Abstract
Hemolytic disease of the fetus and newborn (HDFN) is a condition characterized by a decreased lifespan of fetal red blood cells caused by maternally produced allospecific antibodies transferred to the fetus during pregnancy. The antibodies bind to the corresponding blood group antigens on fetal red blood cells and induce hemolysis. Cell-free DNA derived from the conceptus circulates in maternal blood. Using next-generation sequencing (NGS), it can be determined if this cell-free fetal DNA encodes the corresponding blood group antigen that is the target of the maternal allospecific antibodies. This determination carries no risk to the fetus. It is important to determine if the fetus is at risk of hemolysis to enable timely intervention. Many tests for blood groups are based solely on the presence or absence of a single nucleotide polymorphism (SNP). Antenatal determination of fetal blood group by NGS analysis holds advantages over polymerase chain reaction (PCR) determination based on allele specific amplification.
Maternal immunological incompatibility with fetal blood groups can have serious consequences for the fetus. Rapid progress is being made to develop next-generation sequencing strategies for evaluating fetal DNA in maternal blood.
The underlying mechanism of hemolytic disease of the fetus and newborn (HDFN) is maternal immunological incompatibility with fetal blood groups leading to the production of maternal antibodies against one or more blood group antigens residing on fetal red blood cells (RBC) (Urbaniak and Greiss 2000; Moise 2008). Maternal IgG antibodies are transferred through the placenta, enter the fetal circulation, and bind to fetal RBCs, leading to a decrease in the lifespan of the fetal red cell. This causes the fetus to produce more RBCs. If the balance of production versus removal of RBCs becomes too unfavorable for the fetus, severe anemia can develop, even leading to fetal demise. Determining if the fetus is at risk of antibody-mediated hemolysis will enable early monitoring and treatment. Some antibodies, such as anti-Kell, also destroy fetal hematopoietic precursors before they get into the circulation, thus giving rise to severe anemia, including thrombocytopenia.
Blood group frequency in the population is one of many factors that determine the incidence of HDFN. Also, some blood groups are more frequently involved in HDFN than others. Antibodies against the D antigen from the Rh blood group have been, and still are, the most common cause of HDFN in RhD positive babies of RhD negative mothers (Nordvall et al. 2009). Efficient prophylaxis, in the form of administration of purified anti-RhD antibodies, is routinely administered to all RhD negative mothers during pregnancy to prevent the mother from producing anti-Rh[D] antibodies. IgG antibodies against other blood groups can also be involved in HDFN, including Kell, c, E, AB0, JK (Kidd), and FY (Duffy) (Moise 2008), but, as yet, there are no interventions to prevent isoimmunization against these antigens. With respect to antenatal fetal blood group determination, the most important blood groups are D, c, E, and K (Scheffer et al. 2011). ABO HDFN is rarely severe (Christensen et al. 2014).
It is important to be able to accurately characterize a given fetus’ blood group to ascertain whether it is at risk of HDFN. This can be performed by allele specific polymerase chain reaction (PCR) amplification of the presumed fetal allele and is performed routinely for the determination of fetal RHD status in maternal blood (Clausen et al. 2013, 2014a,b). Highly specific target sequences exist for the RHD gene, enabling the design of primers to specifically amplify the RHD gene and not the highly homologous RHCE gene. Antenatal determination of blood groups that are defined by a single nucleotide polymorphism (SNP) are technically more demanding to measure using PCR diagnostic assays. This is the reason why we have explored the potential of antenatal blood group determination by next-generation sequencing (NGS).
FETAL BLOOD GROUP TYPING
A number of kits are available for the determination of blood groups based on DNA amplification. These commercial systems use DNA extracted from peripheral blood cells to determine the blood group of the patient or donor. However, determining the fetal blood group from a blood sample collected from a pregnant woman is a very different and difficult task. During pregnancy, the placenta undergoes apoptosis and releases fragmented DNA that circulates as cell-free DNA (cfDNA) in the maternal plasma, together with the much larger amounts released from maternal cells. Early in pregnancy, small amounts of fetally derived cfDNA exist, but the fraction of fetal cfDNA in the maternal circulation increases with advancing gestational age (Finning et al. 2002). Only a small amount of placentally derived template material in a large surplus of maternal cfDNA is available for PCR amplification. Roughly, the fetal fraction will be between 0% and 30% of cfDNA in maternal plasma at 10–20 gestational weeks (Wang et al. 2013). The range is large and is also dependent on other factors such as maternal weight (Bianchi and Wilkins-Haug 2014). Fortunately, at gestational ages of 10–20 wk, the fetal fraction is >2% for the vast majority of pregnant women. Measuring fetal fraction, however, can be challenging and different methods for estimation of the fetal fraction will give somewhat varying results (Lun et al. 2008). It is therefore important to consider the fetal fraction of cfDNA in the overall assay development.
