Abstract
The cytoplasm of prokaryotes contains many molecular machines interacting directly with the chromosome. These vital interactions depend on the chromosome structure, as a molecule, and on the genome organization, as a unit of genetic information. Strong selection for the organization of the genetic elements implicated in these interactions drives replicon ploidy, gene distribution, operon conservation, and the formation of replication-associated traits. The genomes of prokaryotes are also very plastic with high rates of horizontal gene transfer and gene loss. The evolutionary conflicts between plasticity and organization lead to the formation of regions with high genetic diversity whose impact on chromosome structure is poorly understood. Prokaryotic genomes are remarkable documents of natural history because they carry the imprint of all of these selective and mutational forces. Their study allows a better understanding of molecular mechanisms, their impact on microbial evolution, and how they can be tinkered in synthetic biology.
Prokaryotes have highly organized genomes because their DNA interacts directly with molecular machines in the cytoplasm. But their genomes are also very plastic, with high rates of horizontal gene transfer and gene loss.
Prokaryotic cells typically lack a clear physical separation between DNA and the cytoplasm. The cell is therefore a complex network of genetic and biochemical interactions involving the DNA molecules and many cellular processes. The complexity of such interactions is well illustrated by the functioning of Escherichia coli. In fast growing E. coli cells, the replication forks advance bidirectionally and very rapidly from the single origin of replication to the terminus. The cell doubles in 20 min, which is less than the time required to replicate the chromosome (45 to 60 min). This is made possible by up to three simultaneous replication rounds, resulting in the presence of eight replication forks and eight origins of replication per terminus in the cell. As replication proceeds, DNA regions are under different states of replication and are being segregated in function of the growing multiple division septa. Hence, replication, segregation, and cell doubling are tightly linked. In prokaryotes, nascent transcripts are immediately translated by multiple ribosomes and, for certain membrane proteins, integration in the membrane takes place before the end of transcription and translation. Hence, transcription, translation, and protein localization are tightly linked. Exponentially growing cells endure intense gene expression and collisions between the rapid replication fork and the relatively slower RNA polymerases are frequent. These collisions may disrupt both transcription and replication, thereby potentially affecting all of the other above-mentioned cellular processes. Collisions between the replication fork and RNA polymerases may result in the fork’s collapse. Ensuing SOS or SOS-like responses may kick off the transfer of mobile genetic elements to other cells, thereby changing their gene repertoires. The associations between all of these cellular processes through their interactions with the chromosome result in natural selection for genome organization. The strength of selection on a given organizational trait that is as strong as the efficiency of the interaction with the chromosome is important. Because many of the above-mentioned processes are essential, the organization of genomes is under strong selection and the study of genome organization informs about natural history and about cell functioning.
Prokaryotes endure high rates of rearrangement, mutation, deletion, and accretion of genetic material. This leads to trade-offs between selection for genome organization and selection for genetic diversification that drive the evolution of the genome. A full understanding of the organization of genetic elements in the genome (genome organization) and of the structure of DNA molecules in the cell (chromosome structure) therefore requires multidisciplinary approaches, bridging genetics, genomics, biochemistry, and biophysics against a backdrop of evolutionary biology. Several aspects of this topic were reviewed before (Abby and Daubin 2007; Rocha 2008; Kuo and Ochman 2009b; Boussau and Daubin 2010; Touzain et al. 2011; Ptacin and Shapiro 2013). We will focus on recent findings and on aspects for which the evolutionary scope has, in our view, been insufficiently emphasized.
VARIATIONS IN GENOME STRUCTURE
The size of prokaryotic genomes ranges from around 50 kb to more than 13 Mb (Schneiker et al. 2007; Ishii et al. 2013; Tatusova et al. 2015). These genomes are also very compact, with gene density typically approaching 85% (Mira et al. 2001). There is, therefore, a direct proportionality between the size of the genome and the number of encoded proteins. The smallest genomes (<500 kb) correspond to obligatory endosymbionts that have arisen by reduction of larger genomes of free-living bacteria. Some of these genomes have fewer genes than those strictly required for autonomous life in E. coli (McCutcheon and Moran 2012). Larger genomes encode complex metabolic and genetic networks, some of them allowing bacteria to differentiate or regroup into multicellular bodies (Guieysse and Wuertz 2012). Variations in genome size affect cellular functions in different ways. The gene repertoires associated with some housekeeping functions, like translation, show little variation in the known range of genome size. Gene repertoires for other functions are much more variable: smaller genomes are nearly depleted of sensory, transport, communication, and regulatory functions, reflecting narrow environmental ranges (Boussau et al. 2004; Konstantinidis and Tiedje 2004). Importantly, larger genomes are thought to engage much more frequently in horizontal gene transfer (Cordero and Hogeweg 2009) and encode more transposable elements (Touchon and Rocha 2007). Genome size is also positively correlated with the strength of purifying selection acting on protein coding sequences (Kuo et al. 2009). This suggests that natural selection is more efficient in larger genomes, possibly as a result of larger effective population sizes. Very large gene repertoires might in fact require efficient natural selection derived from large effective population sizes, otherwise genes would be rapidly lost by genetic drift. It is thus generally thought that larger genomes correspond to more versatile prokaryotes that are less sexually isolated and in which selection is more efficient.
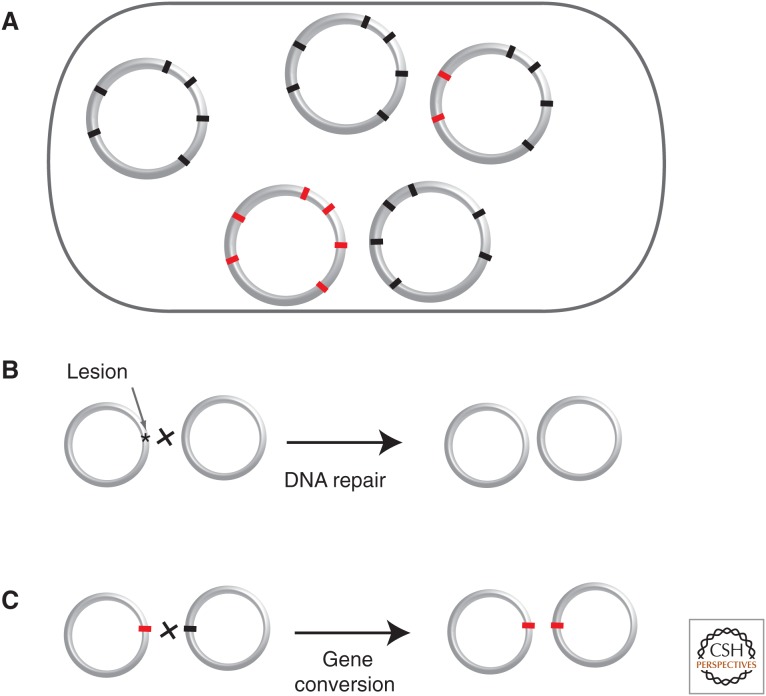
Prokaryotes are often polyploid, with certain species carrying more than 100 copies of the chromosome per cell (Fig. 1). Polyploidy might increase gene dosage in very large cells, in which demand for transcription is very high (Mendell et al. 2008), and facilitate gene expression regulation in endosymbionts (Vinuelas et al. 2011), which have few other means of regulating the rate of gene expression. A large number of identical chromosomes might also diminish the stochastic effects of gene expression, which are exacerbated when there is a single DNA molecule (Soppa 2013). Because sister chromosomes can recombine efficiently by homologous recombination, polyploidy allows transient genetic diversification by reversible heterozygosity (Griese et al. 2011), antigenic variation by recombination (Tobiason and Seifert 2006), and DNA repair (Zahradka et al. 2006). Gene conversion between variants in different chromosomes might also facilitate purifying selection of deleterious alleles (Komaki and Ishikawa 1999; Hildenbrand et al. 2011). Hence, polyploidy might be linked with gene expression, DNA repair, or the efficiency of natural selection. It might also serve to store phosphate for later use (Zerulla et al. 2014). The relative importance of each of these effects remains to be investigated.
