Summary
In Mycobacterium tuberculosis and ampicillin-resistant mutants of Enterococcus faecium, the classical target of β-lactam antibiotics is bypassed by l,d-transpeptidases that form unusual 3→3 peptidoglycan cross-links. β-lactams of the carbapenem class, such as ertapenem, are mimics of the acyl donor substrate and inactivate l,d-transpeptidases by acylation of their catalytic cysteine. We have blocked the acyl donor site of E. faecium l,d-transpeptidase Ldtfm by ertapenem and identified the acyl acceptor site based on analyses of chemical shift perturbations induced by binding of peptidoglycan fragments to the resulting acylenzyme. An NMR-driven docking structure of the complex revealed key hydrogen interactions between the acyl acceptor and Ldtfm that were evaluated by site-directed mutagenesis and development of a cross-linking assay. Three residues are reported as critical for stabilization of the acceptor in the Ldtfm active site and proper orientation of the nucleophilic nitrogen for the attack of the acylenzyme carbonyl. Identification of the catalytic pocket dedicated to the acceptor substrate opens new perspectives for the design of inhibitors with an original mode of action that could act alone or in synergy with β-lactams.
Keywords: Acyl acceptor; β-lactam; Enterococcus faecium; l,d-transpeptidase; peptidoglycan
Introduction
The peptidoglycan polymer (PG) is a giant macromolecule that surrounds the cytoplasmic membrane to protect the bacterium from bursting due to the osmotic pressure of the cytoplasm (Typas et al., 2011). The PG also determines cell shape, intimately participates in cell division, and serves as a scaffold for anchoring various surface polymers. PG is assembled from a disaccharide-pentapeptide subunit, which is polymerized by glycosyltransferases (GTs) (Ostash and Walker, 2005) and cross-linked by transpeptidases (TPs) (Sauvage et al., 2008).
β-lactams are one of the most effective and broadly used family of antibiotics. These drugs inhibit the d,d-transpeptidase activity of enzymes, commonly referred to as penicillin-binding proteins (PBPs), which are responsible for the formation of the most prevalent type of peptidoglycan cross-links (4→3) (Sauvage et al., 2008). A second type of cross-links, the 3→3 cross-links, are formed by l,d-transpeptidases (Ldt) that by-pass the classical PBPs (Mainardi et al., 2000). In Mycobacterium tuberculosis, 80% of the peptidoglycan layer is cross-linked by l,d-transpeptidases (Lavollay et al., 2008) and these enzymes are attractive targets for the development of new drugs for the treatment of multidrug-resistant tuberculosis (Mainardi et al., 2008, Mainardi et al., 2011, Dhar et al., 2015, Hugonnet et al., 2009).
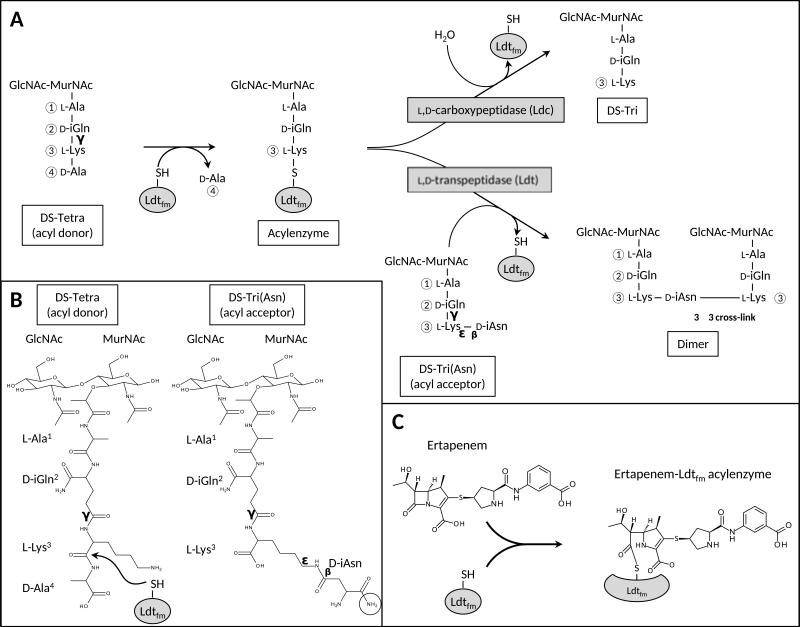
PG cross-linking by Ldts is a two-step reaction (Fig. 1A) (Mainardi et al., 2005). In the first step, the l,d-transpeptidase cleaves the l-Lys3-d-Ala4 peptide bond of the first substrate, the acyl donor, and forms a thioester bond linking the catalytic cysteine residue to the carbonyl of l-Lys3. In the second step, nucleophilic attack of the resulting acylenzyme by the side-chain nitrogen at the third position of the second substrate, the acyl acceptor, leads to formation of a 3→3 cross-link. In Enterococcus faecium, the nucleophilic nitrogen is carried by a d-iso-asparaginyl (d-iAsn) residue leading to formation of l-Lys3→d-iAsn-l-Lys3 cross-links. The l,d-transpeptidase of E. faecium (Ldtfm) is highly specific for acyl donors containing a stem tetrapeptide ending in l-Lys3-d-Ala4 and for acyl acceptors containing a d-iAsn-substituted l-Lys3 at the third position of the stem peptide (Fig. 1B) (Mainardi et al., 2005, Magnet et al., 2007).
Fig. 1. Reactions catalyzed by the l,d-transpeptidase Ldtfm.
A. Two-step cross-linking reaction catalyzed by Ldtfm. In the first step, the active site Cys residue (C442) of Ldtfm attacks the carbonyl of l-Lys3 in the stem tetrapeptide of the acyl donor. This step results in the release of d-Ala4 and the formation of a thioester bond between C442 and the carbonyl of l-Lys3. The resulting acylenzyme is a common intermediate for the l,d-transpeptidation and l,d-carboxypeptidation reactions. In the former reaction, the thioester bond of the intermediate is attacked by the d-iAsn amino group of the acyl acceptor leading to the formation of a muropeptide dimer and release of the enzyme. In the latter reaction, the acylenzyme is hydrolyzed leading to formation of a tripeptide. The two reactions occur in competition.