For blood groups defined by a single base variant, it is technically challenging to design a PCR-based test to specifically amplify a given fetal allele that exists in the maternal peripheral blood in a big surplus of the antithetical maternal allele when using unmodified PCR primers. More genetic variation in primer target regions is needed to achieve a sufficient degree of allele-specific amplification using unmodified PCR primers. Furthermore, the absolute number of template molecules can be very low. A huge number of variations in PCR assays exist, many with the purpose of increasing specificity; these include restriction digests, inhibition of specific allele amplification, among others.
THE POSSIBILITIES OF NGS
To circumvent some of these difficulties, we devised a novel approach focused on NGS for antenatal blood group determination (Rieneck et al. 2013). This approach uses a simple method of unbiased PCR amplification of the two antithetical SNP alleles of a blood group by a primer pair located outside the SNP. This amplifies both alleles, and then counts the number of sequenced SNPs for each of the two alleles. The steps involved in this procedure are outlined in Figure 1. The approach is more laborious and costly than a simple PCR amplification and detection assay. We have focused on antenatal determination of fetal Kell status. The method is still under development at a pace influenced by the fact that the need for determining antenatal Kell status is rare, rendering a very limited number of clinical samples available for analysis. Other blood groups that are occasionally of clinical importance and of relevance for antenatal NGS determination include the Rh antigens c and E, ABO, KEL3, KEL4, JK (Kidd), and FY (Duffy). In addition, similar immunological incompatibilities between fetus and mother of human platelet antigens can cause fetal platelet destruction and intra cranial bleeding, resulting in fetal disability or death. Thus, noninvasive typing of the fetal human platelet antigen (HPA) status is of equal relevance. Overall, the same technique can be applied for these targets as well.
Figure 1.
Flow chart showing the overall procedure for antenatal determination. A number of the crucial steps involved in NGS-based antenatal determination of blood groups are shown.
The whole fetal genome has been made accessible by deep sequencing of cfDNA from maternal blood samples (Lo et al. 2010; Kitzman et al. 2012). This is a very costly procedure but shows the enormous potential of NGS for antenatal diagnostics.
PROCEDURE AND TECHNICAL ASPECTS
In the following section, we describe in more detail selected aspects of this novel approach to antenatal blood group determination, using the Kell blood group as an example (Rieneck et al. 2013).
Even though KEL1 immunized pregnant women are rare (Finning et al. 2007), we have focused on the KEL blood group because HDFN caused by KEL immunization can be clinically severe and can affect hematopoietic stem cells (Nordvall et al. 2009).
Typically, the need for antenatal KEL determination arises in a KEL2 homozygous mother who may carry a KEL1 positive fetus. In maternal plasma, there is only a small fraction of the KEL1 allele relative to the KEL2 allele in the mother’s pool of cfDNA in the plasma. Furthermore, as only small amounts of cfDNA are present in maternal peripheral blood plasma, this tends to make the absolute number of KEL1 alleles very small; early in pregnancy, only a few molecules per mL maternal plasma exist. The fetal allele exists in a large surplus of the maternal allele unless the fetus is KEL1 negative, then at this base position only the KEL2 allele would exist in the maternal pool of cfDNA.
To overcome the difficulties of diagnosing a KEL1 positive fetus by PCR amplification, specifically of the KEL*1 allele, we surmised that NGS could be used for determining the fetal blood group. This methodology affords a number of technical advantages. One important characteristic is that only one set of primers is used to amplify both alleles. The amplification results in amplicons of the same length and composition except for the one base variation that constitutes the SNP. This amplification, therefore, is essentially without bias. After amplification, the amplicons are sequenced by NGS, and by simply counting the numbers of KEL*1- and KEL*2-specific sequences, the fetal KEL blood group can be determined (Rieneck et al. 2013). Importantly, the issue is whether or not KEL*1 alleles can be detected in cfDNA in maternal plasma. To reach a reasonable certainty, a large number of amplicons must be sequenced. This is especially so for a KEL*1 negative fetus. As errors will occur during the sequencing process and the previous PCR amplification, some KEL*1 sequences must be expected even in the situation when the fetus is truly KEL*1 negative. The acceptable level of noise contributed by spurious KEL*1 sequences must be determined empirically. The noise level contributes to establishing a lower level of assay sensitivity. The noise also shows up in deviations from the expected sequence defined by the primers used for PCR amplification of the cfDNA as well as in bases amplified outside primer sequences (Rieneck et al. 2013).
