Abstract
Here we report that the specificity of peptide release in the ribosome on a nonstop mRNA by ArfA and RF2 is achieved by an induced-fit mechanism. Using RF2 that is methylated on the glutamine of its GGQ motif (RF2m), we show that methylation substantially increases the rate of ArfA/RF2-catalyzed peptide release on a nonstop mRNA that does not occupy the ribosomal A site, but has only a modest effect on kcat by the same proteins on longer nonstop mRNAs occupying the A site of the mRNA channel in the ribosome. Our data suggest that enhancement in the kcat of peptide release by ArfA and RF2 under the cognate decoding condition is the result of favorable conformational changes in the nonstop complex. We demonstrate a shared mechanism between canonical and nonstop termination, supported by similarities in the kinetic mechanisms in antibiotic inhibition and methylation-correlated enhancement in the rate of peptide release. Despite these similarities, our data suggest that nonstop termination differs from canonical pathway in the downstream event of recycling.
Keywords: ribosome, quality control, translational termination, nonstop translation, RF2, alternative rescuing factor A
INTRODUCTION
The coupling of transcription and translation in the same cellular compartment enables bacterial cells to replicate rapidly and to respond swiftly to environmental changes. The time required to express protein products of a gene is minimized by having mRNA translation begin before transcription is complete. However, this coupling of transcription and translation poses challenges for protein quality control because bacterial cells cannot monitor the integrity of an mRNA before starting to translate it. Therefore, a ribosome can end up translating a truncated mRNA that has no stop codon. The end result will be a ribosome stalled on an mRNA that cannot elongate or terminate the nascent peptide chain, because its aminoacyl (A) site is unoccupied. This is called nonstop translation.
It is estimated that ∼2%–4% of translations are nonstop translations in Escherichia coli (Ito et al. 2011). This implies that the average ribosome is involved in about five nonstop translation events per cell division cycle (Keiler and Feaga 2014). Thus, stalled ribosomes must be quickly identified and rescued to ensure that both the quantity and the quality of protein synthesis in a cell are not compromised. Bacteria have evolved three mechanisms for releasing stalled ribosomes and resolving nonstop translation events: (1) a trans-translation system involving transfer-messenger RNA and small protein B (tmRNA-SmpB) that rescues ribosomes and targets nonstop mRNAs and nascent polypeptides for rapid degradation; (2) alternative ribosome rescuing factor A and release factor 2 (ArfA/RF2); and (3) alternative ribosome rescuing factor B (ArfB) which releases the nascent peptide and enables stalled ribosomes to recycle. Genetic studies have suggested that nearly all the bacteria species require at least one of the above mechanisms for ribosome rescue in order to survive (Chadani et al. 2010).
RF2, one of the class I protein release factors that participate in normal translational termination, plays a unique role in controlling the quality of the protein synthesis in bacteria. In normal termination, class I release factors in bacteria, RF1, and RF2, bind to the translating ribosome when one of the three near-universal stop codons on an mRNA is encountered in the A site of the small ribosomal subunit. Stop codons are decoded in the ribosome in a manner that is fundamentally different from sense codons. Decoding is done by proteins instead of tRNAs. RF1 and RF2 recognize the three stop codons with overlapping specificity: RF1 recognizes UAG, RF2 recognizes UGA, and both of them recognize UAA (Scolnick et al. 1968). Upon stop codon recognition, class I release factors promote the hydrolysis of peptidyl tRNA in the ribosomal peptidyl (P) site, terminating protein synthesis (Capecchi 1967; Caskey et al. 1968; Scolnick et al. 1968).
The elements of release factors involved in the stop codon recognition and catalysis of peptide release have been revealed using genetics, biochemistry, and sequence analysis. An elegant genetic experiment demonstrated that exchanging a tripeptide motif between RF1 and RF2 (P[A/V]T in RF1; SPF in RF2) switches their respective specificities for UAG and UGA (Ito et al. 2000). A universally conserved tripeptide sequence, GGQ, has been implicated in the hydrolysis of the nascent peptide from P-tRNA (Frolova et al. 1999). Hydroxyl-radical probing has suggested that the SPF and GGQ motifs of RF2 are close to the decoding center and peptidyl transferase center (PTC), respectively, when bound to the ribosome (Wilson et al. 2000). These locations for the decoding (P[V/A]T, SPF) and catalytic (GGQ) motifs of the release factors on a ribosome were confirmed by both cryoEM (Klaholz et al. 2003; Rawat et al. 2003) and low-resolution crystallographic structures (Petry et al. 2005).
Crystal structures of the 70S ribosome complexed with class I release factors and their cognate stop codons, which have been solved at a near-atomic resolution (Korostelev et al. 2008; Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010), have provided us a clear picture of the translational termination in three dimensions. The interactions of RF2 with its cognate stop codons in the decoding center revealed by the crystal structures offers molecular explanations for the specificity of stop codon recognition in the ribosome (Korostelev et al. 2008; Weixlbaumer et al. 2008). For the peptide release, the catalytic GGQ motif in the release factors makes extensive interactions in the PTC (Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010). This catalytic motif is positioned in the PTC by a network of hydrogen bond interactions between highly conserved residues in release factors and nucleotides in the 23S rRNA (Jin et al. 2010). The conformation of the glycines in the GGQ motif facilitates proper placement of the conserved glutamine in the core of the PTC, where it forms a densely packed pocket with the A76 of the peptidyl tRNA, and A2451, C2063, U2585, and U2506 of the 23S rRNA for the catalysis of the peptide release.
Biochemical and structural studies on tmRNA/SmpB-mediated (Ivanova et al. 2004; Neubauer et al. 2012) and ArfB-mediated (Gagnon et al. 2012) pathways involved in rescuing a stalled ribosome in the nonstop translation have provided insight into functions of these ribosome-bound factors in releasing the nascent peptide in the ribosome. Chadani et al. (2010) discovered that ArfA is essential for the viability of E. coli in the absence of tmRNA-mediated trans-translation system using a synthetic lethal screen. The expression of ArfA in E. coli and ArfA-homolog proteins in other bacteria species is regulated by RNase III and tmRNA (Chadani et al. 2011; Garza-Sanchez et al. 2011; Schaub et al. 2012). Class I release factor RF2 is essential for ArfA's function in the cell (Chadani et al. 2012; Shimizu 2012). Remarkably, only the GGQ motif in RF2 is important for the peptide release catalyzed by ArfA and RF2; its SPF motif is dispensable (Chadani et al. 2012).
Despite these exciting advances in our understanding of the translational termination and quality control of protein synthesis, several important questions remain to be answered. The GGQ motif that is required for catalytic activity (Song et al. 2000; Mora et al. 2003) of peptide release is universally conserved. Importantly, the glutamine in this motif is invariably methylated post-translationally at the N5 position. This methylation had been reported to have a noticeable effect on the RF2-dependent but not RF1-dependent release activity in E. coli in vitro (Dincbas-Renqvist et al. 2000). The same modification is also found in Saccharomyces cerevisiae and is required for optimal cell growth (Heurgué-Hamard et al. 2005). The packing role of the side chain of the glutamine for the catalysis of peptide release has been demonstrated biochemically (Shaw and Green 2007), structurally (Jin et al. 2010), and computationally (Trobro and Aqvist 2007, 2009). The enhanced packing of the N5-methylation of the catalytic glutamine in the GGQ motif has been suggested by computational studies (Trobro and Aqvist 2007). It is conceivable that the precise conformation of the glutamine in the ribosome will be influenced by this methylation that normally occurs in vivo. However, the effect of the methylation on the catalysis of the peptide release has not been investigated in most recent kinetic studies, nor has its effects on the termination events involved in the quality control processes been evaluated.