B. Structure of the acyl donor and acceptor. The arrow indicates the position of nucleophilic attack by catalytic C442. The nucleophile of the acyl acceptor is circled. The muropeptides (peptidoglycan fragments) contain a disaccharide (DS) composed of β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). Of note, DS-Tetra is only used as acyl donor since it lacks the side-chain d-iAsn residue (Magnet et al., 2007). Conversely, DS-Tri(Asn) is only used as acyl acceptor since it lacks the C-terminal d-Ala4 residue (Mainardi et al., 2005). Tetra, tetrapeptide, Tri, tripeptide.
C. Acylation of Ldtfm by ertapenem, a β-lactam of the carbapenem class.
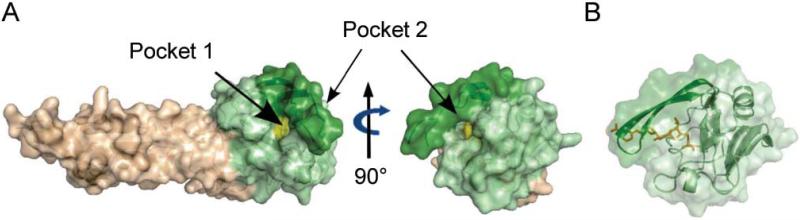
β-lactams are mimics of the acyl donor substrate of PBPs that act as suicide substrates and inactivate the enzymes by acylation of their catalytic serine (Tipper and Strominger, 1965). β-lactams of the carbapenem class similarly inactivate l,d-transpeptidases by acylation of their active site cysteine (Fig. 1C) (Mainardi et al., 2007, Triboulet et al., 2011, Triboulet et al., 2013). Recent NMR and X-ray structures of carbapenem-Ldt acylenzymes have allowed identifying the β-lactam binding pocket and hence the putative acyl donor site in three Ldts including E. faecium Ldtfm (PDB code 3ZGP) (Lecoq et al., 2013) and M. tuberculosis LdtMt1 (PDB code 4JMX) (Correale et al., 2013) and LdtMt2 [PDB code 4GSU (Kim et al., 2013) and 3VYP (Li et al., 2013)]. These structures revealed that the catalytic cysteine is located at the entrance of a narrow tunnel connecting two cavities, which are accessible to the solvent (Pockets 1 and 2; Fig. 2). The carbapenem gets access to the cysteine from Pocket 1 and the β-lactam carbonyl interacts with the backbone amide of the catalytic cysteine. For all l,d-transpeptidases except LdtMt1, the β-lactam nitrogen interacts with a tyrosine hydroxyl group located in the β-sheet separating the two cavities. In some cases, the drug carboxylate group interacts with residues in Pocket 2 but this interaction is more versatile and depends on the nature of the carbapenem side-chain. Together, these data indicate that Pocket 1 is most likely the binding site for the donor stem peptide of the cross-linking reaction catalyzed by l,d-transpeptidases (Lecoq et al., 2013). In contrast, the identity of the binding site for the acyl acceptor is controversial. Biarrotte-Sorin and co-workers proposed that the two paths to the catalytic cysteine are used in l,d-transpeptidases, one for the acyl-donor (Pocket 1) and the other for the acyl acceptor (Pocket 2) (Biarrotte-Sorin et al., 2006). On the contrary, Erdemli and co-workers proposed that both substrates reach the catalytic cysteine from Pocket 1 (Erdemli et al., 2012). Here, we report identification of the determinants for recognition of the acceptor muropeptide within Pocket 2. This information is a key step for the design of new inhibitors active on the l,d-transpeptidases of multidrug-resistant mycobacteria.
Fig. 2. Surface representations of the l,d-transpeptidase Ldtfm.
A. Access to the catalytic cysteine (C442) in the Ldtfm apoenzyme (PDB code 1ZAT). The representation of the catalytic domain of Ldtfm (green) highlights the access to C442 (yellow) from Pockets 1 and 2. The tunnel between Pockets 1 and 2 is formed by a loop that can act like a flap (dark green). The surface representation of the mixed alpha-beta N-terminal domain is shown in beige.
B. Surface and ribbon representation of Ldtfm acylenzyme catalytic domain (PDB code 3ZGP) emphasizing the structure of the flap. Small structural reorganization of a limited number of side chains and/or a limited flexibility of the backbone double-stranded β-sheet is sufficient to open the tunnel between Pockets 1 and 2. This is expected to allow for the release of the cross-linked reaction product without any major structural change.