This NGS approach is very different from the situation in which a single SNP must be determined in a conventional allele-specific PCR amplification using unmodified primers with one base mismatch at the 3′ end of one of the primers. Sufficient allele specificity is difficult to achieve to determine the fetal blood group. Many PCR cycles are needed owing to the low amount of input DNA, the small amounts of plasma DNA, and the relatively low fetal fraction of cfDNA, so over many cycles of amplification the allele specificity falters. Thus, to discern the two alleles, especially in early pregnancy, modifications to the basic PCR amplification must be made. To achieve a higher degree of allele specificity, specialized primers and comprehensive optimization are needed in an effort to increase the allele-specific amplification (Finning et al. 2007). Also, during PCR-based determination, the result is typically generated during the amplification or afterward looking at the total population of amplicons, whereas NGS-based detection is the result of sequencing each molecule after in situ amplification and then counting the number of individual sequences reflecting the presence of one or the other SNP. Thus, NGS offers a higher level of detailed analysis of the amplicons.
The procedure starts with purification of cfDNA from maternal plasma, followed by PCR amplification and purification of the amplicons. The primers are specially designed with appending of sequences needed for anchoring to the chip and priming of DNA synthesis for cluster formation and DNA sequencing. See Table 1, for information on primers we use for sequencing using the MiSeq instrument (Illumina). After adjusting the concentration of the amplicons, they are denatured and sequenced on the MiSeq platform. During sequencing, results are uploaded to BaseSpace, and alignment to a reference sequence is performed with scoring as well as counting of variants. Different software programs exist to call the variants. A sensitive variant caller is important, as the limit of detection would otherwise be determined by this software. Only high quality sequences at Q30 or above are used in the analysis. Phred quality scores Q are defined as a property which is logarithmically related to the base-calling error probabilities P. Q = –10 log10P. Q30 means that the probability of incorrect base call is 1:1000 or, stated differently, that the base call accuracy is 99.9%. A written result that can make up part of a clinical decision for further monitoring or treatment is then made. Some reservations regarding the result are described in a later section. For reliable antenatal determination, a deeper sequencing is required than that used for obtaining a fetal-genome scaffold, although no general consensus exists (Rieneck et al. 2013).
Table 1.
The appended primers used for PCR amplification of KEL amplicons for sequencing on MiSeq
| upstream primer | 5′ | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtagc GTAAATGGACTTCCTTAAACTTTAACCGA 3′ |
| downstream primer | 5′ | CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCATACTGACTCATCAGAAGTCTCAGC 3′ |
The primers amplify only two bases: the KEL SNP and one adjoining base. Upper case in italics are the gene specific part of the primers, upper case appended adaptors and lower case is an ID tag.
STATISTICAL ASPECTS AND ASSUMPTIONS
Some general quantitative considerations are useful to establish the approximate minimum coverage needed to be able to predict with a specified probability whether a given fetus is KEL1 negative.
Ideally, direct probing of the pool of cfDNA in maternal plasma during early pregnancy is desirable. However, this pool is not directly accessible with current technologies owing to the scarcity of the cfDNA molecules carrying the KEL*1 SNP. Thus preamplification is necessary, and afterward the amplified DNA is amenable to sequencing.
It is then important to consider how many molecules (i.e., individual DNA sequences) must be examined to make a reliable determination. In an earlier report of noninvasive sequencing of the entire fetal-genome, an 80-fold coverage was used (Kitzman et al. 2012). This means that every base in the genome sequence is read 80 times. Assuming that there is ∼5% fetal DNA in a maternal plasma sample, one might roughly expect that at any given base position, only four reads would be of fetal origin with only about two copies of each paternal allele being sequenced. In the antenatal situation, a much deeper coverage is needed, especially if the absence of one allele is to be ascertained with high probability.
