplementary Table 4.
The SASP can confer resistance and tumour cell survival following genotoxic chemotherapy60. To determine whether rapamycin suppresses this SASP effect, we measured the viability of prostate tumour cells after treatment with the chemotherapeutic agent mitoxantrone. We cultured the mitoxantrone-treated tumour cells with conditioned media from PSC27 prostate fibroblasts induced to senesce by ionizing radiation in the presence or absence of rapamycin. Conditioned media from senescent fibroblasts substantially attenuated the loss of prostate cancer cell viability caused by mitoxantrone, and this chemoprotection was abolished when the tumour cells were exposed to the conditioned media from senescent fibroblasts that had been treated with rapamycin (Fig. 8e and Supplementary Fig. 8A).
Importantly, rapamycin conferred chemoresistance in vivo. We injected PC3 cells subcutaneously into SCID mice, with or without senescent (ionizing radiation-induced) PSC27 fibroblasts. In contrast to the experiment described in Fig. 8d, we treated the animals, not isolated cells, with mitoxantrone every two weeks three times, with or without rapamycin given every other day. Compared with mitoxantrone alone, rapamycin, administered at a dose that had no single-agent effect, reduced the growth of PC3 xenografts by 30% (Fig. 8f, lanes 2 versus 4). This result could be attributable to direct tumour cell effects, and/or modulation of microenvironmental factors, including the SASP (Fig. 8f and Supplementary Fig. 8D–F). Importantly, tumours that arose from PC3 cells and co-injected PSC27 fibroblasts were substantially larger (compare lanes 1 and 5), and co-administration of rapamycin and mitoxantrone reduced tumour growth by 49% compared with mitoxantrone alone (P < 0.01; Fig. 8f, lanes 6 versus 8; Supplementary Fig. 8D–F).
DISCUSSION
Our findings suggest that rapamycin might postpone ageing and extend lifespan, including suppression of cancer progression by a mechanism that involves selective inhibition of the SASP. The SASP is a strong candidate for contributing to the sterile inflammation that is a hallmark of ageing and many age-related pathologies. In addition, senescent cells accumulate after genotoxic anticancer therapeutics9,49,50,61, and thus may cause or contribute to the treatment resistance and cancer recurrence that frequently follow such therapies. Our discovery that the SASP is regulated by the MTOR pathway, and is suppressed by rapamycin, provides a potential basis for developing rational strategies to reduce the deleterious effects of genotoxic exposures and normal ageing processes (Supplementary Fig. 8G). Further, our data suggest that SASP-reducing interventions need not entail continuous treatment; rather, intermittent treatments may provide substantial benefits.
MTOR inhibitors are in clinical use to prevent organ transplant rejection and restenosis, and as promising anticancer therapies62–64. MTOR inhibitors are thought to act by preventing TOR-driven proliferative responses to nutrients and mitogens. Our data suggest another possibility, at least in mammals: MTOR inhibitors might reduce inflammation caused by senescent cells and the SASP. The SASP includes potent inflammatory cytokines such as IL6, IL8 and monocyte chemoattractant proteins24,47, which can alter tissue environments31,34 and also attract innate immune cells65. It is noteworthy that MTOR inhibition has been shown to extend longevity in species that do not spontaneously develop cancer (such as nematodes), as well as those that do (such as mice)66.
Although rapamycin and other MTOR inhibitors are in clinical use as immunosuppressants (to reduce T-cell maturation) in organ transplant patients, they were recently used in advanced trials as anticancer therapies62–64,67. The immunosuppressive effects of rapamycin might be limited to certain physiological states because it can enhance the ability of mice to survive certain infections68. Thus, immune cells and senescent cells might react differently to MTOR inhibition with regard to the secretion of cytokines and other factors.
In summary, our findings identify suppression of the SASP as a mechanism by which rapamycin might suppress age-related pathologies, including cancer. Understanding how the SASP is controlled and how it might be prevented could provide a basis for developing rational strategies to reduce the deleterious effects of exposure to genotoxins and during the normal process of ageing.