Second, how RF2 catalyzes peptide release on a truncated mRNA in the ribosome remains elusive. In any case, decoding by RF2 appears to be more complicated than that of RF1 (Petropoulos et al. 2014). Furthermore, a so-called “negative-to-positive charge-flip” mutation E167K in RF2 was reported to trigger the peptide release not only at the three conserved stop codons, but also at the sense codons such as UGG, UAC, and UUA (Ito et al. 1998). A similar phenomenon has not been observed in RF1. While much has been learned about the tmRNA-SmpB and ArfB pathways for resolving nonstop translation from the structures of ribosomal complexes with those proteins bound (Gagnon et al. 2012; Neubauer et al. 2012), the molecular interactions that enable the ribosome to sense a nonstop mRNA and recruit ArfA and RF2 for the peptide release are unknown.
Here, we obtained fully methylated RF2 (RF2m) by coexpressing wild-type RF2 and its cognate methyltransferase PrmC under mild lactose induction in E. coli. Using the RF2m obtained by this method, we have measured the kinetics of peptide release on nonstop mRNAs by ArfA/RF2m in E. coli. Our results suggest that ArfA/RF2 catalyze peptide release by the induced-fit mechanism that broadly resembles tRNA-selection in the ribosome. More specifically, ArfA helps induce a fully activated conformation of RF2 to release nascent peptides in the PTC when the ribosome encounters the end of a nonstop mRNA.
RESULTS
To ensure that we accurately elucidated molecular determinants in RF2 that contributed to catalysis of the peptide release in nonstop termination pathways, we used RF2 with fully methylated glutamine in its GGQ sequence for in vitro kinetic assays. Chromosomally expressed RF2 was used for this purpose (Dincbas-Renqvist et al. 2000), but the yield obtained when RF2 is prepared this way is low. More recently, methylated RF2 was obtained by in vitro enzymatic reaction (Kuhlenkoetter et al. 2011). Coexpression of the prmC methylase gene and RF2 was suggested to obtain methylated RF2 (Fei et al. 2010), but results on the methylation status by this method were not reported.
Here we cloned the prmC gene from E. coli as PCR-generated NdeI–XhoI fragments into pET-22b vector with an ampicillin resistance marker, and we also cloned release factor RF2 from E. coli K12 strain into pET-13 with a kanamycin resistance marker (Weixlbaumer et al. 2008; Jin et al. 2010). Next, fully methylated RF2 was obtained by cotransformation of the PrmC and RF2 clones into the E. coli BL21 (DE3) strain and expression with a very mild induction using lactose at 25°C (Supplemental Fig. S1A; Monteiro et al. 2000). RF2 was then purified by affinity, ion-exchange, and size-exclusion chromatography, and the methylation status of RF2 was examined by mass spectrometry (Supplemental Fig. S1B,C). Using this method, we obtained 100% methylated RF2.
ArfA binds to RF2, not RF1, near the decoding center of the ribosome
The catalytic mechanism of ArfA/RF2-mediated termination that bacteria use to resolve stalled ribosomes in nonstop translation is unknown. In the absence of a three-dimensional structure, even the location of the binding site of ArfA on the ribosome remains controversial. It has been suggested that ArfA binds to the 70S, 50S, or, more recently, to the 30S (Chadani et al. 2011; Kurita et al. 2014).
The expression of the wild-type ArfA in the cell is subject to the regulation of RNase III and tmRNA-SmpB activity (Chadani et al. 2011; Garza-Sanchez et al. 2011; Schaub et al. 2012). In the absence of the tmRNA-SmpB system, the C-terminal 17 amino acids truncated protein ArfAΔ17C is produced, and it has been suggested that this is the form of the protein that recruits RF2 to rescue the stalled ribosome (Garza-Sanchez et al. 2011; Schaub et al. 2012). Different truncated ArfA proteins that lack C-terminal 12, 17, 22, 25, and 32 amino acids were constructed, and these variants all showed a similar ribosome-rescuing activity, except for ArfAΔ32C (Chadani et al. 2011). Thus, E. coli ArfAΔ12C was cloned, expressed, and purified for the rest of this study.
The association of ribosomal subunits with ArfAΔ12C was tested in the following experiment. Complexes comprised of 70S ribosome, RF2, ArfAΔ12C, and nonstop mRNAs were first assembled in vitro. The complexes were then passed through two rounds of linear sucrose gradients with different Mg2+ concentrations. The homogenous ribosomal complex of 70S/RF2/ArfAΔ12C was obtained from the first sucrose gradient with 10 mM Mg2+ concentration. The subunits of 70S ribosome were split using the second sucrose gradient with 2 mM Mg2+. Results from Western blot showed that ArfAΔ12C bound to the 70S ribosome (Fig. 1A, Lane 2) and had a stronger affinity for the 30S than for the 50S subunit (Fig. 1A, Lane 4). It was proposed previously that ArfA binds to the 50S subunit of the ribosome (Chadani et al. 2010; Kurita et al. 2014). However, the experiments in question were done using a cell-based assay with the cellular mRNA pool and overexpressed ArfA protein or by incubating ArfA with purified ribosomal subunits, instead of by reconstituting a nonstop system using a truncated mRNA and stoichiometric amount of ArfA/RF2 as we did. These differences may account for the difference between their results and ours. Moreover, our results are consistent with the chemical probing data that were published most recently (Kurita et al. 2014).
FIGURE 1.
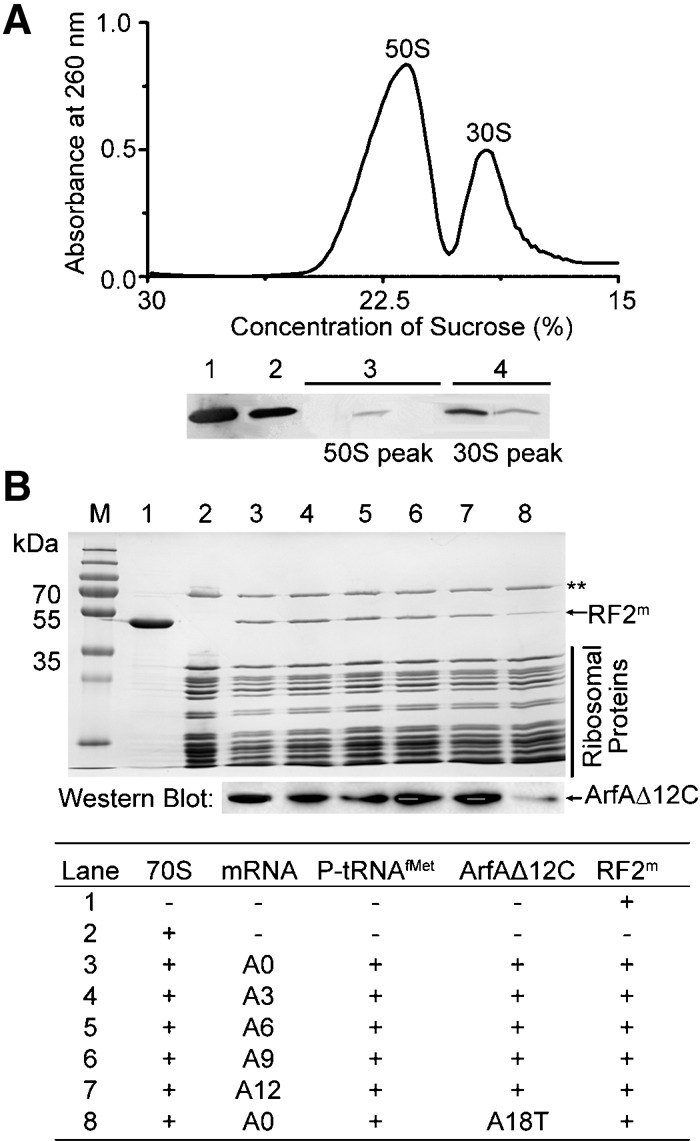
Interaction of ArfA to the ribosome. (A) ArfAΔ12C binds to the 70S ribosome and has a stronger affinity to the 30S subunit. Using antibodies against the His-tag fused to ArfAΔ12C, results from Western blots on His-tagged ArfAΔ12C alone (Lane 1), on 70S pool (Lane 2), on separated subunits 50S (Lane 3), and 30S (Lane 4) pools. (B) Association of ArfAΔ12C and RF2m with the ribosome on a nonstop mRNA. SDS-gel showing components of the nonstop ribosomal complexes after gel filtration. ArfAΔ12C and RF2m bind to the ribosome on different nonstop mRNAs (top gel). Western blot of His-tagged ArfAΔ12C shows that ArfAΔ12C has a similar binding affinity to different ribosomal nonstop complexes (bottom gel). Mutant ArfAΔ12C(A18T) inhibits binding of the protein to the ribosome (Lane 8). (**) Ribosomal protein S1.