Results
Preparation of a stable Ldtfm-ertapenem acylenzyme
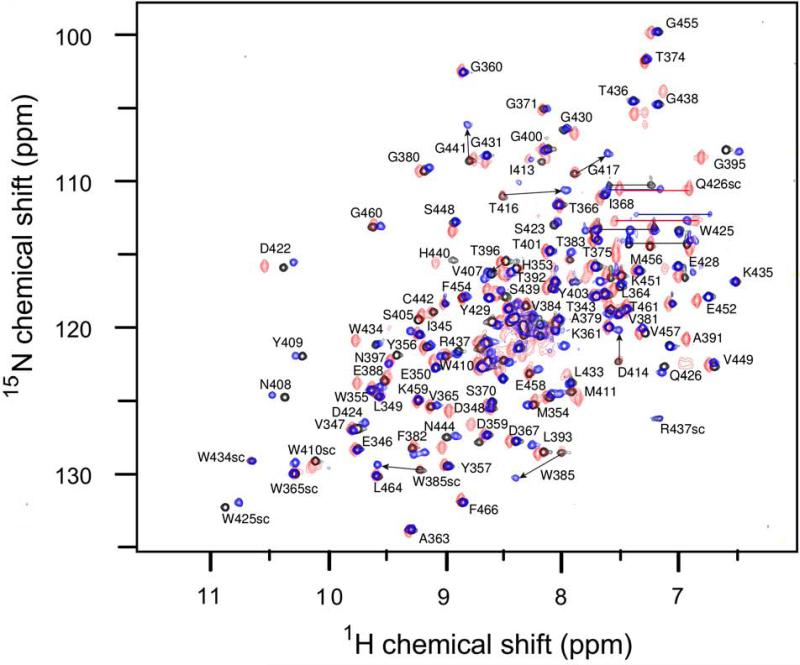
Ldtfm catalyzes in vitro the transpeptidation of peptidoglycan fragments (Fig. 1A) (Mainardi et al., 2005, Magnet et al., 2007). This reaction is too fast to capture the acyl acceptor bound to the acylenzyme for NMR studies. In contrast, adducts resulting from acylation of Ldtfm by β-lactams of the carbapenem class, such as ertapenem (Fig. 1C), are stable for more than 24 hours (Triboulet et al., 2011, Triboulet et al., 2013). We therefore used Ldtfm acylated by ertapenem to investigate the acyl acceptor binding site, as this β-lactam is thought to be a muropeptide mimetic. The initial formation of the acylenzyme was probed by recording 2D 1H-15N HSQC spectra of the 15N-labeled catalytic domain of Ldtfm before and after incubation with 2.2 molar equivalent of ertapenem. Acylation occurred in less than a minute and was clearly established from the characteristic pattern of the amide resonances from residues close to the active site in comparison to the signature of the apoenzyme (Fig. 3) (Lecoq et al., 2014). The stability of the spectroscopic signature over 48 hours confirmed the stability of the acylenzyme over this period (data not shown and Lecoq et al., 2013).
Fig. 3.
Superimposition of the 1H-15N HSQC spectra of wild-type apoenzyme (black), wild-type Ldtfm acylated with ertapenem (blue) and W425A R437A apoenzyme (red). The NMR spectra were collected on a 100 μM solution of 13C,15N-labeled protein in 100 mM sodium buffer containing 300 mM NaCl at pH 6.4 and 25 °C. The overall fingerprint of the protein is not affected by the substitution of amino acids W425 and R437 by Ala indicating that the protein structure remains unchanged. The substituted residues are shown in red. sc indicates side-chain 1H-15N correlations. Correlations are labeled except in the center of the spectrum due to overlaps.
Binding of muropeptides to the Ldtfm-ertapenem acylenzyme
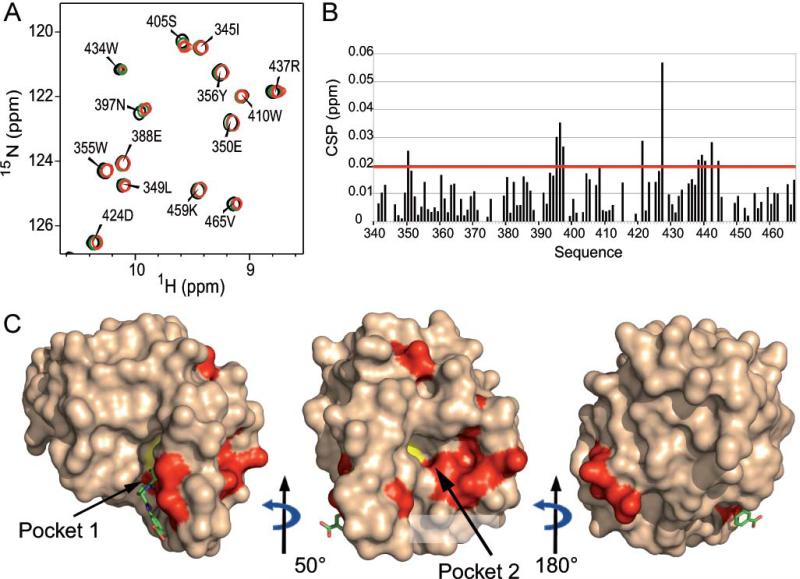
To identify the acyl-acceptor binding site, disaccharide-peptides (muropeptides), obtained from the enzymatic digestion of E. faecium M512 peptidoglycan by muramidases (Mainardi et al., 2000), were progressively added to a freshly prepared 100 μM ertapenem-Ldtfm acylenzyme solution, up to a ~300 molar excess. The acylenzyme remained stable for at least 24 hours after the addition of the muropeptide substrates as shown by its characteristic 1H-15N HSQC signature (Supplementary Fig. S1). However, small but definite chemical shift perturbations (CSPs) were detected in the 1H-15N HSQC spectra collected at different muropeptide:protein ratios (Fig. 4A). These results clearly establish the formation of a non-covalent complex between the acylenzyme and the muropeptides. Furthermore, superimposition of the different spectra showed a gradual shift of a few resonances in response to the increase in muropeptide:protein ratio, suggesting a fast exchange regime between the acylenzyme and the muropeptide:acylenzyme complex with a dissociation constant in the millimolar range.
Fig. 4. Muropeptide titration of Ldtfm acylated with ertapenem.
A. 1H-15N HSQC spectra recorded at 25°C for muropeptide:acylenzyme molar ratios of 0 (black), ~25 (green), and ~300 (red). The small amplitude of the chemical shift perturbations suggests that there is no major structural reorganization of the protein and, in particular, of the flap, upon addition of the muropeptide acceptor.
B. Combined 1HN and 15N chemical shift perturbations (CSPs) measured after addition of ~300 molar equivalents of muropeptides. The red line corresponds to two standard deviations above the mean, calculated over all residues with assigned backbone resonances.
C. Surface representations of the ertapenem-Ldtfm acylenzyme structure (PDB code 3ZGP) (Lecoq et al., 2013). Residues with significant CSP (greater than two standard deviations above the mean) are coloured in red (E350, G395, T396, N397, N408, I420, Q426, R437, G438, S439, G441 and I443). Ertapenem is displayed with green sticks and the catalytic cysteine (C442) is coloured in yellow. Residues impacted by the interaction are localized in the vicinity of C442 in Pockets 1 and 2 and at a remote distance of C442 only in Pocket 2.