METHODS
Cells and cell culture
Fibroblasts were cultured in 3% oxygen in DMEM and 10% fetal bovine serum (FBS), except for PSC27, which were cultured in PSC complete medium (80% MCDB131 (Invitrogen) supplemented with 10% FCS, non-essential amino acids, insulin, dexamethasone, transferrin, selenium and 20% AmnioMax (Life Technologies)) as described previously7,9. The human prostate carcinoma cell lines DU145, PC3 and LNCaP are tumorigenic prostate cancer cell lines derived from prostate cancer metastasis; the cells were obtained from the American Type Culture Collection (ATTC) and cultured in RPMI 1640 and 5% FBS. BPH1 cells are a non-tumorigenic epithelial cell line derived from non-malignant prostatic tissue with benign hyperplasia, immortalized by SV40-LT antigen, and were cultured in DMEM and 10% FBS as described previously69. M12 human prostate cancer cells were derived from a benign prostate epithelium, immortalized with SV40 T antigen and passaged in vivo to generate a metastatic subline designated M12. M12 cells were cultured in RPMI 1640 supplemented with epidermal growth factor (EGF), dexamethasone, insulin, transferrin and selenium as described previously70. Immortalized breast epithelial cells MCF-10A were obtained from the ATTC and cultured in DMEM and 10% FBS, and immortalized breast epithelial cells 184A1 were obtained from M. Stampfer (Lawrence Berkeley National Laboratory, USA) and cultured in DMEM and 10% FBS (refs 13,70). All cells were mycoplasma-free before and during performance of the experiments, as determined by the MycoAlert kit from Lonza.
Reagents
siRNAs that target S6K and lentiviruses encoding shRNAs that target GFP, MTOR, raptor and IL1A were obtained from Open Biosystems, as described in Supplementary Table 1. The 4EBP1 construct was obtained from GeneCopoeia (pReceiverM68-4EBP1). The antibodies used for western blotting are described in Supplementary Table 4. The anti-53BP1 antibody used for immunofluorescence of HCA2 cells was from Bethyl Laboratories (A300-272A) and was used at 1:500 dilution; the anti-53BP1 antibody used for immunofluorescence of prostate fibroblasts was from Abcam (ab36823) and was used at 1:500 dilution. The anti-phospho H2AX (S139) antibody used for prostate fibroblasts was from Abcam (ab2893) and was used at 1:1,000 dilution. Recombinant IL1A (200-LA) was obtained from R&D.
Senescence induction and assessment
Unless noted otherwise, cells were induced to senesce by exposure to 10 Gy ionizing radiation in the form of X-rays or gamma rays as described previously8,9,15,53,54. Briefly, proliferating cells were irradiated for an interval needed to deliver 10 Gy ionizing radiation; control cells (mock irradiated) were placed in the irradiator for an identical interval but remained unirradiated. Alternatively, cells were induced to senesce by replicative exhaustion, expression of oncogenic RAS or MKK6EE delivered by lentiviruses, or incubation with sodium dibutyrate (2 mM) for 1 day or doxorubicin (250 nM) for 3 days. Seven to ten days later, cells were scored for senescence markers, including growth status (clonogenic assays or BrdU incorporation), SA-β-gal activity and the presence of persistent DNA-damage foci. Clonogenic assays were performed as described in the figure legends. BrdU incorporation was assessed using a commercial kit (BD Biosciences). SA-β-gal staining was performed as described previously9 using a commercial kit (BioVision). DNA-damage foci were assessed by immunostaining for 53BP1 or γH2AX foci, as described above. For DNA-damage foci and SAβ-gal positivity, random fields are shown. BrdU incorporation and DNA-damage foci were quantified using CellProfiler, an open source software program (http://www.cellprofiler.org). SA-β-gal staining was quantified by light microscopy and a researcher that was blinded to the treatments.