Next, the association of ArfAΔ12C with the ribosome carrying a nonstop mRNA in the presence of RF2 and RF1 was tested. As expected, RF2, not RF1, bound to the ribosome in the presence of ArfA (Fig. 1B; Supplemental Fig. S2). In the absence of ArfA, RF2 did not bind to the ribosome when the ribosomal A-site was empty or occupied with a sense codon (Supplemental Fig. S2). Furthermore, our results show that ArfAΔ12C and RF2 associate with the ribosome carrying a nonstop mRNA not only when the ribosomal A-site is empty (Fig. 1B, lane 3), but also, with a similar affinity, when the A-site is occupied with a sense codon (Fig. 1B, lanes 4–7).
In the presence of ArfA, RF2 specifically catalyzes peptide release in the ribosome on a nonstop mRNA
To measure the kinetics of peptide release by ArfA and RF2, we used a peptide release assay similar to the one described by Green and colleagues (Zaher and Green 2009; Petropoulos et al. 2014) with modifications (see Materials and Methods). This assay follows the rate of release of f-[35S]-Met from initiator tRNA f-[35S]-Met-tRNAfMet bound in the P site of the ribosome by RF2. Nonstop translational complexes comprised of ribosome, nonstop mRNA, P-site f-[35S]-Met-tRNAfMet, and ArfAΔ12C were formed using methods similar to one that has been used for the formation of canonical termination complexes (Zaher and Green 2009; Petropoulos et al. 2014). Different concentrations of ArfAΔ12C were tested to ensure that a stable association of ArfAΔ12C to the ribosome was achieved (Fig. 2A). In the buffer conditions that we used, a 2.5-fold excess of ArfAΔ12C was chosen for reconstituting the nonstop ribosomal complex for kinetic studies.
FIGURE 2.
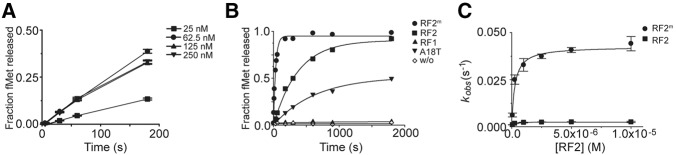
ArfAΔ12C and RF2m promote peptide release on a nonstop mRNA in the ribosome. (A) ArfAΔ12C titrations for establishing reproducible kinetic behavior including formation of a stable nonstop complex and achieving a high reaction end point. (B) Representative time courses of peptide release at 25 nM nonstop ribosomal complexes by fully methylated RF2m (•), unmethylated RF2 (▪), and RF1 (▴) in the presence of ArfAΔ12C, by RF2m and mutant ArfAΔ12C with A18T point mutation (▾) and by RF2m (◊) in the absence of ArfAΔ12C. In every case, the concentrations of proteins are as follows: 62.5 nM ArfAΔ12C, 5 µM RF2m, 5 µM RF2, and 5 µM RF1. (C) Observed rate versus RF2 concentrations showing fits for catalytic rate constant kcat and K1/2 calculations.
Next, titrations of different concentrations of RF2 were carried out to establish conditions where the ribosomes were saturated with RF2 (data not shown). Rate constants for peptide release by RF2 on nonstop mRNAs were measured in triplicate at six different concentrations of RF2, and the average rate constants were plotted versus RF2 concentrations (Fig. 2B,C). The kcat and K1/2 were obtained by fitting the data to the Michaelis–Menten equation and the results are summarized in Table 1. All the data were obtained at 37°C in a buffer containing 10 mM MgCl2 and represent averages from at least three independent experiments using independently purified nonstop complexes. The kcat and K1/2 of fMet-release by RF2m on stop codon UGA obtained in this study were in good agreement with previously published data using chromosomally expressed RF2 from E. coli K12 strain (Dincbas-Renqvist et al. 2000).
TABLE 1.
Rate and binding constants of peptide release for different ribosomal complexes
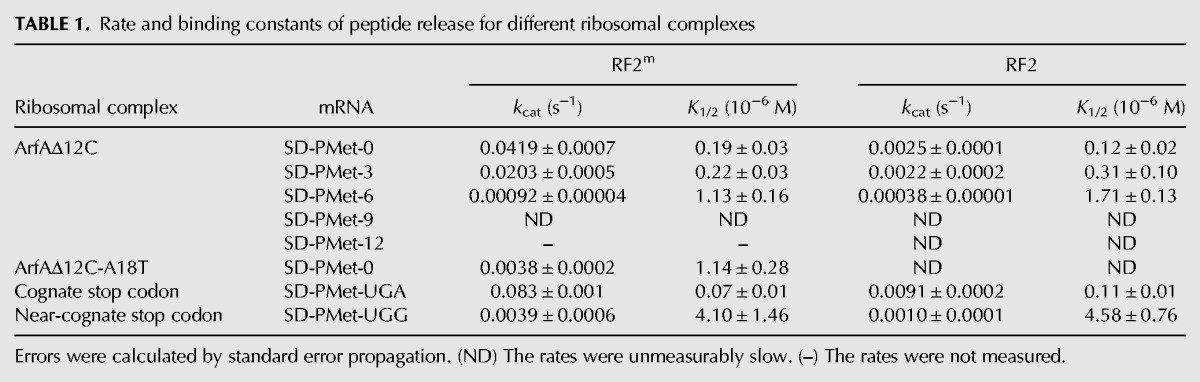
Consistent with the data shown in Supplemental Figure S2, RF2m did not release fMet on a nonstop mRNA in the ribosome when the ArfA was absent (Fig. 2B). Not surprisingly, only RF2, not RF1, catalyzed the peptide release in the presense of ArfA. Remarkably, the kcat by RF2m was ∼20-fold higher than the one obtained using unmethylated protein, although K1/2 remained almost unchanged (Table 1; Fig. 2C). This result suggests that N5-methylation of the glutamine in the GGQ motif significantly stimulates peptide release in ArfA/RF2-mediated termination. We observed a similar change of kcat and K1/2 results by glutamine N5-methylation in RF2 on a cognate stop codon (an ∼10-fold increase in kcat, Table 1). These observations suggest that ArfA emulates a cognate stop codon UGA in the decoding center when it interacts with RF2, and this interaction leads to a fully induced state of RF2 for the subsequent peptide release over 75 Å away in the PTC.