An important aspect of using NGS for antenatal blood group determination is that the unmodified primers used for PCR amplification of the target alleles result in a rather unbiased amplification. The SNPs, which constitute the two alleles, in this case KEL*1 and KEL*2, are both amplified. We assume that this occurs with the same efficiency, as the amplicons will have the same length when the primers are the same and amplification conditions optimized. Note that the amplicons only differ at one base position from each other. This means that after the PCR amplification, the fraction of the respective alleles in the circulating plasma of the mother remains unchanged among the amplicons. This is an important and plausible assumption. If, for instance, there is a slight bias against the KEL*1 allele in the method, a change of primer design would be necessary. We have not yet experimentally measured any potential bias systematically.
The true fraction of fetal cfDNA is almost never known. It shows considerable variation in different pregnant women for the same gestational age (Wang et al. 2013). However, reasonable assumptions can be made based on other investigations, and in week 20, the fraction of fetal cfDNA in maternal plasma can be assumed to be >2% in the vast majority of pregnant women (Wang et al. 2013). To obtain reliable results, a large number of amplicons must be sequenced. If, for instance, one wants to be 99% sure not to miss a KEL1 positive fetus in a maternal blood sample with 4% fetal cfDNA, the formula for the binomial distribution can be adjusted to give the theoretical minimal number of cfDNA molecules, under ideal conditions, to be sequenced as
| (1) |
where n is the number of molecules carrying a KEL SNP, α is the probability of not finding a KEL*1 SNP, and p is the fraction of fetal cfDNA possibly containing the KEL1 allele.
Thus, setting α = 0.01 and the fetal fraction at 2%, in theory it can be calculated from Equation 1 that
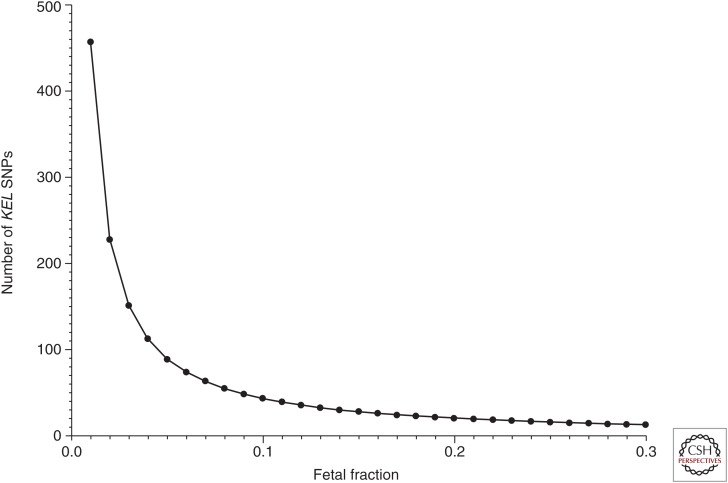
gives more than 228 independent cfDNA molecules from maternal plasma that must be investigated and found negative for KEL*1, before it can be predicted with 99% probability that the fetus is KEL1 negative. In Figure 2 the relationship is shown between n and the fetal fraction to theoretically reach a 99% probability of determining a KEL1 negative fetus correctly.
Figure 2.
Importance of fetal fraction. The number of KEL SNPs that must be examined and found KEL*1 negative to designate the sample as KEL*1 negative with a 99% probability, are given as a function of the fetal cfDNA fraction.
However, in reality a larger number of cfDNA molecules need to be investigated, as deviations from an ideal situation may arise during steps 1 to 4 (Fig. 1). For example, during the PCR amplification, errors can occur with erroneous incorporation of bases. This may also happen during the sequencing process, which is preceded by an on-chip in situ amplification step in which further errors occur. Using proof reading DNA polymerases with low error incorporation is important to keep the overall sensitivity as high as possible. Only sequences of high quality will be used in the bioinformatics analysis. For the MiSeq, we strive to obtain KEL coverage of at least 10,000 reads. We have not yet measured the variance of individual runs.