Figure 1.
Traits associated with polyploidy in prokaryotes. The presence of multiple copies of replicons, and particularly chromosomes, has been proposed to confer several advantages. (A) It allows distributing gene expression through the entire cytoplasm in very large cells. It allows heterozygosity. In the presence of several replicons, the ratio between replicons allows gene expression regulation. (B) It allows repair by homologous recombination with other similar replicons. (C) Recombination between similar replicons also allows gene conversion and allelic exchange. Heterozygosity is indicated using distinct colors (red and black).
Around 5% of the complete genomes in GenBank/EMBL/DDBJ contain more than one type of chromosome (not to be confounded with polyploidy). Such genomes typically have chromosomes of very different sizes. The larger chromosome encodes most essential and highly expressed genes. Smaller chromosomes are also called secondary chromosomes or chromids (Harrison et al. 2010), and vary widely in size and persistence in bacterial lineages. Genomes of the genus Burkholderia carry one, two, or three chromosomes suggesting rapid changes in genome architecture (Mahenthiralingam et al. 2005). On the other hand, Vibrio and closely related genera systematically carry two chromosomes of very different size (Okada et al. 2005), showing that multiple chromosomes can be stably kept for hundreds of millions of years. The reasons for the existence of multiple chromosomes are unclear. Genes on secondary chromosomes are gained and lost at higher rates, and their sequences also evolve slightly faster relative to homologs in the larger chromosome. This has led to suggestions that secondary replicons might favor evolvability (Cooper et al. 2010). Additionally, it has been observed that replicon fusions in Vibrio lead to lower growth rates and more frequent dimer formation (Val et al. 2012). The systematic presence of two chromosomes in the genus of Vibrio, typically very fast-growing bacteria, led to suggestions that multiple chromosomes facilitate rapid bacterial growth and the management of chromosome dimerization in large genomes. Yet, some of fastest growing bacteria do not have multiple chromosomes and the largest genomes only have one chromosome. The reason(s) behind the existence of multiple chromosomes is therefore still an open subject of research.
Plasmids are the most common extra-chromosomal replicons and some genomes carry more than 20 such elements (Casjens et al. 2000). A few prophages are also extrachromosomal (Ravin 2011). Plasmids carry genes for their propagation and maintenance in the cell, but also host-adaptive genetic information (de la Cruz and Davies 2000; Rankin et al. 2011). There is no very clear distinction between large plasmids (megaplasmids) and secondary chromosomes. In principle, replicons should only be named plasmids when they lack essential genes. However, gene essentiality is rarely known experimentally at the moment of labeling a replicon as a plasmid or a chromosome. Furthermore, the definition of essentiality is controversial because some plasmid-encoded key traits, like nitrogen fixation in rhizobiales (Masson-Boivin et al. 2009) and virulence in many pathogens (Rankin et al. 2011), are not essential for growth in the laboratory but are effectively essential for the ecology of the bacterium in the environment. For example, the deletion of two megaplasmids encompassing >3 Mb (45%) of the Sinorhizobium meliloti genome produces a viable mutant that is highly impaired in terms of metabolic and mutualistism-associated functions (diCenzo et al. 2014). The largest plasmids known to encode homologs of E. coli essential genes are not conjugative and have nucleotide and codon compositions close to the chromosome, suggesting they are becoming domesticated as secondary chromosomes (Harrison et al. 2010; Smillie et al. 2010). The process of domestication has been poorly studied. It might involve a first step of plasmid stabilization (e.g., because of the presence of highly adaptive traits preventing plasmid segregation). Plasmid carriage can be costly and one might expect selection for translocation of the adaptive genes from the plasmid to the chromosome ultimately leading to plasmid loss. However, experimental evolution shows that plasmids and hosts can rapidly evolve to decrease and even erase this cost (Bourma and Lenski 1988). The long-term coevolution of the plasmid and the chromosome inevitably results in occasional translocation of chromosomal genes to the plasmid (and vice versa). As a result, certain traits start requiring the presence of both replicons to be expressed, further tightening the genetic link between the plasmids and the chromosome. As the number of exchanges between replicons accumulates, plasmids may acquire essential genes and effectively become fixed in the bacterial lineage. Hence, plasmids under selection for long periods of time are potential targets for domestication into secondary chromosomes (Touchon et al. 2014). This might explain why some secondary chromosomes replicate and segregate using plasmid-like mechanisms (Egan et al. 2005).
THE MAP OF THE CELL IS IN THE CHROMOSOME
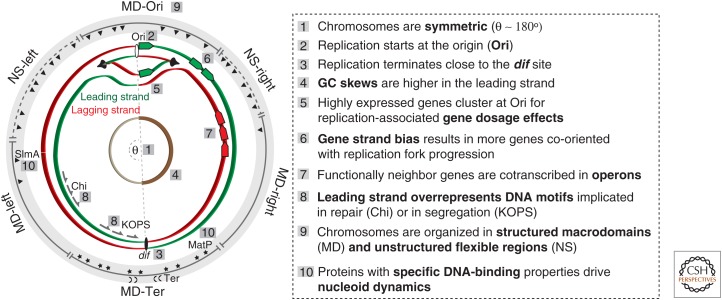
The observation of nonrandom gene-distribution patterns suggests that the organization of genetic elements in the chromosome and the structure of the chromosome are intimately linked with cell organization (Fig. 2) (Danchin and Henaut 1997; Ptacin and Shapiro 2013). In E. coli, intrachromosomal recombination assays revealed a chromosome organized into four macrodomains and two large unstructured regions (Valens et al. 2004). In newly replicated cells, the macrodomains around the origin (Ori) and terminus (Ter) of replication are localized near opposite cell poles leading to a linear arrangement of the genetic information in the cell (Niki et al. 2000). The sequence determinants of the macrodomains are unknown, with the exception of the Ter macrodomain that is organized by one DNA-binding protein (MatP) (Mercier et al. 2008), and insulated from the neighboring chromosomal regions by another (YfbV) (Thiel et al. 2012). The DNA sequence-specific association of proteins, like MatP, FtsK (Stouf et al. 2013), and SlmA (Tonthat et al. 2013), facilitate chromosome orientation in the function of replication and segregation in the cell. Macrodomains differ in the types of genes they encode and are associated with clusters of functionally neighbor genes that respond in concert, from a transcriptional point of view, to nucleoid perturbations (Scolari et al. 2011). The link between genome organization and chromosome structure might also drive the evolutionary rate of genes, because the density of point mutations is higher in the regions of higher superhelicity of the E. coli chromosome (Foster et al. 2013), and horizontally transferred genes accumulate in Ter-proximal macrodomains.
Figure 2.
Elements of genome organization. The cellular processes that interact with the chromosome shape its organization.