An A18T mutation in the ArfA has been reported to affect the peptide release but leave the ribosome association of RF2 unchanged (Shimizu 2012). To study the effect of this mutation on the kinetic profile of peptide release, we made a mutant protein ArfAΔ12C(A18T) by site-directed mutagenesis and examined its behavior. The A18T mutant could bind to the RF2 in the ribosome, although the interaction appeared to be somewhat compromised, as suggested by a fainter band in the binding experiment (Fig. 1B, lane 8). Methylation at the glutamine in the GGQ sequence makes a difference in the peptide release by RF2 that has been recruited to the ribosome by the mutant. With an unmethylated RF2, the rate constant of the fMet release was too low to be measured accurately. However, with the N5-methylated RF2m, the catalytic rate constant dropped more than 11-fold, and binding was compromised by about sixfold as a result of the point mutation in the ArfA (Table 1). Furthermore, the end point of the release reaction was decreased by about half by the mutant when compared to the one by the wild-type ArfAΔ12C (Fig. 2B). These results indicate that the A18T mutation in the ArfA might inhibit the formation of a fully induced conformation of RF2. The observed effect was most likely due to an interruption of important hydrophobic interactions between the two proteins as a result of the A18T point mutation.
Occupancy level of mRNA channel in the ribosome influences ArfA/RF2-mediated peptide release in nonstop translation
Sensing the empty mRNA channel in the A-site by external proteins that bind to the ribosome was suggested to be essential to resolving nonstop translation (Gagnon et al. 2012; Neubauer et al. 2012). However, it has been shown that the mRNA channel does not need to be empty for the ribosome rescue to occur (Ivanova et al. 2004; Shimizu 2012). Consistent with these results, we have found that ArfAΔ12C and RF2m bind to the ribosome with similar affinity when the mRNA channel in the A-site of the 30S subunit is occupied (Fig. 1B). Using single-turnover kinetic experiments, we studied the kinetics of the peptide release on nonstop mRNAs with varying lengths in the mRNA channel (Table 1; Fig. 3). Our results showed a drastic decrease in the kcat of the fMet release when the number of nucleotides occupying the mRNA channel downstream from the P site increased. mRNAs with 9 nucleotides (nt) following the P site showed almost no fMet release by ArfAΔ12C and RF2m in the ribosome under the experimental conditions that we used. The rate constant dropped by half when only one triplet codon was added (termed A3 mRNA) compared to the A0 mRNA; but it decreased by ∼45-fold when two triplet codons were added (termed A6 mRNA). In contrast to the changes in kcat, K1/2 showed a mild change with about sixfold increase on A6 compared to A0 mRNA.
FIGURE 3.
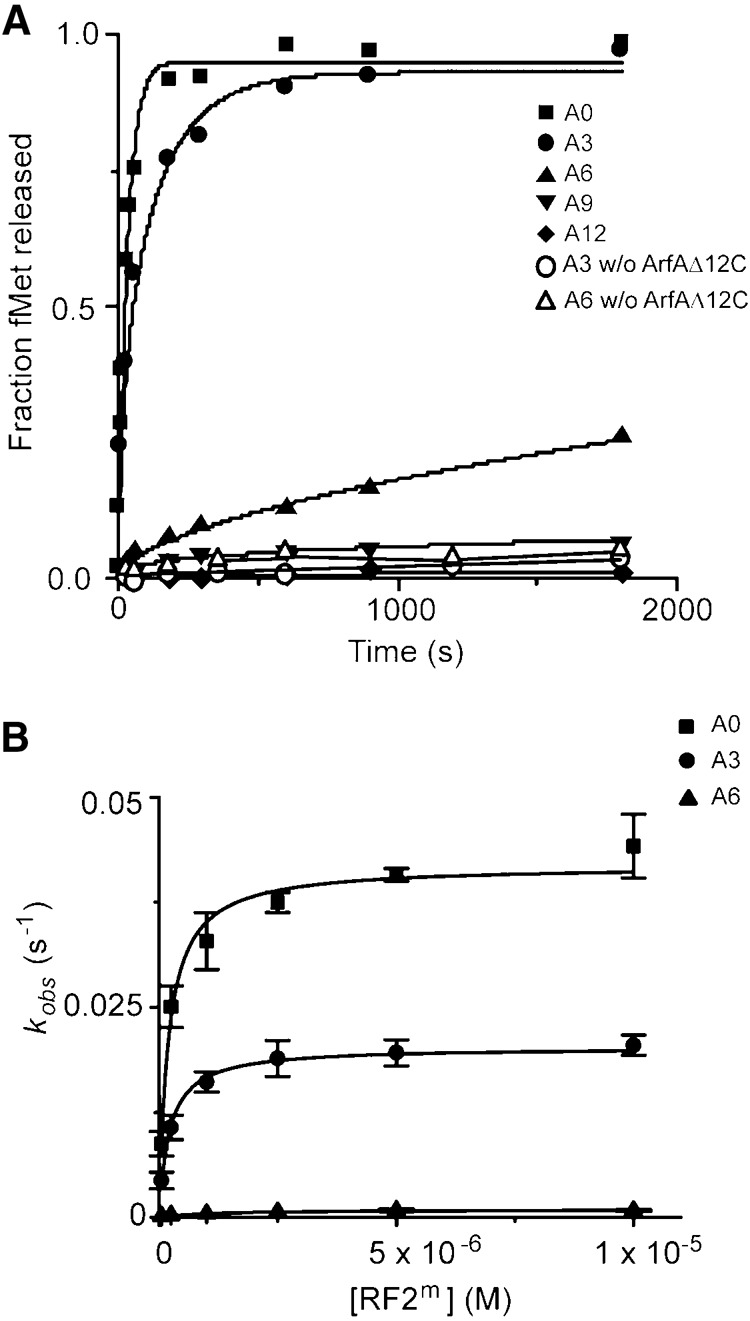
Peptide release by ArfAΔ12C and RF2m on different nonstop mRNAs in the ribosome. (A) Representative time courses of peptide release at 25 nM ribosomes by 62.5 nM ArfAΔ12C and 5 µM RF2m on nonstop mRNAs A0 (▪), A3 (•), A6 (▴), A9 (▾), and A12 (♦). Time courses of peptide release at 25 nM ribosomes by 5 µM RF2m on nonstop mRNAs A3 (O) and A6 (Δ) in the absence of ArfAΔ12C were also shown. (B) Observed rate versus RF2m concentrations showing fits for catalytic rate constant kcat and K1/2 calculations for A0 (▪), A3 (•), and A6 (▴) mRNAs.
Furthermore, methylation in the GGQ motif of the RF2 only mildly influenced the rate constant of peptide release on A6 mRNA in the ribosome. As shown in Table 1, compared to about a 20-fold increase on A0, kcat of the fMet release increases by 10-fold on A3 and only 2.4-fold on A6 mRNA when RF2 is methylated in the GGQ motif, similar to what we have observed in the changes of rate constants by the methylation of RF2 on a near-cognate stop codon in canonical termination. Assuming the rate-limiting step of the pathway is not changed due to the length of mRNAs or the N5-methylation in the glutamine, a straightforward explanation of this observation is that nucleotides downstream from the P site inhibit the formation of a fully activated conformation of ArfA and RF2, which results in a sub-optimal placement of the glutamine in the PTC for the peptide release.