A NOTE ON PCR PRIMERS
The KEL primers are designed so that both KEL*1 and KEL*2 are amplified, with the underlying assumption that there is no bias in the PCR amplification. The primers are carefully designed so that no other or only very rare SNPs are found in the primer target sequence and there is high amplification efficiency. If a primer target sequence encompasses a variant that gives rise to a mismatch with a primer, obviously, a biased PCR amplification would be the result. If the amplification was biased against the KEL*1 allele, even as a 5′ end mismatch, significant change of the original ratio of KEL*1/KEL*2 molecules may occur, which could result in a false negative result. Our original primers (Rieneck et al. 2013) had a gene specific part with appended adaptors for the DNA sequencing on Genome Analyzer IIx (GAIIx) (Illumina, San Diego, CA). This approach obviated the need for an extra PCR amplification to append the adaptors streamlining the process, but it resulted in very long PCR primers. We have since migrated the assay to the MiSeq platform, which necessitated a modification (Table 1) of the original primers used for GAIIx.
COMMUNICATION OF RESULTS AND CAUTION
We have primarily been working on the KEL blood group because of its clinical importance and the difficulty of antenatal determination of fetal KEL status. Originally, we made the first measurements on the Illumina GAIIx. After migration of the assay to the MiSeq platform, we only had a few clinical samples, but will continue to gather experience with this approach as we expand our repertoire. We have chosen an overall cut off at <0.5% KEL*1 sequences. If 0.5% or more KEL*1 sequences are detected, the result of the antenatal KEL determination will be given as a prediction that the fetus is KEL1 positive with high probability. The level of 0.5% has been chosen pragmatically, taking into consideration the many sources of noise (Rieneck et al. 2013). It should undoubtedly be possible to increase the sensitivity. Furthermore, because of the small and variable fractions of fetal cfDNA in maternal plasma, we do not encourage use of our NGS-based antenatal determination before gestational week 10; however, with a few modifications this should certainly be possible and also clinically highly relevant. We give the result as positive or negative or indeterminate with the percentage of KEL*1 sequences of all examined KEL sequences and indicate the coverage level together with a caution that the test is still under development. More experience with actual clinical samples is needed to precisely establish the cut off levels. Finally, we recommend that a sample be taken 1–3 wk after the first and tested independently. As part of gathering information on the accuracy of the predictions, we ask for a postnatal serological KEL determination of the newborn. Recommendations as to preanalytical handling of the clinical samples for NGS-based antenatal determination are also given to the clinicians (Wong et al. 2013). We recommend that the sample be stored at room temperature and forwarded for analysis as soon as possible. Storage of the sample releases more maternal DNA from cells and dilutes the amount of fetal cfDNA present (Barrett et al. 2011).
c AND E TARGETS OF THE RHCE
We are currently working to include the c and E defining SNPs in the RHCE gene in the panel of blood group determining SNPs to be detected by the NGS strategy. These SNPs pose special problems, as the genes encoding RHD and RHCE are highly homologous. The cfDNA fragments, in general, are on average only about 166 bp (maternal) or 143 bp (fetal) long (Lo et al. 2010). Thus, only short amplicons can be expected to be amplified with good efficiency. This means that the PCR primers cannot be located far away from a SNP that defines c. As D and C (antithetical to c) are identical over ∼180 bp segment of the genome that includes a c-defining SNP and only differ from c in five SNP positions spread rather evenly in this segment, primer design is difficult. Furthermore, as we use a proof reading DNA polymerase a 3′ single base mismatch to bias the primers for c will be of little use. It is preferable to use higher coverage and bioinformatically sort out the c reads from a sequencing reaction. The sensitivity of antenatal c determination would consequently be reduced. Similar considerations for antenatal E determination exist. These design considerations are made on the basis of the canonical c and D sequences from RefSeq, but in the population other SNPs exist that should be taken into consideration.
Other general PCR primer design aspects also need to be taken into consideration. As always with PCR-based assays, meticulous primer design and use of a high number of templates is important. In cases in which a pregnant woman is D negative, i.e., when the RHD gene is absent, the sensitivity of c and E detection would increase as compared with a situation in which the mother carries one or two copies of the RHD gene. Because of the frequency of allele coupling, most often, the c and E alleles are found in D negative mothers.