Genome organization linked to chromosome structure can drive developmental processes. Sporulation in Bacillus subtilis is regulated by the differential expression of two σ factors, one in the forespore (σF) and one in the mother cell (σK). The expression of σK is restricted to the mother cell because its expression requires the excision of a phage-like element from the genome, which occurs only in the mother-cell chromosome (Stragier et al. 1989). The sporulation septum bisects the B. subtilis cell asymmetrically, and initially only 30% of the chromosome is in the forespore compartment. At this stage, Ter-proximal regions are in two copies in the mother cell and absent from the forespore, thereby resulting in the absence of a σF repressor in the forespore. This allows expression of σF specifically in the forespore and further differentiation between the two cells (Frandsen et al. 1999). Translocation of the repressor to Ori-proximal regions abolishes the expression of the σF factor. Hence, regulatory dependencies can, in some cases, be traced from the order of genes in the chromosome.
The association between asymmetric division and chromosome structure has been thoroughly studied in Caulobacter crescentus. This chromosome is organized longitudinally along the cell following the Ori–Ter axis (Viollier et al. 2004), as it seems to be the case for other bacteria (Wang and Rudner 2014). The chromosome of C. crescentus shows 23 chromosomal interaction domains whose boundaries are associated with highly expressed genes (Le et al. 2013). Hence, the cellular organization of the C. crescentus chromosome recapitulates the genome map, and its chromosomal domains are determined by the order of genes in the genome. The transcripts of C. crescentus tend to remain physically close to the respective genes in the chromosome even after transcription termination (Montero Llopis et al. 2010). Proximal translation of these transcripts could facilitate the folding of heteromeric protein complexes encoded in neighboring operons. The close spatial association between protein complexes and the corresponding transcription units might result in the functional compartmentalization of bacterial cells, especially for large machineries that do not diffuse freely in the cytoplasm (Parry et al. 2013).
REPLICATION, RECOMBINATION, AND SEGREGATION
Bacterial chromosomes endure selection for replication symmetry, with origin and terminus separated by ∼180° in circular replicons, so that both forks complete replication synchronously. Chromosomal inversions lead to poorly growing bacteria that require the presence of specific recombination and segregation functions (Esnault et al. 2007; Lesterlin et al. 2008; Matthews and Maloy 2010). Replication forks advance rapidly on the chromosome displacing attached molecules, changing DNA modifications, and perturbing local and global nucleoid structures. As DNA replication is the key mechanism allowing transmission of heritability, the interactions of the molecules involved in this process with the chromosome are expected to be under very strong selection. This and the mechanistic asymmetries of replication drive large-scale organization of the genome (Fig. 2).
Presence of multiple replication forks in fast-growing bacteria produces a transient replication-associated gene dosage effect that leads to selection of highly expressed genes near the origin of replication (Couturier and Rocha 2006). Atypical genome configurations are not exception. Highly expressed genes concentrate near the multiple origins of replication of certain archaeal chromosomes (Andersson et al. 2010). The larger Vibrio chromosome enjoys stronger replication-associated gene dosage effects than the secondary chromosome and accumulates most highly expressed genes near its origin of replication (Dryselius et al. 2008). Replication-associated gene dosage effects can be efficiently counteracted by genetic regulation, so that lowly expressed genes can still be accommodated near the origin of replication (Block et al. 2012). Interestingly, the temporal pattern of gene expression in E. coli corresponds to the order of genes in the Ori–Ter axis of the genome (Couturier and Rocha 2006; Sobetzko et al. 2012).
The asymmetric functioning of the replication fork, producing a leading and a lagging strand, drives two broad organizational traits: GC skews and gene strand bias. The two DNA strands are replicated asymmetrically leading to different nucleotide composition in each strand (GC skews reviewed in Frank and Lobry 1999 and Touchon and Rocha 2008). Genes downstream from the replication fork may be transcribed by RNA polymerases in the same direction as the fork, leading eventually to co-oriented collisions, or in the opposite direction, leading to head-on collisions. The latter are much more deleterious and present a challenge to the integrity of the chromosome (Pomerantz and O’Donnell 2010; Srivatsan et al. 2010), and natural selection favors genes transcribed codirectionally with the replication fork (leading strand genes). This effect is very strong for essential genes, but also significant for highly expressed genes and large operons (Rocha and Danchin 2003; Omont and Kepes 2004; Price et al. 2005a). Intriguingly, certain categories of weakly expressed or even silent genes, such as prophages and other horizontally transferred genes, are highly abundant in the leading strand (Campbell 2002; Hao and Golding 2009). On the other hand, regulatory functions are more frequent in the lagging strand than expected (Mao et al. 2012). A number of models have been proposed to explain why selection against head-on collisions causes gene strand bias. They explain the deleterious effects of head-on collisions based on their effect on replication stalling (Mirkin and Mirkin 2007), mutagenesis (Srivatsan et al. 2010; Paul et al. 2013), production of truncated transcripts (Rocha and Danchin 2003), and induction of genome rearrangements (reviewed in Bermejo et al. 2012; Merrikh et al. 2012). It was suggested that bacterial genes under positive or diversifying selection might be in the lagging strand to enjoy increased mutagenesis associated with head-on collisions (Paul et al. 2013), but this has been contended on empirical and theoretical grounds (Chen and Zhang 2013).
The intimate link between replication and recombination (Michel et al. 2004) and segregation (Niki et al. 2000) also drives strand bias of motifs associated with these processes (reviewed in Touzain et al. 2011). Notably, leading strands are enriched in Chi motifs, which in a number of bacteria are involved in regulating the activity of RecBCD or the analogous AddAB complex in the early stages of homologous recombination (Halpern et al. 2007). FtsK-orienting polar sequence (KOPS) motifs are involved in chromosome segregation by FtsK. Their frequency increases regularly with the proximity to the terminus of replication and their strong overrepresentation in the leading strand provides information on the chromosome polarity to the segregation machinery (Bigot et al. 2005). The localization of motifs in certain regions of the chromosome is likely to constraint genome rearrangements and horizontal gene transfer (Hendrickson and Lawrence 2006).
OPERONS AND BEYOND
The majority of genes in prokaryotes are expressed under the form of polycistronic units called operons (Jacob and Monod 1961), including from two to dozens of genes (average ∼3–4 genes) (Zheng et al. 2002). The organization of genes in operons is a compact way of regulating gene expression because genes in the same operon are expressed at more similar rates than random pairs of genes (Sabatti et al. 2002; Price et al. 2006). Pairs of contiguous genes in operons are highly conserved showing rearrangement rates orders of magnitude lower than other interoperonic pairs (de Daruvar et al. 2002; Rocha 2006; Moreno-Hagelsieb and Janga 2008). Larger genomes tend to have fewer genes in operons, shorter and less conserved operons, and many more transcription factors (Cherry 2003; Minezaki et al. 2005; Nunez et al. 2013). Importantly, the number of transcription factors in a genome, once controlled for genome size, is negatively associated with operon conservation (Nunez et al. 2013). These observations could be explained by the existence of a trade-off between the advantages of individual gene regulation, requiring transcription factors, and coregulation of several genes by a single operon, constraining the expression of each individual gene. Large genomes have more complex genetic networks and many more different transcription factors than small genomes. This might explain increased selection for operons in small genomes.
Genes in operons often encode physically interacting proteins (Mushegian and Koonin 1996; Huynen et al. 2000) or functional neighbors (Rogozin et al. 2002). For example, operons often encode enzymes of consecutive steps in metabolic pathways (Zaslaver et al. 2006), which has been proposed to reduce stochastic stalling of metabolism at low-expression levels (Kovacs et al. 2009). Transcription factors tend to be encoded at the edges of operons, and autorepressors are often the first genes in an operon (Rubinstein et al. 2011). The systematic association between functionally related genes in operons allows the use of guilt-by-association methods to characterize unknown function genes (Overbeek et al. 1999; Moreno-Hagelsieb and Janga 2008). Genomic colocalization of genes expressed at the same moment further contributes to link the organization of genetic loci with the subcellular location of certain physiological processes by way of chromosome structure. For example, colocalization of highly expressed genes leads to transcription foci in the cell with a high concentration of active RNA polymerases (Cagliero et al. 2013).