Finally, end points of the fMet release reaction by ArfA/RF2m decreased when the number of nucleotides increased (Fig. 3A), and the end point deficiency for the peptide release could not be overcome by raising the concentrations of Mg2+ (Supplemental Fig. S3A) for A6 and A9 mRNAs. This is a strong indication that conformations of the nonstop complexes formed on longer nonstop mRNAs (A6 and A9 mRNAs) and that of the complex formed on A0 mRNA are different.
Peptide release by RF2m on a truncated mRNA in the ribosome is sensitive to viomycin and paramomycin
To further probe the nature of ArfA/RF2-mediated decoding on a nonstop mRNA, we studied the kinetic behavior of the process in the presence of antibiotics paromomycin and viomycin. Both antibiotics bind in the decoding center and are known to inhibit the translocation step of protein synthesis. The effect of paromomycin on peptide release was studied in canonical termination. Paromomycin belongs to the aminoglycoside antibiotic family; it binds in the major groove of helix 44 (h44) in the decoding center of the ribosome and promotes A1492 and A1493 to “flip out” the h44 helix, thereby stabilizing a conformation of the decoding center in a state that would only adopt when the correct codon–anticodon interactions take place. While promoting the tRNA binding and miscoding events, paromomycin binds competitively to the ribosome with RF1 (Youngman et al. 2007), hence it inhibits the translation termination.
Viomycin is one of tuberactinomycin antibiotics; it binds at the junction formed by h44 of the 16S rRNA and helix 69 (H69) from the 23S rRNA (Stanley et al. 2010). Viomycin inhibits translocation by stabilizing A-tRNA binding to the ribosome, leaving functions of EF-G, including its association to the ribosome and GTP hydrolysis, largely unaltered (Modolell and Vázquez 1977; Yamada et al. 1978). In translational termination, H69 interacts with the RF2 and h44 in the 30S subunit, establishing a connection between the decoding center and PTC. Therefore, it is possible that viomycin would inhibit peptide release, but experimental evidence is required to confirm this argument.
To study the effect of these antibiotics on peptide release catalyzed by ArfA/RF2 in a nonstop complex, we formed the nonstop complexes in the presence of the antibiotics and examined the rate of fMet release by RF2m. Our results showed that under a high concentration (500 µM), both antibiotics inhibited peptide release with viomycin, showing a more pronounced effect (Fig. 4A). We also obtained the same results for the RF2m-catalyzed peptide release on stop codon UGA (Fig. 4B).
FIGURE 4.
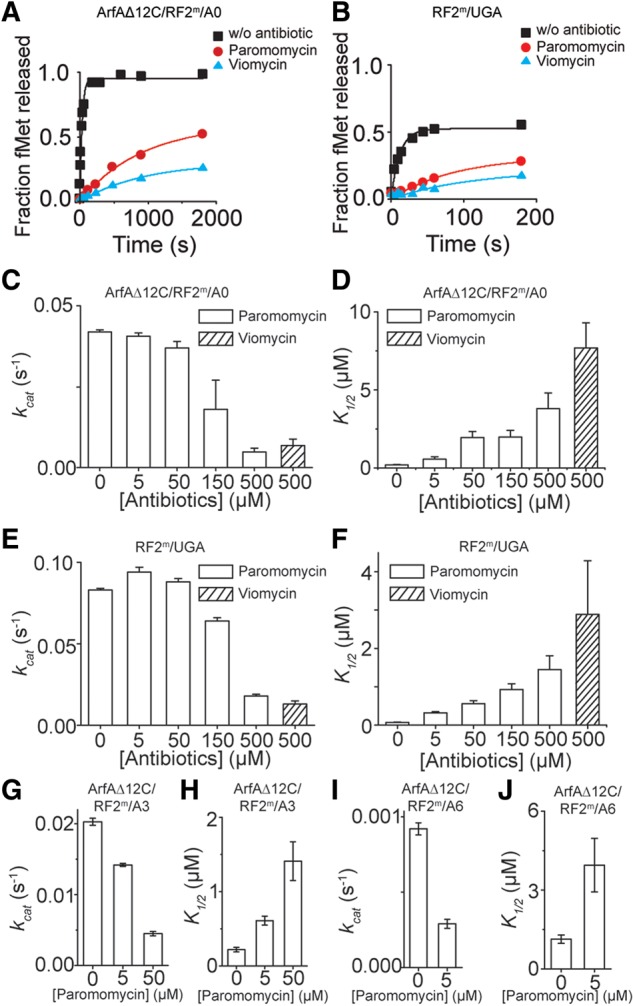
The effects of antibiotics viomycin and paromomycin on peptide release. Representative time courses of peptide release: (A) on A0 nonstop mRNA at 25 nM ribosomes by 62.5 nM ArfAΔ12C and 5 µM RF2m; (B) on stop codon UGA at 25 nM ribosomes by 5 µM fully methylated RF2m without antibiotics, and with 500 µM paramomycin or 500 µM viomycin bound. (C–J) Catalytic rate constants for peptide release in the absence or presence of antibiotics at different concentrations. kcat and K1/2 of peptide release by ArfAΔ12C/RF2m on A0 nonstop mRNA (C,D), by RF2m on the stop codon UGA (E,F), by ArfAΔ12C/RF2m on A3 mRNA (G,H), and on A6 mRNA (I,J) with different concentrations of antibiotics.
We further investigated the inhibitory effects of various concentrations (0–150 µM) of paromomycin on different nonstop complexes (Fig. 4C–J). Our results showed that when the concentration of paromomycin was below 150 µM, the kcat of peptide release by ArfAΔ12C/RF2m in the ribosome on A0 nonstop mRNA stayed nearly the same, but K1/2 increased as the concentrations of paromomycin increased (Fig. 4C,D). Similar degrees of the change in kinetic profiles represented by kcat and K1/2 were seen on stop codon UGA (Fig. 4E,F), supporting competitive binding of paromomycin and RF2. Thus, our data suggest a similar effect of the two antibiotics on canonical and nonstop termination pathways.
However, peptide release in the ribosome on A3 and A6 nonstop mRNAs showed increased sensitivity to paromomycin (Fig. 4G–J). The rates of peptide release decreased and K1/2 increased (Fig. 4G–J), indicating different conformations of the nonstop complexes on A3 and A6 mRNAs compared to the one formed on A0 mRNA. The same effect of paromomycin on peptide release at a near-cognate codon was seen by Green and colleagues (Youngman et al. 2007).
Combining the binding results obtained in the previous section, our data strongly suggest that ArfA/RF2 decode a nonstop mRNA and hydrolyze the nascent peptide from the P-site tRNA by an induced-fit mechanism.
The end points of release reactions in the viomycin- and paromomycin-inhibited ribosomes were decreased (Fig. 4A,B), suggesting changes of ribosomal conformation as a result of the binding of the antibiotics. Importantly, in contrast to the peptide release in the nonstop complexes on A6 and A9 mRNAs, elevating concentrations of Mg2+ helped to recover the compromised end points and led to an increased fraction of peptide release on the viomycin-bound ribosome with A0 mRNA bound (Supplemental Fig. S3B). This observation indicated that the antibiotic-bound ribosomes were trapped in a less active state for the peptide release. Addition of the magnesium ions helped the ribosome to recover from the less-active state, presumably through stabilizing RNA structures, and helped the system transit into an active state, thereby faciliating the progression of the hydrolytic reaction of peptide release.