RNA isolation and PCR
Cell cultures were homogenized in Trizol (Invitrogen) and total RNA was isolated using the RNeasy Kit (Qiagen). The RNA was quantified and amplified using the MessageAmp RNA amplification kit (Ambion). For prostate fibroblasts, cDNA was generated by reverse transcription, and quantitative PCR analyses were performed in triplicate using an Applied Biosystems 7700 sequence detector with ~5 ng of cDNA, 1 µM designated primer pairs and SYBR Green PCR master mix (Applied Biosystems). Alternatively, for all other fibroblasts, cDNA was generated from total RNA using the Cells-to-CT kit (Ambion), and real-time quantitative PCR was performed also in triplicate using the Roche Universal Probe Library (UPL), both according to the manufacturer’s instructions. All primer sequences or primer-probe combinations for the UPL are listed in Supplementary Table 3. The mean cycle threshold (Ct) for each gene was normalized to levels of RPL13A or tubulin in the same sample (delta Ct). Unpaired two-sample t-tests were used to determine differences in mean delta Ct values between treatment groups. The fold change was calculated by the delta-delta Ct method (fold = 2ddCt).
Western blot analyses and immunofluorescence
Western blotting and immunofluorescence were performed as described previously7,9,53,54. Briefly, for western blot analyses, cultured cells were lysed in SDS sample buffer (50 mM Tris HCl, pH6.8, 100 mM dithiothreitol, 2% SDS, 10% glycerol), and proteins were separated on 4–12% precast polyacrylamide gels (NuPAGE; Life Technologies) by electrophoresis and transferred to nitrocellulose membranes. The membranes were blocked using 5% non-fat milk in PBS-T, and then incubated with the primary antibodies described in Supplementary Table 2. After washing, the membranes were incubated with the secondary antibodies and washed again. Signals were detected using an enhanced chemiluminescence detection kit (Pierce or GE Healthcare), according to the supplier’s instructions. For immunofluorescence, cells were cultured and treated in chamber slides, fixed in neutral 10% formalin, and permeabilized with PBS containing 0.5% Triton X-100. After washing, the slides were blocked using PBS containing 1% BSA and 4% serum from a species that corresponded to the secondary antibody used for detection. After washing, the slides were incubated with primary antibodies, washed again, incubated with secondary antibodies and mounted with slow-fade gold (Molecular Probes) containing DAPI (to visualize nuclei). Cells were viewed by fluorescence microscopy and images were acquired for analysis using Spotfire software (Diagnostics Instruments).
NF-κB transcription assays
NF-κB transcription activity was determined using a reporter construct containing the firefly luciferase gene under the control of NF-κB-binding sites as described previously11,15. Briefly, the reporter construct was introduced into cells along with a control vector containing Renilla luciferase under the control of a CMV promoter, both using lentiviral vectors. After selection for viral infection, the cells were either maintained as non-senescent or were induced to senesce by ionizing radiation; the cells were then treated with vehicle or rapamycin as described in the text. Seven days after treatment, the cells were lysed and the lysates were analysed for luciferase activity using a luminometer and a commercial kit (Promega) and substrates for either firefly or Renilla luciferase, according to the supplier’s instructions. Firefly luciferase activity was normalized to cell number or Renilla luciferase activity.
Antibody arrays
Conditioned media for antibody array analyses were prepared by washing approximately 2 × 106 cells 2 to 3 times with PBS, and incubating them in serum-free medium for 24 h. The conditioned media were collected in a centrifuge tube, and the cells remaining on the dish were counted to normalize conditioned medium volumes for cell number. The conditioned media were clarified by brief centrifugation, diluted with serum-free medium to a concentration equivalent to 1.5 × 106 cells per 1.2 ml, and applied to the antibody arrays (Raybiotech; AAH-CYT-G1000-8) as described previously9,15 and as recommended by the supplier. The signals were detected using a GenePix 4200A Professional microarray scanner and were analysed using LI-COR Odyssey software. Signals were averaged and displayed as described in the figure legend.
ELISAs
Conditioned media were prepared by incubating cells for 24 h in serum-free medium as described above; cells remaining on the dish were counted for normalization purposes. The conditioned media were diluted to contain the equivalent of equal numbers of cells and then analysed using the AlphaLISA IL-6 Immunoassay kit and reagents, following the procedures described by the manufacturer (Perkin Elmer; AL223F).