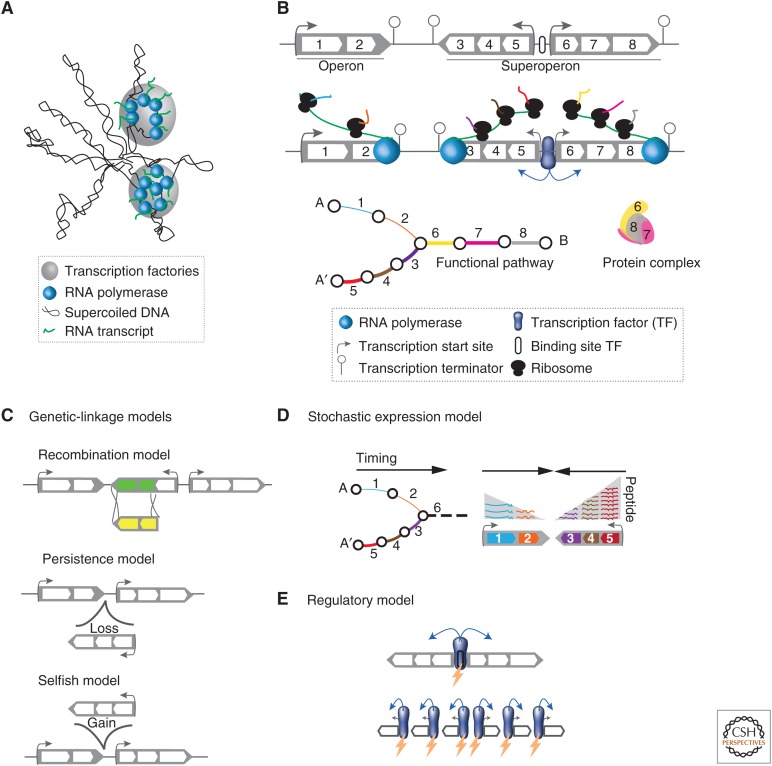
Despite the impact of operons in the regulation of gene expression and of the constraints they impose on the organization of the bacterial chromosome, there is no consensus yet on why operons are formed and conserved. A number of models have been proposed to explain operon formation based on the effects of genetic linkage, stochastic gene expression, and gene regulation (Fig. 3). Genetic linkage could favor physical clustering of coevolving genes to avoid breaking coadaptive changes by recombination (recombination model) (Stahl and Murray 1966) or to lower the cost of large genetic deletions (persistence model) (Fang et al. 2008). Operons could also facilitate the horizontal transfer of coregulated functional modules (selfish operon model) (Lawrence and Roth 1996). Most prokaryotic cells are small and most genes in the genome are expressed at relatively low levels. These conditions lead to important stochasticity in gene expression (Elowitz et al. 2002). Operons could minimize shortfall or waste in gene expression because cotranscription and translational coupling synchronize the expression of the different components of the same functional module (stochastic expression models) (Swain 2004; Lovdok et al. 2009; Sneppen et al. 2010; Ray and Igoshin 2012). By definition, operons are sets of genes under the control of a single transcription start site. This arrangement concentrates selection pressure for regulatory sequences in a single region. This could favor the optimization of the associated DNA motifs and protect them against mutation pressure (regulatory models) (Price et al. 2005b; Lynch 2006).
Figure 3.
The genetic organization of gene expression. (A) Schematic representation of the transcription factory model, in which multiple active RNA polymerases are concentrated at discrete sites in the nucleoid. (B) Bacterial genes are organized into operons, which group in superoperons. In addition to being physically close in the genome, genes in operons are cotranscribed, coregulated, and encode proteins involved in the same functional pathway or protein complex. Superoperons are not cotranscribed, but may share regulatory regions. (C) Schematic representation of three models aiming at explaining the formation and conservation of operons based on genetic linkage. (D) Transcription of genes in a single transcript is expected to diminish gene expression noise and ensure more precise stoichiometry. It also allows responding optimally to demand for a given pathway when the first genes to be transcribed are those starting the functional pathway. (E) Operons place several genes under the same regulatory region that is thus subject to more efficient selection.
Comparative studies have presented arguments against some of these models. Notably, both the recombination and the persistence model explain gene clustering but not cotranscription, and the former may not be compatible with the small size of recombination tracts typically observed in bacteria (Kennemann et al. 2011). The selfish operon model may explain the formation of operons of frequently transferred genes, but fails to explain why essential genes are more often found in both ancient and recent operons (Pal and Hurst 2004; Price et al. 2005b) and why larger genomes enduring more horizontal transfer have fewer and less conserved operons. Models based on minimization of gene expression noise fail to explain why operons of highly expressed genes, the ones for which gene expression noise is less important, are the most highly conserved (Nunez et al. 2013). Regulatory models seem more in accordance with the available data. Nevertheless, other models might contribute to explain the formation and conservation of certain kinds of operons (e.g., the selfish model for frequently transferred genes, the persistence model for essential genes, and models invoking gene expression stochasticity for lowly expressed genes).
Recent data suggests that the organization of transcription at the genomic level is more plastic than previously thought because of frequent alternative transcription start sites (Cho et al. 2009), chromosome structure (Bryant et al. 2014), and supraoperonic organization (Lathe et al. 2000; Warren and ten Wolde 2004; Hershberg et al. 2005). It is therefore possible that intraoperonic, operonic, and supraoperonic gene organization represent a continuum of scales of transcriptional organization. Indeed, gene clustering in prokaryotic genomes extends beyond operons. Functionally neighbor operons are often encoded in neighboring regions of the genome, leading to strong patterns of operon pairs conservation (Korbel et al. 2004). There is also evidence of large-scale clustering (Bailly-Bechet et al. 2006; Fritsche et al. 2012) and of periodic organization (Junier et al. 2012) of genes encoding neighboring housekeeping functions or genes expressed at similar levels. Several hypotheses were proposed to explain supraoperonic gene organization. These involve horizontal gene transfer, chromosome structure, gene regulation, and mRNA management. The selfish operon model is based on the idea that the success of horizontal gene transfer is higher when it includes neighboring functions that can constitute functional modules. When functional modules are very large, they require multiple contiguous operons, and this might explain the clustering of genes encoding virulence factors or antibiotic resistance in genomic islands of pathogens (Dobrindt et al. 2004; Juhas et al. 2009). Operons encoding closely associated functions are expressed at the same time. If these operons are encoded close in the genome, they are likely to be close in the nucleoid. Genomic colocalization of coexpressed operons might be favored because their expression would require opening the same nucleoid region (Jin et al. 2013). Proteins often participate in different cellular processes and, therefore, are functional neighbors of many different proteins. Distances between operons might reflect a compromise between the gene expression requirements of these different processes, leading to complex patterns of gene clustering at supraoperonic levels (Yin et al. 2010). The presence of different functionalities in different species might thus produce a variety of genetic architectures of functionally related genes. Finally, operons physically close in the chromosome show correlated patterns of mRNA degradation (Selinger et al. 2003; Montero Llopis et al. 2010). The genomic colocalization of operons with similar patterns of mRNA demand and degradation might also favor supraoperonic gene organization. Further studies will be needed to understand how organizational traits beyond the operon reflect or constrain the evolution of genetic networks and chromosome structure.