Effects of class II release factor RF3 on the ArfA/RF2-mediated peptide release in the ribosome
Since our results thus far suggested a shared mechanism of peptide release on a nonstop mRNA by ArfA/RF2 and the release on a stop codon by RF2, we asked whether they also share the same immediate downstream event. In canonical translational termination, after the peptide release, in some species of bacteria including E. coli, a class II release factor and also a translational GTPase, RF3, binds to the ribosome in the GTP state and accelerates the disassociation of the RF1 or RF2. Finally, GTP hydrolysis on RF3 leads to the subsequent release of RF3 from the ribosome. The function of RF3 in translational termination was proposed by Ehrenberg and colleagues (Zavialov et al. 2001, 2002). Recent biochemical (Koutmou et al. 2014; Peske et al. 2014) and structural studies (Jin et al. 2011) showed that RF3 with GTP bound (RF3·GTP) tightly associates with the ribosome in its rotated state, promoting the disassociation of class I release factors RF1 or RF2.
However, counter-intuitively, RF3·GTP inhibited the peptide release by ArfA and RF2 when RF2 was present in excess (single-turnover peptide release), even though ArfA/RF2-mediated termination is mechanistically similar to the RF2-mediated termination at a stop codon, as shown in Figure 5. At a sub-stoichiometric concentration of RF2 (multiple turnover peptide release), the presence of RF3 did not promote peptide release by RF2 (Supplemental Fig. S4).
FIGURE 5.
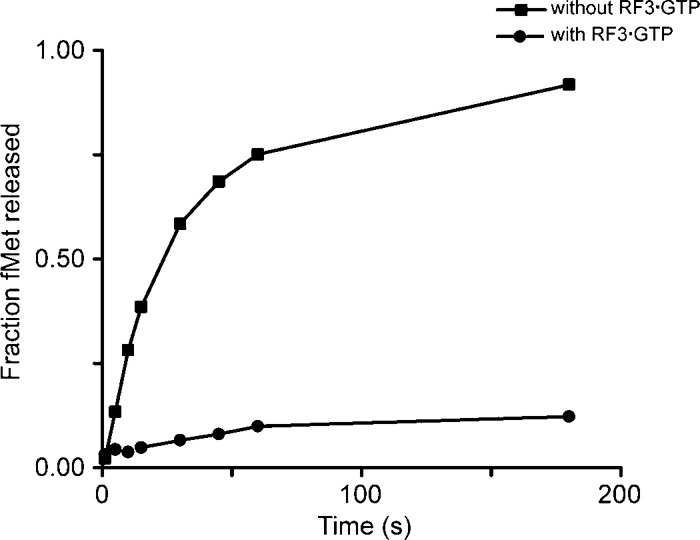
RF3·GTP inhibits the peptide release on a nonstop mRNA by ArfAΔ12C and RF2m in the ribosome. Time courses were obtained by incubating 25 nM release complexes with 62.5 nM ArfAΔ12C and 1 µM RF2m in the absence (▪) or presence (•) of 15 µM RF3·GTP.
RF3·GTP binds to the ribosome in the hybrid state (Jin et al. 2011), but RF2 catalyzes peptide release in the classical state (Weixlbaumer et al. 2008; Jin et al. 2010). One possible explanation for the failure to stimulate peptide release is that unlike the termination at a stop codon by RF2, after the peptide release on a nonstop mRNA by ArfA/RF2, the presence of ArfA “locks” the ribosome and stabilizes the ribosome in a defined state, most likely a classical state, thereby preventing RF3·GTP from sampling a rotated state. In contrast, normal peptide release in the P site “unlocks” the ribosome, allowing the ribosomal subunits to rotate relative to each other, thus offering an opportunity for RF3-GTP to sample different states of the ribosome.
DISCUSSION
Bacterial class I release factor RF2, plays a unique function in the translational quality control. In resolving a nonstop translation, in addition to the tmRNA and SmpB, RF2 interacts with alternative ribosome rescuing factors to rescue the stalled ribosome.
In this study, we investigated the mechanism of peptide release on nonstop mRNAs in the ribosome by RF2 and alternative ribosome rescuing factor A in E. coli. We first reconstituted a well-defined in vitro system using purified components, and verified that ArfA binds to the 30S and only recruits RF2 in the ribosome. RF1 was not recruited to a nonstop mRNA under all experimental conditions that we tested. We showed in the absence of ArfA, RF2 did not catalyze the peptide release on the nonstop RNA. ArfA substantially promotes the binding of the RF2 to the ribosome and the catalytic rate of the reaction of peptide release.
An induced-fit mechanism for ArfA/RF2-catalyzed peptide release
Using RF2 with methylated glutamine in the GGQ motif (termed RF2m in this study), we studied the mechanism of ArfA/RF2-catalyzed peptide release in a nonstop translation. Several lines of evidence established that, ArfA and RF2 catalyzed the peptide release on a nonstop mRNA by an induced-fit mechanism.
First, we demonstrated that the more occupied mRNA channel downstream the ribosomal P site, the more drastic decrease of the rate constants of the peptide release (Fig. 3), although both ArfA and RF2 could bind to the ribosome with a similar affinity (Fig. 1). In general, peptide release by RF2m on A3, A6, and A9 is characterized by rapid decreases in the rate constant and moderate increases in the concentration of RF2m required to achieve half of the kcat (K1/2). The kcat of the release reaction catalyzed by ArfA/RF2m decreased by 50% on the A3 mRNA, and it dropped to a much lower level that it was not measurable under our experimental conditions when A9 mRNA was present. On the A6 mRNA, the length of which is in between the A3 and A9, the overall kinetics of the release reaction resembled the one by RF2 on a near-cognate stop codon in canonical termination.
Second, the end points of the peptide release on A6 and A9 mRNAs decrease substantially compared with the ones on the A0 and A3 mRNAs, indicating different conformations of the enzyme–substrate complexes on the mRNA variants when both ArfA and RF2 bind to the ribosome.
Third, in the presence of ArfA, RF2 catalyzes the peptide release on a nonstop mRNA in the same way as RF2 does on a cognate stop codon. As shown in Table 1, kinetic parameters obtained from our experiments, kcat and K1/2 of the fMet released by ArfA/RF2m on the A0 mRNA in the ribosome, were similar to those by RF2m on a cognate stop codon UGA. Furthermore, the rates of peptide release in both systems, either catalyzed by ArfA/RF2 on a nonstop mRNA or promoted by RF2 on a stop codon UGA, showed a similar degree of response to the N5-methylation of the glutamine in GGQ motif of the RF2. This modification leads to ∼20-fold increase in the kcat of the release reaction by RF2 on the A0 mRNA in the presence of ArfA, leaving the binding of RF2 to the nonstop complex largely unchanged. An ∼10-fold change in the kinetics was observed on the peptide release by RF2 at the stop codon UGA. These results suggested that the two release pathways responded to the changes in the PTC in the same way. Assuming the rate-limiting step in the release pathway is not changed by the modification, our data indicated that N5-methylation of the glutamine positions this residue in an optimal position for the catalysis in the PTC of the ribosome. Although methylation of the glutamine in the GGQ motif substantially increases the rate of peptide release on cognate stop codons, it has almost no effect on the rate constant catalyzed by the same protein on near-stop codons. The same effect of the modification on the rate of peptide release by ArfA/RF2 was observed on A3 and A6 mRNA, which suggested that an occupied mRNA channel downstream from the P site in the ribosome compromises an induction of a fully active state of the RF2.