Detection of cell surface-bound IL1A
Cell surface-bound IL1A was detected using a FITC-conjugated IL1A antibody (FAB200F from R&D) and analysed by flow cytometry using a FACSaria flow cytometer (BD) and software as described previously11. Briefly, 2–3 × 105 cells were washed with PBS, detached from the culture dish using EDTA/PBS (without trypsin), and collected by gentle centrifugation. The cell pellets were washed with cold PBS containing 0.5% BSA, blocked with PBS containing human nonspecific IgG and incubated in the dark and cold with the FITC-conjugated antibody (10 µl per 105 cells). After washing twice with PBS, the cells were suspended in PBS for flow cytometry analysis.
Polysome analyses
Before collection, cells were incubated for 20 min in serum-free DMEM containing rapamycin or dimethylsulphoxide (DMSO) supplemented with 50 µg ml−1 cycloheximide. The cells were then lysed in solubilization buffer (300 mM NaCl, 50 mM Tris HCl pH 8.0, 10 mM MgCl2, 1 mM EGTA, 200 µg ml−1 heparin, 400 U ml−1 RNasin, 1 mM phenylmethylsulphonyl fluoride, 0.2 mg ml−1 cycloheximide, 1% Triton X-100), collected by scraping into 1.5 ml centrifuge tubes and placed on ice. The lysates were clarified by centrifugation at 16,000g for 10 min, and loaded onto sucrose gradients. For each sample, lysates from 3 culture plates, each containing approximately 5 × 106, were pooled. The gradients were assembled in ultracentrifuge tubes (Beckman) and consisted of sucrose dissolved in high-salt resolving buffer (HSRB: 140 mM NaCl, 25 mM Tris HCl pH 8.0, 10 mM MgCl2) and five successive layers, each containing 1.67 ml HSRB plus sucrose at 50%, 40%, 30%, 20% and 10% w/v concentrations, respectively. The gradients were sealed with Parafilm, incubated at room temperature for 2 h, and stored at −80 °C overnight. The gradients were thawed at 4 °C for 1 h before loading. Cell lysates were loaded onto the gradients, spun at 245,000g for 2 h at 4 °C and stored on ice. After centrifugation, 20 fractions, each containing 0.55 ml and 1.5 ml TRI LS reagent (Sigma), were collected and frozen in liquid nitrogen. Optical density (OD) at 254 nm was monitored to verify gradient quality. Representative examples of the OD traces are shown in the figures.
RNA purification from polysomes and subsequent qPCR analyses
Gradient fractions were thawed on ice, and supplemented with 200 ng of luciferase mRNA (Promega) to control for the efficiency of RNA recovery. Each fraction was extracted with 200 µl of 1-bromo-3-chloropropane. One millilitre of the aqueous phase was transferred to a tube containing 25 µg of glycogen (Ambion), and precipitated using 1 ml of isopropanol. After collecting the precipitate by centrifugation, the pellet was washed in 75% ethanol. The pellets were suspended in 200 µl of water, purified using RNEasy MinElute columns (Qiagen), and suspended in 50–100 µl of RNAse-free water. qPCR reactions were performed using the Roche Universal Probe Library as described above. To control for RNA recovery, Ct values were corrected against the Ct value for luciferase mRNA. For each gene, the transcript levels present in each fraction were further normalized against the total signal from all fractions from the same gradient, and expressed as percentage of total signal. On the basis of the positions of the peaks on the OD (254 nm) traces, fractions 1–12 were designated non-translated and fractions 13–20 were designated translated.
Tumour cell migration, invasion, proliferation and tumorigenicity
Tumour cell migration and invasiveness were assessed in culture using Transwells (Cultrex 24-well Cell Migration Assay plates) containing a porous (8 µm pore size) membrane uncoated (for the migration assay) or coated (for the invasion assay) with a 0.5X solution of basement membrane extract and the indicated conditioned media in the bottom portion of the well. After 24 h, migrating or invading cells on the bottom side of the porous membranes were stained and quantified by absorbance as recommended by the supplier. Epithelial cell proliferation was determined using the MTT assay. For tumorigenicity assays in mice, cell preparations and assessments of tumour volumes were performed as described previously8,13. Briefly, we prepared 106 fibroblasts, irradiated (10 Gy) or mock irradiated, and treated them with DMSO or rapamycin for 8 d. We combined these fibroblasts with 106 tumour cells and injected the suspension of mixed cells subcutaneously into 12-week-old male SCID mice obtained from Taconic. Xenograft growth was assessed at weekly intervals by ultrasound. The animals were euthanized 8 weeks later. On resection, the tumour volumes (V) were again measured as described above. Alternatively, tumour cells were injected into animals, which were treated or not with mitoxantrone or rapamycin as described in the text and figure legends, and tumour volumes were assessed as described above. The Institutional Animal Care and Use Committee (IACUC) of the Buck Institute for Research on Aging and the Fred Hutchinson Cancer Research Center reviewed and approved all of the animal protocols and procedures.