VARIATIONS IN GENE REPERTOIRES IN THE LIGHT OF GENOME ORGANIZATION
The previous sections showed that a large number of organizational traits are under selection in the genomes of prokaryotes. Organizational traits are strongly affected by genome rearrangements, because a single rearrangement can render chromosomes asymmetric, break operons, and disrupt chromosome domains. Spontaneous rearrangement rates are high, often of the order of genomic mutation rates (Sun et al. 2012), but divergent genomes show remarkably few fixed large rearrangements (Rocha 2006). This is consistent with the view that most large rearrangements are deleterious and removed by purifying selection. Because replication and associated mechanisms are responsible for the organization of the chromosome at very large scales, rearrangements breaking chromosome symmetry or gene strand biases are highly counterselected (Eisen et al. 2000; Tillier and Collins 2000; Mackiewicz et al. 2001; Liu et al. 2006; Darling et al. 2008). Genetic elements favoring rearrangements, such as DNA repeats, are also counterselected, especially when their location leads to particularly deleterious changes (Achaz et al. 2003). This suggests a trade-off between selection for the organization of genomes and selection for their diversification by intrachromosomal recombination.
The gene repertoires of prokaryotes evolve extremely fast (Tettelin et al. 2008; Kuo and Ochman 2009b; Polz et al. 2013), and acquisitions of new genes occur mostly by horizontal gene transfer (Treangen and Rocha 2011). Most incoming DNA is rapidly lost as it corresponds to genetic information that is either deleterious or of no adaptive value (Kuo and Ochman 2009a; Koskiniemi et al. 2012; Lee and Marx 2012). The size of the bacterial genome results from the equilibrium between the rates of acquisition and deletion of genetic material. Mobile genetic elements have a key role in horizontal transfer (Frost et al. 2005). For example, the E. coli O157:H7 strain encodes 25% more genes than the standard MG1655 strain and includes 18 prophages and several plasmids encoding most of the strain’s virulence factors (Perna et al. 2001; Ogura et al. 2007). An analysis of 20 complete E. coli strains showed that the pangenome is four times larger and the core genome two times smaller than the average genome of the species (Touchon et al. 2009). How can such massive influx of genetic material be compatible with the above-mentioned principles of genomic organization?
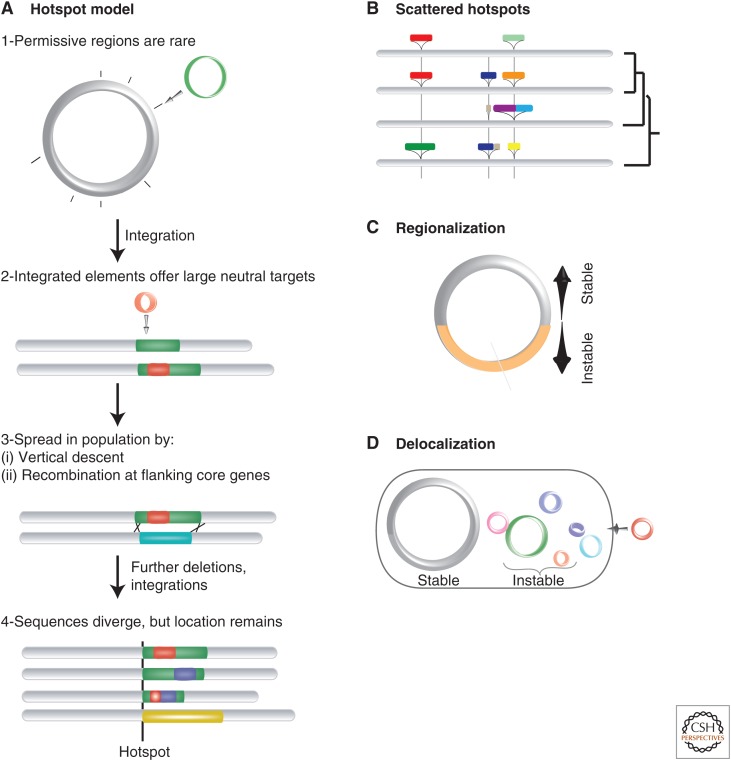
Although systematic studies of this question are still unavailable, a number of different patterns have been observed. Most of the accessory E. coli genome is found in a very small number of loci—integration hotspots—that are conserved among strains and even among species (Touchon et al. 2009). The origin and maintenance of these hotspots is probably the combined result of integration biases and natural selection (Fig. 4). Some hotspots are located next to genetic elements that are frequently targeted by mobile genetic elements for integration in the chromosome, like tRNAs (Williams 2002). Yet, the lack of such genes in many hotspots suggests the presence of other evolutionary mechanisms, possibly involving selection. For example, large integrations might provide a neutral ground for further insertions and deletions, thereby promoting the creation of hotspots. Hotspots might be subsequently transferred to other cells and incorporated in the chromosome by double homologous recombination at the flanking core genes (Schubert et al. 2009). Interestingly, hotspots are not randomly distributed in genomes. They are typically intergenic and tend to accumulate in regions closer to the terminus of replication and in secondary chromosomes (Okada et al. 2005; Andersson et al. 2010; Flynn et al. 2010). Some genomes have only a few very large regions that accommodate most of the accessory genomes (Fig. 4). The two chromosome arms of Streptomyces make up half of the genome and accumulate most of gene gain and loss in the species (Bentley et al. 2002; Choulet et al. 2006). In other genomes, most gene flux is confined to extrachromosomal mobile genetic elements. In Borrelia burgdorferi, the chromosome is very stable, whereas a large plasmid pool accommodates most of the repeats involved in antigenic variation (Casjens et al. 2000; Qiu et al. 2004). There are, therefore, different ways of reconciling strong selection for genome organization and for sequence diversification. Usually, they involve the confinement of genetic plasticity to certain regions of the genome, inside or outside the main chromosome, which preserves the organization of the regions encoding essential and highly expressed genes.
Figure 4.
Balancing between genome organization and diversification. (A) Schematic representation of a model for the formation and evolution of integration hotspots. (B) Hotspots can be scattered in the chromosome. (C) Some chromosomes show very large regions in which most genetic diversification takes place. (D) Some chromosomes are very stable and most genetic diversification takes place in plasmids.
CONCLUDING REMARKS
Recent advances have provided a much clearer view of genome organization and chromosome structure. The latter, given experimental hurdles, has for the moment concerned only a few model species. It would be most interesting to understand the evolution of chromosome structure and its coevolution with genome organization. This would help to identify the chromosome structural features that constrain and are constrained by genome organization. These studies might facilitate the identification of chromosomal domains and the mechanisms underlying their formation.
Some integrative mobile elements are hundreds of kilobases long and this must have some effect on the chromosome structure and on genome organization. Prophages are more frequent closer to the terminus of replication and they encode DNA motifs that match the local concentration of these motifs in the bacterial chromosome (Bobay et al. 2013). This suggests that mobile elements select for motifs that allow their seamless integration in the bacterial genome. Future work will hopefully unravel how the chromosome accommodates these and other mobile elements with little or no impact on fitness.