Identification of the acceptor binding site and mapping of residues in contact with the substrate
To localize the muropeptide acyl-acceptor binding site, the combined 1HN and 15N CSP of each amide resonance was calculated for the largest muropeptide:protein ratio and reported in the histogram showing the perturbation induced by muropeptides along the protein sequence (Fig. 4B). Protein residues, that showed a combined CSP greater than two standard deviations above the mean (red line in Fig. 4B), were considered as significantly affected by the interaction with muropeptides and were sketched on the structure of the acylenzyme (Fig. 4C). These residues were found to mainly concentrate in the vicinity of the catalytic cysteine and in Pocket 2.
Construction of a model for the interaction of the acyl acceptor with the ertapenem-Ldtfm acylenzyme
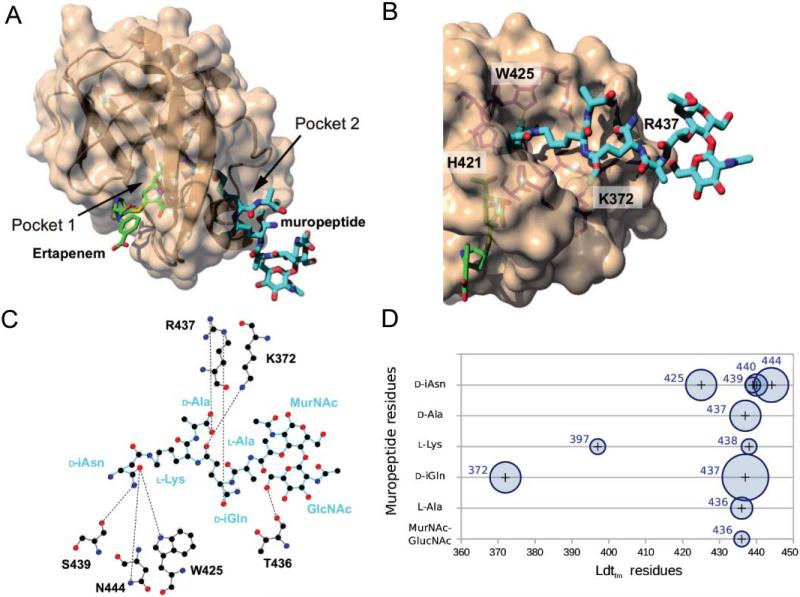
An NMR-driven structural model of the Ldtfm acylenzyme:muropeptide complex was calculated with the CNS structure calculation software (Brunger, 2007), in which the NMR experimental data (CSPs) were introduced as ambiguous interaction restraints using the HADDOCK protocol (de Vries et al., 2010). In this approach, the structure of a disaccharide-tetrapeptide acyl-acceptor generated in silico (see Experimental Procedures) is docked onto the NMR structure of the acylenzyme (Lecoq et al., 2013), considering the two biomolecules as rigid bodies. During the initial minimization process, residues of the protein, identified as perturbed by the muropeptides in the CSP analysis (Fig. 4B and 4C), are considered to potentially interact with all atoms of the acyl acceptor. Structures of the complex were further refined using online HADDOCK parameters. Even though the muropeptide always localized in Pocket 2 of Ldtfm in a well-defined position, introduction of a 2.5 Å-distance restraint between the sulfur atom of the catalytic cysteine and the amino nitrogen of d-iAsn improved the initial convergence. This constraint, which is consistent with the l,d-transpeptidation mechanism, was kept in the final calculation, that led to a cluster of 40 structures (Supplementary Table S1). A representative model from the cluster, corresponding to the structure of lowest energy, is depicted in Fig. 5A and 5B.
Fig. 5. Binding of the acyl acceptor to Pocket 2 of Ldtfm.
A. Lowest energy structure obtained by NMR data-driven docking of the muropeptide DS-Tetra(d-iAsn) into the ertapenem-Ldtfm acylenzyme structure (PDB code 3ZGP) (Lecoq et al., 2013). Ertapenem mimicking the acyl donor and the acceptor muropeptide are shown in green and cyan, respectively. DS-Tetra(d-iAsn), GlcNAc-MurNAc-l-Ala1-d-iGln2-l-Lys3(d-iAsn)-d-Ala4.
B. Surface representation highlighting the acyl acceptor in Pocket 2 (cyan, in sticks) and contacts with protein residues.
C. Schematic representation of the H-bond network (dashed lines) in the ertapenem-Ldtfm:acyl acceptor complex established with LigPlot+.
D. Schematic representation of hydrogen-bond interactions in the 5 model complexes of lowest energy. These models are selected within the cluster with the best HADDOCK score. The horizontal and vertical axes sketch the protein sequence (residue numbers) and the muropeptide residues, respectively. Circles represent intermolecular interactions between residues of the protein and of the muropeptide. These interactions may involve one or several hydrogen bonds. The surface of the circles is proportional to the number of structures showing the interaction among the five structures with the best HADDOCK scores. The number of structures showing interaction ranges from one (e.g. T436 with GlcNAc) to five (e.g. R437 with d-iGln).