FUTURE MODIFICATIONS
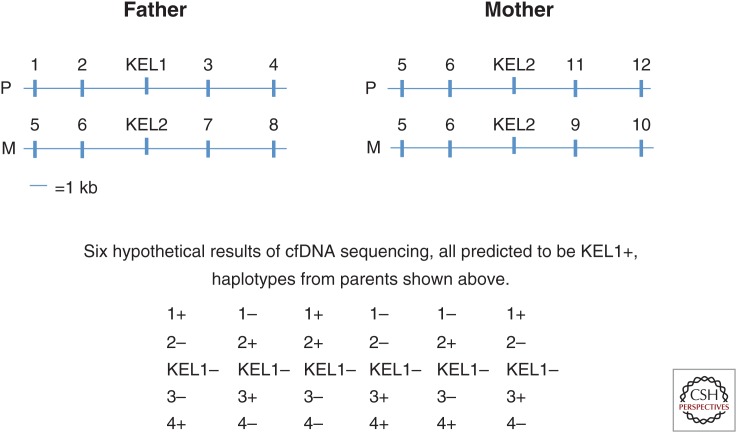
To increase the reliability of detection of fetal blood group antigens using antenatal-based NGS methodology, we are contemplating further technical modifications. The approach is based on first obtaining information on the haplotypes of both parents from high molecular weight DNA extracted from living cells, and then establishing which unique SNPs are closely coupled to the paternal gene in question, as well as which unique SNPs would not be coupled to the paternal allele. The parental haplotypes can be established by using, e.g., Moleculo technology (Illumina, San Diego, California) or possibly by Nanopore sequencing (Oxford Nanopore technology, Oxford). The approach is shown in Figure 3.
Figure 3.
Genotyping assessed by proxi sequencing (GAPS). Hypothetical situation illustrating part of the homologous chromosomes 7 from the father and mother with haplotypes showing closely coupled SNPs indicated by numbers. Paternal-specific variants coupled to KEL*1 detected in cfDNA from the mother, predict a KEL*1 positive fetus, even though the KEL*1 itself may not be detected. Only a fraction of paternal-specific alleles of the correct haplotype need to be detected. Detection of any nonmaternal allele(s) can be informative as to inheritance of KEL*1 or KEL*2 and may serve as control for the presence of fetal DNA. P, paternal chromosome; M, maternal chromosome.
After establishing the parental haplotypes, unique SNPs closely coupled to the paternal KEL*1 allele are selected and PCR amplified from short fragments of cfDNA from the maternal plasma. Subsequently, the amplified DNA is sequenced. Other target SNPs closely coupled to KEL*1 could substitute for the paternal KEL*1 allele detection, and the prediction of a KEL1 positive fetus can be based on detection, not only of the KEL*1 SNP itself but on a number of closely coupled unique SNPs. Alternatively, the detection of the KEL*1 SNP can be corroborated by detection of a number of SNPs closely coupled to the KEL*1 SNP that are specific for the father. If the fetus is actually KEL1 negative, the SNPs not coupled to KEL1 and specific to the father can indicate absence of this allele. Depending on the specific requirements for deeming a test result positive, this would potentially increase sensitivity, specificity, and robustness of the assay. This method, however, is labor intensive, and one could argue that it may not be attractive to diagnose a blood group merely on the basis of genetic coupling. On the other hand, the coupling is so close that crossing over in the father’s gametes is highly unlikely. We have named this modification “genotyping assessed by proxi sequencing” (GAPS).
Also to be considered is the concurrent use of an extra gene specific pair of primers to minimize the risk of amplification bias because of unexpected genetic variation of the first primer target sequences.
CONCLUDING REMARKS
We have developed an NGS-based test for determination of antenatal blood groups in the fetus. This is a new area. Despite experience with targeted-sequencing, including targeted-sequencing for noninvasive prenatal testing (NIPT) and for tumor mutations, further exploration into the use of NGS for antenatal blood group determination is needed. Although it is still expensive to run NGS analyses, the cost is continually falling. It is a realistic assumption that more and more DNA-based diagnostic tests in the future will be performed using sequencing.
There are many different variations of the described assay that have not been explored. Because of the need for very deep coverage, and hence high costs, it is likely that there will still be some time before nontargeted NGS assays will be routinely used for antenatal blood group determination.
If no KEL*1 sequences are found, it is important that a large enough number of amplicons are sequenced to ascertain that the number of KEL*1 sequences in maternal plasma is below the detection level. Despite genetic variation, reliable amplification might be obtained by more than one primer pair with nonoverlapping target sequences. Amplifications and several controls can be built into the assay.
We have migrated the original assay to the MiSeq platform, but scarcity of clinical samples is challenging the process of accumulating experience and integrating the assays into clinical practice.
NGS technology has many practical applications in biology and medicine, and the whole area is buzzing with activity. Results roll in with ever increasing speed, and the technology itself is under rapid development. We believe that NGS is well suited for noninvasive antenatal determination of fetal blood group; however, more experience is needed.