Many compositional patterns have been observed in genomes, including variations in intra- and intergenomic G+C composition (Muto and Osawa 1987; Daubin and Perriere 2003), GC skews (Lobry 1996), or the pervasive AT richness of horizontally transferred genes (Lawrence and Ochman 1997; Daubin et al. 2003). So far, the precise molecular mechanisms behind these patterns have remained elusive (Rocha et al. 2006; Hershberg and Petrov 2010; Hildebrand et al. 2010; Raghavan et al. 2012). Yet, these mechanisms have a very important impact in sequence evolution, because they affect substitution rates (Lee et al. 2012), codon usage bias (Novembre 2002), mobility and expression of mobile elements (Dorman 2014), horizontal transfer (Doyle et al. 2007), and amino acid composition (Lobry 1997). Shifts in compositional patterns are also likely to complicate evolutionary analyses (Galtier and Gouy 1995).
Statistical approaches to bacterial population genomics have focused on the study of nucleotide substitutions in the core genome. This allows understanding phylogenetic and epidemiological patterns (Parkhill and Wren 2011). However, to understand how bacteria adapt, one must also study the population patterns of gene gain and loss. Some recent works have started to put forward population genetics techniques to study genetic mobility (Baumdicker et al. 2012; Collins and Higgs 2012; Lobkovsky et al. 2013). Further work is required to model the dynamics of gene repertoires, compare it with neutral processes, and highlight which new genes are effectively adaptive.
Organizational patterns can be used to predict genetic features, like origins of replication (Lobry 1996) or transcription units (Salgado et al. 2000), and physiological traits, such as optimal growth temperature (Zeldovich et al. 2007) or minimal doubling times (Vieira-Silva and Rocha 2010). Large-scale engineering projects of bacterial genomes benefit from using known genome organization rules (Kepes et al. 2012). Such projects have already provided important clues on the constraints acting on the evolution of prokaryotic genomes, sometimes with surprising results. For example, although natural linear E. coli chromosomes have not been observed, E. coli’s chromosome can be artificially linearized with no effect on growth (Cui et al. 2007) and even split into two linear chromosomes with only a slight growth defect (Liang et al. 2013). Linear chromosomes can also be circularized (Volff et al. 1997). Laboratory manipulations have shown that chromosomes can double in size in a small number of events (Itaya et al. 2005), be split in multiple chromosomes (Itaya and Tanaka 1997), and multiple chromosomes can be merged into one (Val et al. 2012). The effects of these dramatic structural modifications are as small as the rules of genome organization are respected. This opens the possibility of developing synthetic bacteria that can be made to evolve under new sets of physiological or ecological constraints to unravel how chromosome structure and genome organization coevolve.
ACKNOWLEDGMENTS
We thank laboratory members who have participated in our work on this topic. Work in our laboratory is funded by a European Research Council Grant (EVOMOBILOME No. 281605).
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Abby S, Daubin V. 2007. Comparative genomics and the evolution of prokaryotes. Trends Microbiol 15: 135–141. [DOI] [PubMed] [Google Scholar]
- Achaz G, Coissac E, Netter P, Rocha EPC. 2003. Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics 164: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Pelve EA, Lindeberg S, Lundgren M, Nilsson P, Bernander R. 2010. Replication-biased genome organisation in the crenarchaeon Sulfolobus. BMC Genomics 11: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M, Danchin A, Iqbal M, Marsili M, Vergassola M. 2006. Codon usage domains over bacterial chromosomes. PLoS Comput Biol 2: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumdicker F, Hess WR, Pfaffelhuber P. 2012. The infinitely many genes model for the distributed genome of bacteria. Genome Biol Evol 4: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. 2012. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell 45: 710–718. [DOI] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F. 2005. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. Embo J 24: 3770–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block DH, Hussein R, Liang LW, Lim HN. 2012. Regulatory consequences of gene translocation in bacteria. Nucleic Acids Res 40: 8979–8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, Rocha EPC, Touchon M. 2013. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol 30: 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourma JE, Lenski RE. 1988. Evolution of a bacteria/plasmid association. Nature 335: 351–352. [DOI] [PubMed] [Google Scholar]
- Boussau B, Daubin V. 2010. Genomes as documents of evolutionary history. Trends Ecol Evol 25: 224–232. [DOI] [PubMed] [Google Scholar]
- Boussau B, Karlberg EO, Frank AC, Legault BA, Andersson SG. 2004. Computational inference of scenarios for α-proteobacterial genome evolution. Proc Natl Acad Sci 101: 9722–9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JA, Sellars LE, Busby SJ, Lee DJ. 2014. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res 42: 11383–11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliero C, Grand RS, Jones MB, Jin DJ, O’Sullivan JM. 2013. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res 41: 6058–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AM. 2002. Preferential orientation of natural lambdoid prophages and bacterial chromosome organization. Theor Popul Biol 61: 503–507. [DOI] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. 2000. A bacterial genome in flux: The twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35: 490–516. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J. 2013. Why are genes encoded on the lagging strand of the bacterial genome? Genome Biol Evol 5: 2436–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JL. 2003. Genome size and operon content. J Theor Biol 221: 401–410. [DOI] [PubMed] [Google Scholar]
- Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 27: 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Aigle B, Gallois A, Mangenot S, Gerbaud C, Truong C, Francou FX, Fourrier C, Guerineau M, Decaris B, et al. 2006. Evolution of the terminal regions of the streptomyces linear chromosome. Mol Biol Evol 23: 2361–2369. [DOI] [PubMed] [Google Scholar]
- Collins RE, Higgs PG. 2012. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol Biol Evol 29: 3413–3425. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. 2010. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput Biol 6: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. 2009. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci 106: 21748–21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier E, Rocha E. 2006. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol 59: 1506–1518. [DOI] [PubMed] [Google Scholar]
- Cui T, Moro-oka N, Ohsumi K, Kodama K, Ohshima T, Ogasawara N, Mori H, Wanner B, Niki H, Horiuchi T. 2007. Escherichia coli with a linear genome. EMBO Rep 8: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin A, Henaut A. 1997. The map of the cell is in the chromosome. Curr Opin Genet Dev 7: 852–854. [DOI] [PubMed] [Google Scholar]
- Darling AE, Miklos I, Ragan MA. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet 4: e1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Perriere G. 2003. G+C3 structuring along the genome: A common feature in prokaryotes. Mol Biol Evol 20: 471–483. [DOI] [PubMed] [Google Scholar]
- Daubin V, Lerat E, Perriere G. 2003. The source of laterally transferred genes in bacterial genomes. Genome Biol 4: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Daruvar A, Collado-Vides J, Valencia A. 2002. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J Mol Evol 55: 211–221. [DOI] [PubMed] [Google Scholar]
- de la Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: Lessons from bacteria. Trends Microbiol 8: 128–133. [DOI] [PubMed] [Google Scholar]
- diCenzo GC, MacLean AM, Milunovic B, Golding GB, Finan TM. 2014. Examination of prokaryotic multipartite genome evolution through experimental genome reduction. PLoS Genet 10: e1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2: 414–424. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. 2014. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 75C: 1–11. [DOI] [PubMed] [Google Scholar]
- Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315: 251–252. [DOI] [PubMed] [Google Scholar]
- Dryselius R, Izutsu K, Honda T, Iida T. 2008. Differential replication dynamics for large and small Vibrio chromosomes affect gene dosage, expression and location. BMC Genomics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Fogel MA, Waldor MK. 2005. Divided genomes: Negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Heidelberg JF, White O, Salzberg SL. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol 1: 11.11– 11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. [DOI] [PubMed] [Google Scholar]