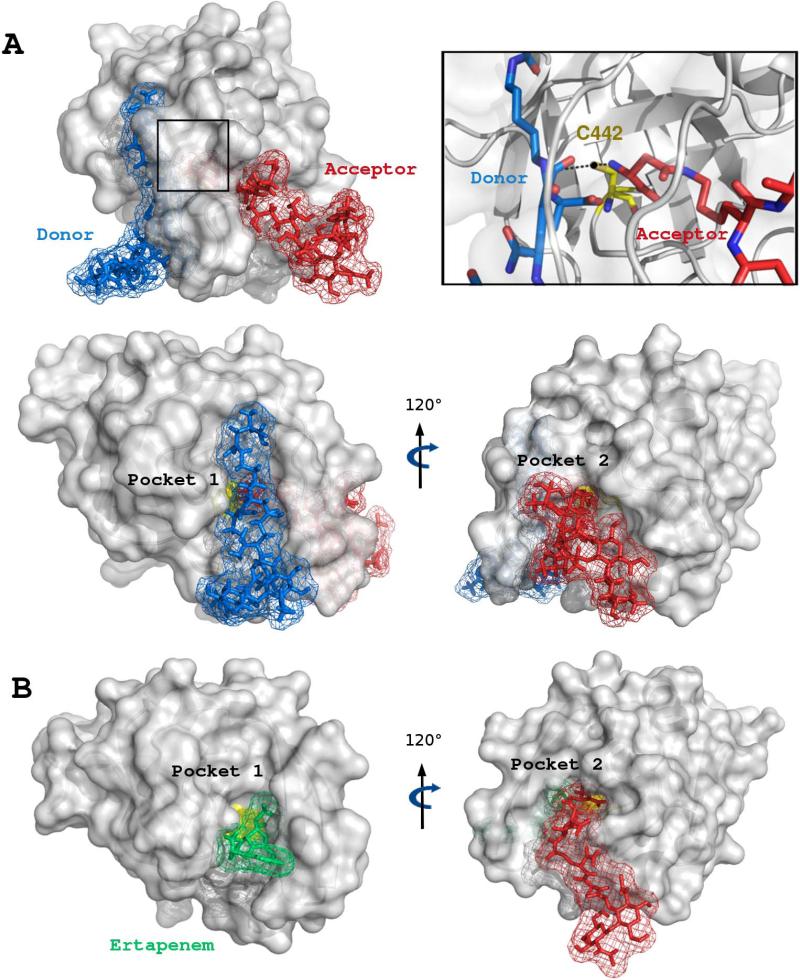
In order to confirm the specificity of Pockets 1 and 2 for the acyl donor and acceptor, respectively, modelling was also performed with two disaccharide-tetrapeptides substituted by a d-iAsn residue (Fig. 6A). This muropeptide can act both as a donor and an acceptor. Introduction of spatial constraints specific of the Ldtfm-donor (C442-Lys3) and Ldtfm-acceptor (C442-d-iAsn) interactions showed that the acyl donor approaches the catalytic cysteine from Pocket 1, whereas the acyl acceptor binds to Pocket 2 (see Supplementary Table S2 for statistical convergence). These results confirmed the specificity of Pockets 1 and 2 for the donor and for the acceptor in the absence of ertapenem. Altogether, the ab initio model depicted in the lower part of Fig. 6A and the experimentally-derived model with ertapenem (Fig. 5 and Fig. 6B) also show that the steric hindrance caused by the positioning of the donor in Pocket 1 prevents the acceptor from accessing to the catalytic cysteine by the same pocket. Set side-by-side, the two models evidence the diminished occupancy of Pocket 1 with the antibiotic in comparison to its occupancy with the muropeptide. The latter observation indicates that binding of ertapenem in Pocket 1 is unlikely to impair binding of the acyl acceptor in Pocket 2.
Fig. 6. Binding of the acyl donor and acceptor to Ldtfm.
A. Modelling of the complex formed by the binding of two DS-Tetra(d-iAsn) muropeptides to Ldtfm. An energy minimization was run with the structures of Ldtfm (PDB code 1ZAT) and of two identical muropeptides, DS-Tetra(d-iAsn), which can act as a donor and as an acceptor in the cross-linking reaction. In order to assign a donor role to one of the two muropeptides, a distance restraint was introduced between the sulfur of the catalytic cysteine (C442) and the carbonyl carbon of l-Lys3. In order to assign an acceptor role to the other muropeptide, a distance restraint was introduced between the sulfur of C442 and the nitrogen of the amino group of d-iAsn. Following energy minimization, the distance constraint involving C442 and l-Lys3 led to the localisation of the muropeptide into Pocket 1 (blue). Conversely, the C442-d-iAsn constraint led to the localisation of the muropeptide into Pocket 2 (red). The upper-left panel shows a front view of the two cavities separated by the flap. The upper-right panel shows an enlargement of the C442 environment with the distance restraints indicated by dotted lines. The occupancy of each individual pocket by the muropeptide is illustrated in the two lower panels.
B. NMR data-driven model of the DS-Tetra(d-iAsn) muropeptide docked onto the ertapenem-Ldtfm acylenzyme. The surface representation is shown in the same orientation as the lower part of panel A. These views show that ertapenem (in green) provides a smaller steric hindrance than the donor in Pocket 1.
DS-Tetra(d-iAsn), GlcNAc-MurNAc-l-Ala1-d-iGln2-l-Lys3(d-iAsn)-d-Ala4.
Candidate interactions stabilizing the acyl acceptor within the Ldtfm catalytic cavity
In the model with ertapenem depicted in details in Fig. 5, the peptide stem of the acceptor is mainly stabilized in Pocket 2 by a series of hydrogen bonds (Fig. 5C). The relevance of these interactions in the stabilization of the complex was analyzed based on their persistence in the 5 structures of lower energy (Fig. 5D). In Pocket 2, W425 is likely to critically contribute to the orientation of the nucleophilic nitrogen of the acceptor by establishing a hydrogen bond with the oxygen of the carboxamide of d-iAsn. S439 and N444 may be of assistance to this orientation process. Additionally, K372 and R437 form several hydrogen bonds with the γ-carbonyl and α-carboxamide of d-iGln, respectively. These interactions are likely to stabilize the conformation of the tetrapeptide stem within Pocket 2. Together, these results led to the identification of the acceptor binding site of Ldtfm and of candidate enzyme residues potentially involved in binding of the acceptor substrate.
Assay of the cross-linking activity of Ldtfm and derivatives obtained by site-directed mutagenesis
The role of Ldtfm residues inferred from the structural model was assessed by determining the cross-linking activity of derivatives obtained by site-directed mutagenesis. Chemical shift perturbation assays were used to show that impaired enzyme activity was not due to important modification of the protein conformation (Fig. 3). A linear tetrapeptide (DS-Tetra) and a branched tripeptide [DS-Tri(Asn)] were used as substrates since these muropeptides are exclusively used as acyl donor and acceptor, respectively (Fig. 1A and 1B). This led to formation of a single peptidoglycan dimer [DS-Tri(Asn)-DS-Tri], which was not further polymerized. The only side reaction was the hydrolysis of the l-Lys3-d-Ala4 peptide bond of the acyl donor to form a tripeptide (DS-Tri) (Fig. 1A).