Statistical analyses
All data that show error bars are presented as mean ± s.e.m. The significance of difference in the mean values was determined using two-tailed Student’s t test unless otherwise mentioned, and normal distribution was assumed for all of these. P < 0.05 was considered significant. All calculations were performed using GraphPad Prism software. Every cell culture experiment was reproduced at least twice independently. For each experiment, the number of samples and replicates is indicated in the figure legends. No randomization or blinding was used for the animal treatment experiments and subsequent data collection. Eight to ten mice were used per condition for each in vivo experiment; the experiments were done once. SA-β-gal quantification was performed blinded. No animals were excluded from analysis. Sample sizes were selected on the basis of previous experiments; no statistical method was used to predetermine sample sizes. The experiments were not randomized, and investigators were not blinded during experiments and outcome assessments, unless noted otherwise. All in vivo experiments were conducted on animals of the same age and gender and therefore equal variance was assumed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Rogers, B. Kennedy, G. Lithgow, S. Melov and members of the Campisi laboratory (Buck Institute) for comments and discussions, I. Coleman for assistance with figure preparation and data analysis, and R. Strong (U Texas Health Science Center) for providing the rapamycin encapsulated chow. This work was supported by grants from the National Institutes of Health (NIH) AG045288 to R.-M.L., Hillblom Medical Foundation (CP), NIH T32 training grant AG000266-16 to K.A.W.-E., NIH grants CA143858, CA155679 and CA071468 to C.C.B., NIH grants AG032113 and AG025901 to P.K., DOD-PCRP grant PC111703 and National Natural Science Foundation of China 81472709 to Y.S., NIH grants CA164188, CA165573 and CA097186 and the Prostate Cancer Foundation to P.S.N., and NIH grants AG09909 and AG017242 to J.C. and AG041122 to J.C. and Y.I.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
R.-M.L., Y.S., A.V.O., C.K.P., A.F., L.Z., S.C.C., A.R.D., K.A.W.-E., S.L., C.L., M.D. and P.L. acquired and interpreted the data. M.J. analysed rapamycin concentrations. G.B.H. and Y.I. provided pathological assessment. R.-M.L., Y.S., C.C.B., P.-Y.D., P.K., P.S.N. and J.C. designed experiments and interpreted data. R.-M.L., Y.S., P.-Y.D., P.K., P.S.N. and J.C. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapahi P, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim. Biophys. Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campisi J, d’Adda di F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 6.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 7.Bavik C, et al. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 2006;66:794–802. doi: 10.1158/0008-5472.CAN-05-1716. [DOI] [PubMed] [Google Scholar]
- 8.Coppe JP, et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS ONE. 2010;5:e9188. doi: 10.1371/journal.pone.0009188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 11.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1α is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc. Natl Acad. Sci. USA. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krtolica A, Parrinello S, Lockett S, Desprez P, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl Acad. Sci. USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 15.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasto S, et al. Inflammation, ageing and cancer. Mech. Ageing Dev. 2009;130:40–45. doi: 10.1016/j.mad.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 17.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 18.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franceschi C, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 22.McElhaney JE, Effros RB. Immunosenescence: what does it mean to health outcomes in older adults? Curr. Opin. Immunol. 2009;21:418–424. doi: 10.1016/j.coi.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freund A, Orjalo A, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16:238–248. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 26.Beausejour CM, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binet R, et al. WNT16B is a new marker of cellular senescence that regulates p53 activity and the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2009;69:9183–9191. doi: 10.1158/0008-5472.CAN-09-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang BD, et al. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc. Natl Acad. Sci. USA. 2002;99:389–394. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppe JP, Kauser K, Campisi J, Beausejour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 2006;281:29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- 32.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novakova Z, et al. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene. 2010;29:273–284. doi: 10.1038/onc.2009.318. [DOI] [PubMed] [Google Scholar]