- Esnault E, Valens M, Espeli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Rocha EP, Danchin A. 2008. Persistence drives gene clustering in bacterial genomes. BMC Genomics 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn KM, Vohr SH, Hatcher PJ, Cooper VS. 2010. Evolutionary rates and gene dispensability associate with replication timing in the archaeon Sulfolobus islandicus. Genome Biol Evol 2: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Hanson AJ, Lee H, Popodi EM, Tang H. 2013. On the mutational topology of the bacterial genome. G3 (Bethesda) 3: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N, Barák I, Karmazyn-Campelli C, Stragier P. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev 13: 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. 1999. Asymmetric patterns: A review of possible underlying mutational or selective mechanisms. Gene 238: 65–77. [DOI] [PubMed] [Google Scholar]
- Fritsche M, Li S, Heermann DW, Wiggins PA. 2012. A model for Escherichia coli chromosome packaging supports transcription factor-induced DNA domain formation. Nucleic Acids Res 40: 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol 3: 722–732. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. 1995. Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci 92: 11317–11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol Lett 323: 124–131. [DOI] [PubMed] [Google Scholar]
- Guieysse B, Wuertz S. 2012. Metabolically versatile large-genome prokaryotes. Curr Opin Biotechnol 23: 467–473. [DOI] [PubMed] [Google Scholar]
- Halpern D, Chiapello H, Schbath S, Robin S, Hennequet-Antier C, Gruss A, El Karoui M. 2007. Identification of DNA motifs implicated in maintenance of bacterial core genomes by predictive modeling. PLoS Genet 3: 1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Golding GB. 2009. Does gene translocation accelerate the evolution of laterally transferred genes? Genetics 182: 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Lower RP, Kim NK, Young JP. 2010. Introducing the bacterial “chromid”: Not a chromosome, not a plasmid. Trends Microbiol 18: 141–148. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. 2006. Selection for chromosome architecture in bacteria. J Mol Evol 62: 615–629. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Yeger-Lotem E, Margalit H. 2005. Chromosomal organization is shaped by the transcription regulatory network. Trends Genet 21: 138–142. [DOI] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet 6: e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. 2011. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol 193: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W III, Bork P. 2000. Predicting protein function by genomic context: Quantitative evaluation and qualitative inferences. Genome Res 10: 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T. 2013. Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol 79: 5013–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M, Tanaka T. 1997. Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc Natl Acad Sci 94: 5378–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M, Tsuge K, Koizumi M, Fujita K. 2005. Combining two genomes in one cell: Stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc Natl Acad Sci 102: 15971–15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C, Zhou YN. 2013. Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chem Rev 113: 8662–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33: 376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier I, Herisson J, Kepes F. 2012. Genomic organization of evolutionarily correlated genes in bacteria: Limits and strategies. J Mol Biol 419: 369–386. [DOI] [PubMed] [Google Scholar]
- Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, et al. 2011. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci 108: 5033–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepes F, Jester BC, Lepage T, Rafiei N, Rosu B, Junier I. 2012. The layout of a bacterial genome. FEBS Lett 586: 2043–2048. [DOI] [PubMed] [Google Scholar]
- Komaki K, Ishikawa H. 1999. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol 48: 717–722. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci 101: 3160–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Jensen LJ, von Mering C, Bork P. 2004. Analysis of genomic context: Prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol 22: 911–917. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Sun S, Berg OG, Andersson DI. 2012. Selection-driven gene loss in bacteria. PLoS Genet 8: e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs K, Hurst LD, Papp B. 2009. Stochasticity in protein levels drives colinearity of gene order in metabolic operons of Escherichia coli. PLoS Biol 7: e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. 2009a. Deletional bias across the three domains of life. Genome Biol Evol 1: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. 2009b. The fate of new bacterial genes. FEMS Microbiol Rev 33: 38–43. [DOI] [PubMed] [Google Scholar]
- Kuo CH, Moran NA, Ochman H. 2009. The consequences of genetic drift for bacterial genome complexity. Genome Res 19: 1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe WC, Snel B, Bork P. 2000. Gene context conservation of a higher order than operons. Trends Biochem Sci 25: 474–479. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. 1997. Amelioration of bacterial genomes: Rates of change and exchange. J Mol Evol 44: 383–397. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR. 1996. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics 143: 1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Marx CJ. 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet 8: e1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci 109: E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. 2008. Asymmetry of chromosome replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet 4: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Baek CH, Katzen F. 2013. Escherichia coli with two linear chromosomes. ACS Synth Biol 2: 734–740. [DOI] [PubMed] [Google Scholar]
- Liu GR, Liu WQ, Johnston RN, Sanderson KE, Li SX, Liu SL. 2006. Genome plasticity and ori-ter rebalancing in Salmonella typhi. Mol Biol Evol 23: 365–371. [DOI] [PubMed] [Google Scholar]
- Lobkovsky AE, Wolf YI, Koonin EV. 2013. Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 5: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13: 660–665. [DOI] [PubMed] [Google Scholar]
- Lobry JR. 1997. Influence of genomic G+C content on average amino-acid composition of proteins from 59 bacterial species. Gene 205: 309–316. [DOI] [PubMed] [Google Scholar]
- Lovdok L, Bentele K, Vladimirov N, Muller A, Pop FS, Lebiedz D, Kollmann M, Sourjik V. 2009. Role of translational coupling in robustness of bacterial chemotaxis pathway. PLoS Biol 7: e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2006. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol 60: 327–349. [DOI] [PubMed] [Google Scholar]
- Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S. 2001. Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biol 2: interactions1004.1–1004.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3: 144–156. [DOI] [PubMed] [Google Scholar]
- Mao X, Zhang H, Yin Y, Xu Y. 2012. The percentage of bacterial genes on leading versus lagging strands is influenced by multiple balancing forces. Nucleic Acids Res 40: 8210–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C, Giraud E, Perret X, Batut J. 2009. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol 17: 458–466. [DOI] [PubMed] [Google Scholar]
- Matthews TD, Maloy S. 2010. Fitness effects of replichore imbalance in Salmonella enterica. J Bacteriol 192: 6086–6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26. [DOI] [PubMed] [Google Scholar]
- Mendell JE, Clements KD, Choat JH, Angert ER. 2008. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci 105: 6730–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O. 2008. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135: 475–485. [DOI] [PubMed] [Google Scholar]
- Merrikh H, Zhang Y, Grossman AD, Wang JD. 2012. Replication–transcription conflicts in bacteria. Nat Rev Microbiol 10: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Grompone G, Florès MJ, Bidnenko V. 2004. Multiple pathways process stalled replication forks. Proc Natl Acad Sci 101: 12783–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Nishikawa K. 2005. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res 12: 269–280. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet 17: 589–596. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. 2007. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Janga SC. 2008. Operons and the effect of genome redundancy in deciphering functional relationships using phylogenetic profiles. Proteins 70: 344–352. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. 1996. Gene order is not conserved in bacterial evolution. Trends Genet 12: 289–290. [DOI] [PubMed] [Google Scholar]
- Muto A, Osawa S. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci 84: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223. [PMC free article] [PubMed] [Google Scholar]
- Novembre JA. 2002. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol 19: 1390–1394. [DOI] [PubMed] [Google Scholar]