In the presence of equimolar concentrations (30 μM) of the acyl donor and acceptor, wild type Ldtfm catalyzed formation of the muropeptide dimer (l,d-transpeptidase activity) and of DS-Tri (l,d-carboxypeptidase activity) with similar efficiencies (Table 1). The ratio of the two activities increased linearly with the concentration of the acceptor in the 10 to 90 μM range. The observed absence of saturation of the enzyme by the acyl acceptor at 90 μM is commensurate with the relatively high dissociation constant (mM range) reported above for NMR-based analyses of the binding of the acceptor to the Ldtfm-ertapenem acylenzyme (Supplementary Fig. S2). Thus, binding of ertapenem to Pocket 1 does not appear to artefactually impair binding of the acyl acceptor in Pocket 2 in agreement with the results of the modelling experiments presented above (Fig. 6).
Table 1.
Impact of amino acid substitutions on Ldtfm activitiesa.
|
l,d-transpeptidase |
l,d-carboxypeptidase |
||||
|---|---|---|---|---|---|
| Substitution | Turnoverb | %c | Turnoverb | %c | Ratiod |
| None | 1,800 ± 620 | 100 | 2,400 ± 900 | 100 | 1.3 |
| R437A | 460 ± 82 | 26 | 3,400 ± 1,100 | 140 | 7 |
| K372A | 130 ± 16 | 7 | 1,800 ± 500 | 77 | 14 |
| K372A R437A | 37 ± 7 | 2 | 1,100 ± 200 | 45 | 29 |
| W425A | ≈ 0.69 | ≈ 0.04 | 430 ± 18 | 18 | 630 |
Ldtfm was incubated with muropeptides DS-Tetra (30 μM) and DS-Tri(Asn) (30 μM) to determine competitive formation of dimer (l,d-transpeptidase activity) and DS-Tri (l,d-carboxypeptidase activity).
Mean ± SD from a minimum of three determinations (×106 s−1).
Relative to native Ldtfm.
Ratio of turnover numbers (l,d-carboxypeptidase over l,d-transpeptidase).
The K372A and R437A substitutions, alone or in combination, had little impact on the l,d-carboxypeptidase activity (relative activities ranging from 45 to 140%). In contrast, substitution K372A led to a 14-fold reduction in the l,d-transpeptidase activity (from 100 to 7%). A further 4-fold decrease was observed for the combination of substitutions K372A and R437A (from 7 to 2%). Alone, the latter substitution resulted in a similar 4-fold decrease (from 100 to 26%). These results indicate that R372A, and to a lesser extent R437A, impaired interaction of Ldtfm with the acceptor, without interfering with formation of the acylenzyme. In agreement, our structural model (Fig. 5) predicts that these residues are located remotely from the catalytic C442 in the acceptor pocket and should therefore not participate in the hydrolytic pathway. Residue W425 is located at the bottom of the acceptor pocket, closer to C442 than K372 and R437. The W425A substitution almost completely abolished the l,d-transpeptidase activity but produced a much more moderate effect on the l,d-carboxypeptidase activity (0.04% versus 18% residual activities). This single substitution leads to a 500-fold decrease in the l,d-transpeptidase to l,d-carboxypeptidase turnover ratio indicating that the substitution preferentially impaired the l,d-transpeptidase activity. In the model depicted in Fig. 5C, W425 establishes a hydrogen bond with the d-iAsn residue that carries the nucleophilic amine of the cross-linking reaction. This interaction may directly participate in the positioning of this group for catalysis. A minor impact on the first step of the reaction may be accounted for by the close proximity of W425 and the catalytic C442 residue.
Discussion
Peptidoglycan transpeptidases are attractive and validated targets for antibacterial drug development, as demonstrated by successful use of antibiotics of the β-lactam family in the past seven decades. These targets include the classical PBPs in most bacterial pathogens (Zapun et al., 2008). These targets also include l,d-transpeptidases, which are the predominant cross-linking enzymes in M. tuberculosis (Lavollay et al., 2008, Kumar et al., 2012), Mycobacterium abscessus (Lavollay et al., 2011) and Clostridium difficile (Peltier et al., 2011). In addition, one of the five l,d-transpeptidase paralogues of M. tuberculosis has a critical role in virulence (Gupta et al., 2010) and inhibition of these enzymes by certain β-lactams results in rapid cytolysis and elimination of persisters (Dhar et al., 2015).
Most structural studies of PBPs and l,d-transpeptidases have focused on the interaction of β-lactams with the acyl donor site (Sauvage et al., 2008, Zapun et al., 2008). Consequently, little is known on the acceptor site. The specificity of PBPs for the acceptor substrate has only been indirectly assessed in vivo based on genetic manipulation of the structure of peptidoglycan precursors (Arbeloa et al., 2004, Bellais et al., 2006). These studies led to the conclusion that PBPs tolerate important variations in the structure of both the acyl acceptor and the acyl donor. In contrast, in vitro cross-linking assays showed that l,d-transpeptidases tolerate little variation in the donor and essentially no variation in the acceptor (Magnet et al., 2007). Here we show that the acceptor site of transpeptidases is amenable to structural analyses by NMR based on irreversible acylation of the enzyme by a β-lactam, which occupies the donor site, and titration of the acceptor site by muropeptides. The approach is applicable to both PBPs and l,d-transpeptidases.