Fourth, we demonstrated that the canonical and nonstop termination pathways responded to the perturbations of antibiotics viomycin and paromomycin in the same way. We showed that viomycin and paromomycin competed with RF2 to bind to the ribosome, with viomycin showing a more pronounced effect. As suggested by almost the same degrees of changes in kcat and K1/2, the kinetic profiles of the release reactions catalyzed by ArfA/RF2m on the A0 mRNA and by RF2m on the UGA stop codon were very similar. Under 150 µM paromomycin, we demonstrated that the antibiotic did not affect the kcat of peptide release by ArfAΔ12C/RF2m on A0 nonstop mRNA, whereas the K1/2 were increased ∼10-fold, suggesting a competitive binding model of paromomycin and RF2 on ribosome in the cognate decoding conditions.
However, different kinetic profiles were obtained in peptide releases on A3 and A6 mRNAs, characterized by a decrease in the kcat and simultaneous increase in the K1/2 in the presence of paromomycin. The same kinetic profiles were seen in an earlier study on peptide release by RF1 on the near-cognate stop codon UCA/UAC codon (Youngman et al. 2007). These results suggest that A3/A6 mRNAs resemble the near-cognate decoding condition, and the stalled ribosome likely adopts a different conformation with longer mRNAs compared to the one when A0 mRNA is bound.
Taken together, our data support ArfA/RF2-catalyzed peptide release in the stalled ribosome by an induced-fit mechanism; ArfA/RF2m induce an active conformation which ultimately leads to optimal placement of the GGQN5-Me in the PTC to catalyze peptide release.
An induced-fit mechanism has been shown to be important for the tRNA selection in translational elongation (Pape et al. 1999) and stop codon recognition and peptide release in termination (Youngman et al. 2007). Here we showed that the same mechanism is used for the translational quality control. Termination of a protein synthesis when problematic translation occurs is an essential quality control mechanism that contributes to the accuracy and fidelity of the translation in all living cells. While different in the level of complexity and the details of molecular interactions, a release factor-like protein is involved in the quality control in all kingdoms of life. An important shared feature for all these processes is that the signal that intervention is required to originate in the decoding center of the ribosome and the actions that result in peptide release occur in the large ribosomal subunit. Therefore, it is not surprising that the molecular mechanism underlying this process is likely to be an induced-fit for every organism.
Conformation of GGQN5-Me in the PTC of the ribosome
Based on the conclusion above, it is reasonable to gain insights into the molecular interactions of ArfA and RF2 on a nonstop mRNA in the ribosome from those of a cognate stop codon and RF2 in the canonical translational termination. It is demonstrated that highly specific protein–RNA interactions between the decoding loop of the RF2 and the stop codon determined the specificity of the stop codon recognition (Korostelev et al. 2008; Weixlbaumer et al. 2008). These interactions must be replaced by specific protein–protein interactions between ArfA and RF2.
In the PTC, the backbone of the GGQ motif of the RF2 is likely positioned the same way as the ones seen in the previous structures. Although eukaryotic, archeal, and bacterial class I release factors share little sequence or structural identity, the GGQ motif that is required for catalytic activity (Song et al. 2000; Mora et al. 2003) of the peptide release is universally conserved. In the crystal structure, the backbones of the two conserved glycines adopt a conformation that is not possible for any other amino acid (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010), explaining why their mutation results in loss of the catalytic activity of the release factor (Zavialov et al. 2002; Shaw and Green 2007). The conformation of the two glycines is obviously important to optimally place the glutamine in the GGQ motif in the PTC.
The glutamine is important for the peptide release. The main chain of this residue is critically involved in the catalysis. In all the canonical termination complexes, the main chain amide group is within hydrogen bond distance to the catalytic water, suggesting that it directly coordinates the water for the hydrolytic reaction in releasing the nascent peptide from the P-tRNA. Furthermore, it has been demonstrated that a substitution of a glutamine by a proline abolished the peptide release activity by RF2 without deforming the overall conformation of the GGQ motif in the PTC of the ribosome (Santos et al. 2013). In addition to the main chain amide group, the side chain of the glutamine has been implicated in providing important packing for specifically selecting a water molecule as the nucleophile for the release reaction, as proposed from a seminal biochemical investigation (Shaw and Green 2007). Consistent with the results, in the crystal structure of a release complex with a nonhydrolysable tRNA-peptide analog that represents the translational state right before the peptide release (Jin et al. 2010), conformation of the glutamine suggests that it creates a tightly packed pocket to accommodate a water molecule for the catalysis of the peptide release. Finally, molecular dynamic simulations showed the side chain of the glutamine made an entropic contribution to the hydrolytic reaction (Trobro and Aqvist 2009).
In line with the results obtained by earlier investigations, our kinetic data supported the view that N5-methylation of the glutamine in the GGQ motif positions the catalytic glutamine in an optimal position for the catalysis in the PTC. The packing of the catalytic pocket by class I release factors in the PTC, conceivably, would be further facilitated by methylation of the glutamine. The observation that a Q240E mutant (Q240 is the catalytic glutamine in the structure) has reasonable peptide release activity suggests a role for the carbonyl oxygen of Q240 in coordinating the catalytic water (Shaw and Green 2007). Therefore, methylation at the N5 could help to position the amine away from the catalytic center and thus orient the carbonyl oxygen of the Q240 toward the water molecule for an optimal attack on the ester bond.
Effects of RF3 on the peptide release by ArfA/RF2m and in the ribosome
The class II release factor RF3 in the GTP-binding state promotes the recycling of the class I release factors on the ribosome after the nascent peptide is released (Zavialov et al. 2001; Koutmou et al. 2014; Peske et al. 2014). Therefore, the presence of RF3-GTP increases the catalytic efficiency of the peptide release catalyzed by RF1 or RF2, but leaving the rate of the hydrolytic reaction unchanged in canonical termination. In contrast, RF3 does not promote recycling of the RF2 and ArfA in the nonstop complex. Instead, it inhibits the peptide release by ArfA and RF2 when RF2 is present in excess. These observations suggest that while mechanistically resembling a canonical termination on a cognate stop codon by RF2, ArfA/RF2-mediated termination on a nonstop mRNA likely proceeds toward a different downstream pathway that does not involve RF3 for immediate recycling.
MATERIALS AND METHODS
Ribosome preparation, translation initiation, and release factors
Ribosomes from E. coli MRE600 (a kind gift from Huang, R.H.) were purified as previously described (Shaw and Green 2007). E. coli IF1, IF2, and IF3 were overexpressed in BL21 (DE3) cells and purified with Ni-NTA column (Wolfrum et al. 2003). N-terminal His-tags in these proteins were removed by HRV-3C protease during purifications. E. coli Methionine-tRNA synthetase (MetRS) and methionyl-tRNA formyltransferase (FMT) were overexpressed in BL21 (DE3) cells and purified (Schmitt et al. 1996).
To obtain fully methylated RF2m, N-terminally His-tagged RF2 was coexpressed with its cognate methyltransferase (PrmC) in E. coli BL21 (DE3) strain under induction with 0.5% lactose at 25°C (Monteiro et al. 2000). N-terminally His-tagged RF3 from E. coli was expressed using the T5 vector pET-30. Both protein release factors were purified on a Ni-NTA affinity column, followed by ion-exchange chromatography and gel filtration (Jin et al. 2010, 2011). RF3·GTP was formed by incubating 15 µM RF3 with 10 mM GTP at 37°C for 10 min.