ACKNOWLEDGMENTS
We thank Birgitte Bundgaard for expert technical assistance and Erik Holst for great help with valuable statistical considerations.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Barrett AN, Zimmermann BG, Wang D, Holloway A, Chitty LS. 2011. Implementing prenatal diagnosis based on cell-free fetal DNA: Accurate identification of factors affecting fetal DNA yield. PLoS ONE 6: e25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi DW, Wilkins-Haug L. 2014. Integration of noninvasive DNA testing for aneuploidy into prenatal care: What has happened since the rubber met the road? Clin Chem 60: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen RD, Nussenzveig RH, Yaish HM, Henry E, Eggert LD, Agarwal AM. 2014. Causes of hemolysis in neonates with extreme hyperbilirubinemia. J Perinatol 34: 616–619. [DOI] [PubMed] [Google Scholar]
- Clausen FB, Jakobsen TR, Rieneck K, Krog GR, Nielsen LK, Tabor A, Dziegiel MH. 2013. Pre-analytical conditions in non-invasive prenatal testing of cell-free fetal RHD. PLoS ONE 8: e76990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen FB, Steffensen R, Christiansen M, Rudby M, Jakobsen MA, Jakobsen TR, Krog GR, Madsen RD, Nielsen KR, Rieneck K, et al. 2014a. Routine noninvasive prenatal screening for fetal RHD in plasma of RhD-negative pregnant women—2 years of screening experience from Denmark. Prenat Diagn 34: 1000–1005. [DOI] [PubMed] [Google Scholar]
- Clausen FB, Damkjær MB, Dziegiel MH. 2014b. Noninvasive fetal RhD genotyping. Transfus Apher Sci 50: 154–162. [DOI] [PubMed] [Google Scholar]
- Finning KM, Martin PG, Soothill PW, Avent ND. 2002. Prediction of fetal D status from maternal plasma: Introduction of a new noninvasive fetal RHD genotyping service. Transfusion 42: 1079–1085. [DOI] [PubMed] [Google Scholar]
- Finning K, Martin P, Summers J, Daniels G. 2007. Fetal genotyping for the K (Kell) and Rh C, c, and E blood groups on cell-free fetal DNA in maternal plasma. Transfusion 47: 2126–2133. [DOI] [PubMed] [Google Scholar]
- Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, et al. 2012. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 6: 137ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, et al. 2010. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 8: 61ra91. [DOI] [PubMed] [Google Scholar]
- Lun FM, Chiu RW, Chan KC, Leung TY, Lau TK, Lo YM. 2008. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem 54: 1664–1672. [DOI] [PubMed] [Google Scholar]
- Moise KJ. 2008. Fetal anemia due to non-Rhesus-D red-cell alloimmunization. Semin Fetal Neonatal Med 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Nordvall M, Dziegiel M, Hegaard HK, Bidstrup M, Jonsbo F, Christensen B, Hedegaard M. 2009. Red blood cell antibodies in pregnancy and their clinical consequences: Synergistic effects of multiple specificities. Transfusion 49: 2070–2075. [DOI] [PubMed] [Google Scholar]
- Rieneck K, Bak M, Jønson L, Clausen FB, Krog GR, Tommerup N, Nielsen LK, Hedegaard M, Dziegiel MH. 2013. Next-generation sequencing: Proof of concept for antenatal prediction of the fetal Kell blood group phenotype from cell-free fetal DNA in maternal plasma. Transfusion 53: 2892–2898. [DOI] [PubMed] [Google Scholar]
- Scheffer PG, van der Schoot CE, Page-Christiaens GC, de Haas M. 2011. Noninvasive fetal blood group genotyping of rhesus D, c, E and of K in alloimmunised pregnant women: Evaluation of a 7-year clinical experience. BJOG 118: 1340–1348. [DOI] [PubMed] [Google Scholar]
- Urbaniak SJ, Greiss MA. 2000. RhD haemolytic disease of the fetus and the newborn. Blood Rev 14: 44–461. [DOI] [PubMed] [Google Scholar]
- Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. 2013. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn 33: 662–666. [DOI] [PubMed] [Google Scholar]
- Wong D, Moturi S, Angkachatchai V, Mueller R, DeSantis G, van den Boom D, Ehrich M. 2013. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem 46: 1099–1104. [DOI] [PubMed] [Google Scholar]