Using E. faecium Ldtfm as a model, we show that the acylenzyme formed with carbapenem remains stable upon muropeptide addition. This allowed mapping the chemical shift perturbations specifically induced by the binding of muropeptides to the acceptor site. Based on these structural data we propose a model in which the acyl acceptor bind to Pocket 2 which is distant from the antibiotic binding cavity (Fig. 4 and 5). Interestingly, a model built independently from the chemical shift perturbations led to the identification of the same binding Pockets for the acyl donor and acyl acceptor (Fig. 6). The interaction model of the acyl acceptor with Ldtfm was validated by site-directed mutagenesis of candidate residues and assay of the residual cross-linking activity of the resulting Ldtfm derivatives. Of note, our l,d-transpeptidase assay relies on determination of the l,d-carboxypeptidase activity of Ldtfm, a side reaction which provides an internal control for the efficacy of the first step of the cross-linking reaction (i.e. formation of the acylenzyme). Thus, any decrease in the l,d-transpeptidase to l,d-carboxypeptidase ratio of activities identifies impaired interactions with the acyl acceptor in the second step of the reaction. Using these approaches, we identified amino acid substitutions that exclusively (R372A and R437A) or preferentially (W425A) impaired the l,d-transpeptidase activity of Ldtfm (Table 1). The latter substitution had the largest impact on Ldtfm activity and led to a 500-fold decrease in the l,d-transpeptidase to l,d-carboxypeptidase ratio. Based on these data K372 and R437 are predicted to stabilize the conformation of the acyl acceptor by forming hydrogen bonds with the γ-carbonyl and α-carboxamide of d-iGln at the 2nd position of the stem peptide, respectively (Fig. 5C). Residue W425 forms a hydrogen bond with the α-carboxamide of d-iAsn and is predicted to orientate the α-amino group of d-iAsn for nucleophilic attack of the acylenzyme carbonyl.
Mapping of the acceptor site opens new avenues for the design of inhibitors with an original mode of action. Such drugs are unlikely to be affected by modifications of the target that confers resistance to β-lactams since distinct sites are involved in the binding of the donor and acceptor substrates. Furthermore, drugs targeting the acceptor site may act in synergy with β-lactams.
Experimental Procedures
Production and Purification of Ldtfm
A 13C,15N–labeled protein containing the catalytic domain of Ldtfm (residues 341 to 466) was produced in Escherichia coli BL21 (DE3) cells harbouring plasmid pETTEVΩldtfm and purified, as previously described (Lecoq et al., 2014, Lecoq et al., 2013).
Muropeptides preparation and l,d-transpeptidase assay
The disaccharide-peptides used as the acyl donor (DS-Tetra) and acceptor [DS-Tri(Asn)] in the transpeptidase assay were purified (Arbeloa et al., 2004) from the peptidoglycan of Enterococcus faecalis BM4314 (Bouhss et al., 2002) and E. faecium M512 (Mainardi et al., 2000), respectively. The concentration of the disaccharide-peptides was determined by amino acid analysis after acidic hydrolysis with a Hitachi autoanalyser (Mengin-Lecreulx et al., 1999). Formation of 3→3 cross-links was tested in 60 μL of phosphate buffer (15 mM, pH 7.0) containing Ldtfm (2.5 to 50 μM), (DS-Tetra) (30 μM), and [DS-Tri(Asn)] (30 μM). The reaction was allowed to proceed at 37 °C, aliquots (10 μL) were withdrawn at various times (15 min to 2 h), and the reaction was stopped by the addition of 2 μL of 10% trichloroacetic acid. Muropeptides were desalted (ZipTipC18, Millipore) and analyzed by electrospray mass spectrometry in the positive mode (Qstar Pulsar I, Applied Biosystem), as previously described (Arbeloa et al., 2004). Muropeptides used in the NMR interaction experiment were produced from unlabeled E. faecium M512 peptidoglycan (Mainardi et al., 2000).
NMR Spectroscopy
A 100 μM 13C,15N–Ldtfm sample in 100 mM sodium phosphate buffer containing 100 mM NaCl at pH 6.4 (buffer A) and 2.2 molar equivalent of ertapenem (INVANZ) was used to prepare the acylenzyme for the NMR study. One aliquot of 10 μL (~25 molar equivalent with respect with the protein), 4 aliquots of 20 μL, and a last aliquot of 15 μL of the unlabeled muropeptides stock-solution (155 mg/mL in buffer A) were successively added to the protein sample. 1H-15N HSQC spectra were recorded at 25 °C on a 600 MHz Agilent Direct Drive spectrometer after each muropeptide addition using a triple resonance cryogenic probe. Data were processed with NMRPipe and analyzed with CcpNMR in which the published acylenzyme assignments (Lecoq et al., 2014, Lecoq et al., 2013) were initially transferred.
Ldtfm/ muropeptide docking
Models of muropeptide docked onto Ldtfm were built with “The HADDOCK web server for data-driven biomolecular docking” of HADDOCK2.1 (de Vries et al., 2010) using CNS1.2 (Brunger, 2007) for the structure calculations. The initial structural model for muropeptide DS-Tetra(Asn) was built from a disaccharide and a modified tetrapeptide motif generated within the GlyCaNS (http://haddock.chem.uu.nl/glycans/) and PRODRG softwares, respectively. A patch for the topology and parameter files was used to connect the two fragments through the lactoyl group of MurNAc leading to the initial PDB structure for the muropeptide.
For the NMR data-driven modelling of the muropeptide acceptor into the ertapenem Ldtfm acylenzyme, the coordinates of the acylenzyme were taken from the NMR structure [PDB code 3ZGP (Lecoq et al., 2013)]. The docking was performed and analyzed with default HADDOCK parameters except a clustering cutoff of 3.0 Å and random removal of restraints turned off. Protein residues E350, G395, T396, N397, N408, I420, Q426, R437, G438, S439, G441 and I443 that were identified through CSP analysis were declared as ambiguous interaction restraints. Non-bonded interactions were calculated with the OPLS force field using a cutoff at 6.5 Å. The HADDOCK score was used to rank the generated models, which were further analyzed within the Pymol software. The model of lowest energy was examined with LigPlot+v.1.4 to extract the interaction map between the muropeptide and the protein.