- 34.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimri GP, et al. A novel biomarker identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedelnikova OA, et al. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 38.Castro P, Giri D, Lamb D, Ittmann M. Cellular senescence in the pathogenesis of benign prostatic hyperplasia. Prostate. 2003;55:30–38. doi: 10.1002/pros.10204. [DOI] [PubMed] [Google Scholar]
- 39.Collado M, et al. Tumor biology: senescent in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 40.Ding G, et al. Tubular cell senescence and expression of TGF-β1 and p21(WAF1/CIP1) in tubulointerstitial fibrosis of aging rats. Exp. Mol. Path. 2001;70:43–53. doi: 10.1006/exmp.2000.2346. [DOI] [PubMed] [Google Scholar]
- 41.Erusalimsky JD, Kurz DJ. Cellular senescence in vivo: its relevance in ageing and cardiovascular disease. Exp. Gerontol. 2005;40:634–642. doi: 10.1016/j.exger.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Martin JA, Buckwalter JA. The role of chondrocyte senescence in the pathogenesis of osteoarthritis and in limiting cartilage repair. J. Bone Joint Surg. Am. 2003;85:106–110. doi: 10.2106/00004623-200300002-00014. [DOI] [PubMed] [Google Scholar]
- 43.Matthews C, et al. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ. Res. 2006;99:156–164. doi: 10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- 44.McGlynn LM, et al. Cellular senescence in pretransplant renal biopsies predicts postoperative organ function. Aging Cell. 2009;8:45–51. doi: 10.1111/j.1474-9726.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 45.Paradis V, et al. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum. Pathol. 2001;32:327–332. doi: 10.1053/hupa.2001.22747. [DOI] [PubMed] [Google Scholar]
- 46.Roberts S, Evans EH, Kletsas D, Jaffray DC, Eisenstein SM. Senescence in human intervertebral discs. Eur. Spine J. 2006;15:312–316. doi: 10.1007/s00586-006-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt CA. Cellular senescence and cancer treatment. Biochim. Biophys. Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt CA, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 50.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–1883. [PubMed] [Google Scholar]
- 51.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 52.Ogryzko VV, Hirai TH, Russanova VR, Barbie DA, Howard BH. Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol. Cell. Biol. 1996;16:5210–5218. doi: 10.1128/mcb.16.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodier F, et al. Persistent DNA damage signaling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodier F, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl Acad. Sci. USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Apsel B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsyan A, et al. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 58.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl Acad. Sci. USA. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demidenko ZN, et al. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 60.Sun Y, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat. Med. 2012;18:1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang BD, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 62.Sheiban I, et al. Next-generation drug-eluting stents in coronary artery disease: focus on everolimus-eluting stent (Xience V) Vasc. Health Risk Manage. 2008;4:31–38. doi: 10.2147/vhrm.2008.04.01.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ciuffreda L, Di Sanza C, Incani UC, Milella M. The mTOR pathway: a new target in cancer therapy. Curr. Cancer Drug Targets. 2010;10:484–495. doi: 10.2174/156800910791517172. [DOI] [PubMed] [Google Scholar]
- 64.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 67.Huang S, Bjornsti MA, Houghton PJ. Rapamycins: mechanism of action and cellular resistance. Cancer Biol. Ther. 2003;2:222–232. doi: 10.4161/cbt.2.3.360. [DOI] [PubMed] [Google Scholar]
- 68.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 69.Hayward SW, et al. Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1. In Vitro Cell. Dev. Biol. Anim. 1995;31:14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 70.Bae VL, et al. Metastatic sublines of an SV40 large T antigen immortalized human prostate epithelial cell line. Prostate. 1998;34:275–282. doi: 10.1002/(sici)1097-0045(19980301)34:4<275::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.