- Nunez PA, Romero H, Farber MD, Rocha EP. 2013. Natural selection for operons depends on genome size. Genome Biol Evol 5: 2242–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Ooka T, Asadulghani, Terajima J, Nougayrede JP, Kurokawa K, Tashiro K, Tobe T, Nakayama K, Kuhara S, et al. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol 8: R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Iida T, Kita-Tsukamoto K, Honda T. 2005. Vibrios commonly possess two chromosomes. J Bacteriol 187: 752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omont N, Kepes F. 2004. Transcription/replication collisions cause bacterial transcription units to be longer on the leading strand of replication. Bioinformatics 20: 2719–2725. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci 96: 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Hurst LD. 2004. Evidence against the selfish operon theory. Trends Genet 20: 232–234. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW. 2011. Bacterial epidemiology and biology—Lessons from genome sequencing. Genome Biol 12: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry BR, Surovtsev IV, Cabeen MT, O’Hern CS, Dufresne ER, Jacobs-Wagner C. 2013. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. 2013. Accelerated gene evolution through replication-transcription conflicts. Nature 495: 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529–533. [DOI] [PubMed] [Google Scholar]
- Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Alm EJ, Arkin AP. 2005a. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res 33: 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Huang KH, Arkin AP, Alm EJ. 2005b. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res 15: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Arkin AP, Alm EJ. 2006. The life cycle of operons. PLoS Genet 2: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Shapiro L. 2013. Chromosome architecture is a key element of bacterial cellular organization. Cell Microbiol 15: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, Dunn JJ, Fraser CM, Casjens SR, Luft BJ. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc Natl Acad Sci 101: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Kelkar YD, Ochman H. 2012. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci 109: 14504–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin DJ, Rocha EPC, Brown SP. 2011. What traits are carried on mobile genetic elements, and why? Heredity 104: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV. 2011. N15: The linear phage-plasmid. Plasmid 65: 102–109. [DOI] [PubMed] [Google Scholar]
- Ray JC, Igoshin OA. 2012. Interplay of gene expression noise and ultrasensitive dynamics affects bacterial operon organization. PLoS Comput Biol 8: e1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. 2006. Inference and analysis of the relative stability of bacterial chromosomes. Mol Biol Evol 23: 513–522. [DOI] [PubMed] [Google Scholar]
- Rocha EPC. 2008. The organization of the bacterial genome. Annu Rev Genet 42: 211–233. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A. 2003. Gene essentiality as a determinant of chromosomal organization in bacteria. Nucleic Acids Res 31: 6570–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, Touchon M, Feil EJ. 2006. Similar compositional biases are caused by very different mutational effects. Genome Res 16: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, Szekely LA, Koonin EV. 2002. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res 30: 2212–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein ND, Zeevi D, Oren Y, Segal G, Pupko T. 2011. The operonic location of auto-transcriptional repressors is highly conserved in bacteria. Mol Biol Evol 28: 3309–3318. [DOI] [PubMed] [Google Scholar]
- Sabatti C, Rohlin L, Oh MK, Liao JC. 2002. Co-expression pattern from DNA microarray experiments as a tool for operon prediction. Nucleic Acids Res 30: 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. 2000. Operons in Escherichia coli: Genomic analyses and predictions. Proc Natl Acad Sci 97: 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25: 1281–1289. [DOI] [PubMed] [Google Scholar]
- Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E. 2009. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5: e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolari VF, Bassetti B, Sclavi B, Lagomarsino MC. 2011. Gene clusters reflecting macrodomain structure respond to nucleoid perturbations. Mol Biosyst 7: 878–888. [DOI] [PubMed] [Google Scholar]
- Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res 13: 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74: 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneppen K, Pedersen S, Krishna S, Dodd I, Semsey S. 2010. Economy of operon formation: Cotranscription minimizes shortfall in protein complexes. MBio 1: e00177–e00177-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci 109: E42–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. 2013. Evolutionary advantages of polyploidy in halophilic archaea. Biochem Soc Trans 41: 339–343. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6: e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW, Murray NE. 1966. The evolution of gene clusters and genetic circularity in microorganisms. Genetics 53: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouf M, Meile JC, Cornet F. 2013. FtsK actively segregates sister chromosomes in Escherichia coli. Proc Natl Acad Sci 110: 11157–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Kunkel B, Kroos L, Losick R. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243: 507–512. [DOI] [PubMed] [Google Scholar]
- Sun S, Ke R, Hughes D, Nilsson M, Andersson DI. 2012. Genome-wide detection of spontaneous chromosomal rearrangements in bacteria. PLoS ONE 7: e42639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PS. 2004. Efficient attenuation of stochasticity in gene expression through post-transcriptional control. J Mol Biol 344: 965–976. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O’Neill K, Tolstoy I, Zaslavsky L. 2015. Update on RefSeq microbial genomes resources. Nucleic Acids Res 43: D599–D605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: The bacterial pan-genome. Curr Opin Microbiol 11: 472–477. [DOI] [PubMed] [Google Scholar]
- Thiel A, Valens M, Vallet-Gely I, Espeli O, Boccard F. 2012. Long-range chromosome organization in E. coli: A site-specific system isolates the Ter macrodomain. PLoS Genet 8: e1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. 2000. Genome rearrangement by replication-directed translocation. Nat Genet 26: 195–197. [DOI] [PubMed] [Google Scholar]
- Tobiason DM, Seifert HS. 2006. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol 4: e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. 2013. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci 110: 10586–10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Rocha EP. 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol 24: 969–981. [DOI] [PubMed] [Google Scholar]
- Touchon M, Rocha EP. 2008. From GC skews to wavelets: A gentle guide to the analysis of compositional asymmetries in genomic data. Biochimie 90: 648–659. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bobay LM, Rocha EP. 2014. The chromosomal accommodation and domestication of mobile genetic elements. Curr Opin Microbiol 22: 22–29. [DOI] [PubMed] [Google Scholar]
- Touzain F, Petit MA, Schbath S, El Karoui M. 2011. DNA motifs that sculpt the bacterial chromosome. Nat Rev Microbiol 9: 15–26. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Rocha E. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 7: e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet 8: e1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. 2004. Macrodomain organization of the Escherichia coli chromosome. Embo J 23: 4330–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S, Rocha EPC. 2010. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet 6: e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuelas J, Febvay G, Duport G, Colella S, Fayard JM, Charles H, Rahbe Y, Calevro F. 2011. Multimodal dynamic response of the Buchnera aphidicola pLeu plasmid to variations in leucine demand of its host, the pea aphid Acyrthosiphon pisum. Mol Microbiol 81: 1271–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci 101: 9257–9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Viell P, Altenbuchner J. 1997. Artificial circularization of the chromosome with concomitant deletion of its terminal inverted repeats enhances genetic instability and genome rearrangement in Streptomyces lividans. Mol Gen Genet 253: 753–760. [DOI] [PubMed] [Google Scholar]
- Wang X, Rudner DZ. 2014. Spatial organization of bacterial chromosomes. Curr Opin Microbiol 22C: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren PB, ten Wolde PR. 2004. Statistical analysis of the spatial distribution of operons in the transcriptional regulation network of Escherichia coli. J Mol Biol 342: 1379–1390. [DOI] [PubMed] [Google Scholar]
- Williams KP. 2002. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: Sublocation preference of integrase subfamilies. Nucleic Acids Res 30: 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhang H, Olman V, Xu Y. 2010. Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome. Proc Natl Acad Sci 107: 6310–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443: 569–573. [DOI] [PubMed] [Google Scholar]
- Zaslaver A, Mayo A, Ronen M, Alon U. 2006. Optimal gene partition into operons correlates with gene functional order. Phys Biol 3: 183–189. [DOI] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol 3: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerulla K, Chimileski S, Nather D, Gophna U, Papke RT, Soppa J. 2014. DNA as a phosphate storage polymer and the alternative advantages of polyploidy for growth or survival. PLoS ONE 9: e94819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Szustakowski JD, Fortnow L, Roberts RJ, Kasif S. 2002. Computational identification of operons in microbial genomes. Genome Res 12: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]