For modelling of the complex containing two d-iAsn-substituted disaccharide-tetrapeptide substrates in the Ldtfm, catalytic cavity, the coordinates of the apoenzyme were taken from the X-ray structure [PDB code 1ZAT (Biarrotte-Sorin et al., 2006)]. The docking was performed and analyzed with default HADDOCK parameters. In order to position the reactive groups of the acyl donor and acceptor of the transpeptidation reaction, two independent distance restraints of 2 ± 1 Å were introduced between (i) the sulfur of C442 and the carbonyl carbon of Lys3 and (ii) the sulfur of C442 and the amino nitrogen of d-iAsn. No experimental restraints were used in this calculation.
Supplementary Material
Acknowledgments
This work was supported by the Agence National de la Recherche (ANR), Project CARBATUB (N° ANR 2011 BSV5 024 01) and the National Institute of Allergy and Infectious Diseases (Grants RO1 307 AI046626). ST was supported by the seventh Framework Program of the European Community, Project ORCHID (261378).
References
- Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, et al. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J Biol Chem. 2004;279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- Bellais S, Arthur M, Dubost L, Hugonnet JE, Gutmann L, van Heijenoort J, et al. Aslfm, the d-aspartate ligase responsible for the addition of d-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J Biol Chem. 2006;281:11586–11594. doi: 10.1074/jbc.M600114200. [DOI] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol. 2006;359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Bouhss A, Josseaume N, Severin A, Tabei K, Hugonnet JE, Shlaes D, et al. Synthesis of the l-alanyl-l-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J Biol Chem. 2002;277:45935–45941. doi: 10.1074/jbc.M207449200. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Correale S, Ruggiero A, Capparelli R, Pedone E, Berisio R. Structures of free and inhibited forms of the l,d-transpeptidase LdtMt1 from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2013;69:1697–1706. doi: 10.1107/S0907444913013085. [DOI] [PubMed] [Google Scholar]
- de Vries SJ, Melquiond AS, Kastritis PL, Karaca E, Bordogna A, van Dijk M, et al. Strengths and weaknesses of data-driven docking in critical assessment of prediction of interactions. Proteins. 2010;78:3242–3249. doi: 10.1002/prot.22814. [DOI] [PubMed] [Google Scholar]
- Dhar N, Dubée V, Ballell L, Cuinet G, Hugonnet JE, Signorino-Gelo F, et al. Rapid cytolysis of Mycobacterium tuberculosis by faropenem, an orally bioavailable β-lactam antibiotic. Antimicrob Agents Chemother. 2015;59:1308–1319. doi: 10.1128/AAC.03461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdemli SB, Gupta R, Bishai WR, Lamichhane G, Amzel LM, Bianchet MA. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of l,d-transpeptidase 2. Structure. 2012;20:2103–2115. doi: 10.1016/j.str.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein Ldt(Mt2) is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim J, Im HN, Yoon JY, An DR, Yoon HJ, et al. Structural basis for the inhibition of Mycobacterium tuberculosis l,d-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta Crystallogr D Biol Crystallogr. 2013;69:420–431. doi: 10.1107/S0907444912048998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, et al. Meropenem inhibits d,d-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol. 2012;86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, et al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, et al. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J Bacteriol. 2011;193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq L, Dubee V, Triboulet S, Bougault C, Hugonnet JE, Arthur M, Simorre JP. Structure of Enterococcus faecium l,d-transpeptidase acylated by ertapenem provides insight into the inactivation mechanism. ACS Chem Biol. 2013;8:1140–1146. doi: 10.1021/cb4001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq L, Bougault C, Triboulet S, Dubee V, Hugonnet JE, Arthur M, Simorre JP. Chemical shift perturbations induced by the acylation of Enterococcus faecium l,d-transpeptidase catalytic cysteine with ertapenem. Biomol NMR Assign. 2014;8:339–343. doi: 10.1007/s12104-013-9513-3. [DOI] [PubMed] [Google Scholar]
- Li WJ, Li DF, Hu YL, Zhang XE, Bi LJ, Wang DC. Crystal structure of l,d-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell Res. 2013;23:728–731. doi: 10.1038/cr.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Arbeloa A, Mainardi JL, Hugonnet JE, Fourgeaud M, Dubost L, et al. Specificity of l,d-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J Biol Chem. 2007;282:13151–13159. doi: 10.1074/jbc.M610911200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. Novel mechanism of beta-lactam resistance due to bypass of dd-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, et al. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Hugonnet JE, Rusconi F, Fourgeaud M, Dubost L, Moumi AN, et al. Unexpected inhibition of peptidoglycan l,d-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J Biol Chem. 2007;282:30414–30422. doi: 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. 2008;32:386–408. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Hugonnet JE, Gutmann L, Arthur M. Fighting resistant tuberculosis with old compounds: the carbapenem paradigm. Clin Microbiol Infect. 2011;17:1755–1756. doi: 10.1111/j.1469-0691.2011.03699.x. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Falla T, Blanot D, van Heijenoort J, Adams DJ, Chopra I. Expression of the Staphylococcus aureus UDP-N-acetylmuramoyl- l-alanyl-d-glutamate:l-lysine ligase in Escherichia coli and effects on peptidoglycan biosynthesis and cell growth. J Bacteriol. 1999;181:5909–5914. doi: 10.1128/jb.181.19.5909-5914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Peltier J, Courtin P, El Meouche I, Lemee L, Chapot-Chartier MP, Pons JL. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;286:29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci U S A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet S, Arthur M, Mainardi JL, Veckerle C, Dubee V, Nguekam-Nouri A, et al. Inactivation kinetics of a new target of beta-lactam antibiotics. J Biol Chem. 2011;286:22777–22784. doi: 10.1074/jbc.M111.239988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet S, Dubee V, Lecoq L, Bougault C, Mainardi JL, Rice LB, et al. Kinetic features of l,d-transpeptidase inactivation critical for beta-lactam antibacterial activity. PLoS One. 2013;8:e67831. doi: 10.1371/journal.pone.0067831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.