The 12 C-terminal amino acids removed ArfA (designated as ArfAΔ12C) was cloned as described (Chadani et al. 2010, 2011). ArfAΔ12C was overexpressed with the MBP fusion in BL21 (DE3) cells and purified using Ni-NTA in buffer A (25 mM Tris, pH 7.5, 200 mM NaCl, 0.1 mM PMSF, and 0.1 mM benzamidine). The MBP protein was removed by TEV protease after the purification step.
All E. coli ribosomes and proteins except IF2 were stored in buffer B (50 mM Tris, pH 7.4, 70 mM NH4Cl, 30 mM KCl, 10 mM MgCl2, and 5 mM β-mercaptoethanol) (Zaher and Green 2009; Petropoulos et al. 2014). E. coli IF2 was stored in buffer C (50 mM HEPES, pH 7.6, 1 M NH4Cl, 10 mM MgCl2, and 5 mM β-mercaptoethanol).
Charged tRNA preparation
tRNAfMet was overexpressed and purified (Jin et al. 2010, 2011). Aminoacylation and formylation of tRNAfMet were performed with l-[35S]-methionine (PerkinElmer) using MetRS and FMT as described (Walker and Fredrick 2008) with minor modifications. Briefly, 20 µM tRNAfMet was incubated with 1 µM L-[35S]-methionine, 0.6 mM l-methionine, 12 µM MetRS, 12 µM FMT, 0.6 mM 10-formyltetrahydrofolate, 10 mM ATP, and 0.02 U/µL pyrophosphatase in buffer D (100 mM HEPES-KOH, pH 7.6, 20 mM KCl, 10 mM MgCl2, and 1 mM DTT) at 37°C for 40 min. The resulting fMet-tRNAfMet was then purified by Phenol:chloroform:isoamyl alcohol (25:24:1, v/v) and chloroform extraction followed by ethanol precipitation and dissolved in 2 mM NaOAc, pH 5.2.
mRNA
mRNAs were purchased from Dharmacon (GE, Thermo Scientific) with the various sequences that are suitable for our studies. Sequences of mRNAs used in peptide release assays are listed as following (AUG in bold letters indicates the P-site codon):
SD-PMet-UGA: GGCAAGGAGGAAAAAAUGUGAUACA
SD-PMet-UGG: GGCAAGGAGGAAAAAAUGUGGUACA
SD-PMet-0 (A0): GGCAAGGAGGUAAAAAUG
SD-PMet-3 (A3): GGCAAGGAGGUAAAAAUGAAA
SD-PMet-6 (A6): GGCAAGGAGGUAAAAAUGAAAAAA
SD-PMet-9 (A9): GGCAAGGAGGUAAAAAUG AAAAAAAAA
SD-PMet-12 (A12): GGCAAGGAGGUAAAAAUGAAAAAAAAAAAA
Release factor binding assay
For the formation of a canonical termination complex, release complexes containing 70S ribosome, tRNAfMet and mRNA with AUG and stop codons in the P and A sites, respectively, were formed. These complexes were incubated with fourfold excess of RF2 at 37°C. For the formation of nonstop ribosomal complexes, nonstop mRNAs and twofold excess of ArfAΔ12C were used instead.
For ArfAΔ12C-ribosome association assay, the nonstop ribosomal complex was passed through a 15%–30% sucrose gradient in buffer B at 68,300g in a SW 32 Ti rotor for 15 h. The 70S peak was pooled and then dialyzed against buffer B with 2 mM Mg2+ instead of 10 mM Mg2+. Ribosomal subunits were subsequently separated by a 15%–30% sucrose gradient in buffer B (2 mM Mg2+) at 68,300g in a SW 32 Ti rotor for 18 h. The 50S and 30S peaks were collected and the association of ArfAΔ12C was examined by Western blot using primary and secondary antibodies, monoclonal anti-polyhistidine (Sigma-Aldrich) and anti-Mouse IgG (whole molecule)-Alkaline Phosphatase (Sigma-Aldrich), respectively.
Specificity assay recognitions of a stop codon by RF2 or a nonstop mRNA by ArfAΔ12C/RF2 were performed using size-exclusion chromatography on a Superdex 200 (10/300) column (Amersham/GE Healthcare) and SDS–PAGE. Coomassie-stained SDS gels were used to show binding of RF2 to the ribosome, and Western blot was used to examine the binding of ArfAΔ12C using Anti-Mouse IgG (H+L) Antibody (Human Serum Adsorbed and Peroxidase Labeled, KPL) as the secondary antibody.
Release complex formation
Ribosomal complexes with P-site fMet-tRNAfMet were formed by incubating ribosome (2 µM), mRNA (6 µM), IF1, IF2, IF3 (3 µM each), GTP (2 mM), and fMet-tRNAfMet (3 µM) in buffer B at 37°C for 30 min (Zaher and Green 2009; Petropoulos et al. 2014). The reactions were then pelleted through 0.5 mL of 1 M sucrose cushion in buffer B at 176,800g in a Type 45 Ti rotor for 24 h (Zaher and Green 2009). The resulting complexes were resuspended in buffer B, aliquoted, and stored at −80°C.
Peptide release assay
The kinetic experiment on peptide release was done essentially as described (Freistroffer et al. 2000; Zaher and Green 2009; Petropoulos et al. 2014) with modifications. First, ribosomal termination or nonstop complexes containing E. coli 70S ribosomes, f-[35S]-Met-tRNAfMet and mRNA with AUG codon in the P-site were formed as described in the previous section. The release complexes (25 nM) reacted with an excess amount of RF2 (5 µM) or ArfAΔ12C/RF2 (62.5 nM/5 µM) in buffer B at 37°C. For the assays with antibiotics, the release complex (25 nM) was first incubated with 500 µM antibiotic at 37°C for 10 min (Youngman et al. 2007). The reactions were quenched by adding 5% ice cold trichloroacetic acid at different time points, and precipitants were spun down using 18,000g for 10 min at 4°C to separate f-[35S]-Met from f-[35S]-Met-tRNAfMet. Subsequently, the supernatant was withdrawn and the released f-[35S]-Met was counted in 5 mL of ScintiSafe Econo 1 Cocktail (Fisher Scientific). The maximum releasable fMet (fMetMax) was determined by incubating release complex (25 nM) with 100 µM puromycin (Sigma-Aldrich) at 37°C for 30 sec (Freistroffer et al. 2000). The fraction of f-[35S]-Met was determined by the ratio between the released f-[35S]-Met and fMetMax. The background level of released peptides in the absence of release factor was measured at the same time points and subtracted from each time point in the release reaction.
To determine the kcat and K1/2, 25 nM release complexes were reacted with different concentrations of release factors (50 nM–30 µM) during 1 sec to 6 min depending on the kinetics of the reaction (Freistroffer et al. 2000). The fraction of released fMet (Ft) versus time was plotted and kobs and reaction end point (Fmax) were obtained by fitting the curve to single exponential equation Ft = Fmax · (1 − e−kobs · t). Alternatively, kobs at each concentration was calculated using the equation kobs = −ln(1 − Ft/Fmax)/t, which yielded to the same results. The kcat and K1/2 were obtained by plotting kobs versus different concentrations of the release factor (RF) and fitting the curve using equation kobs = kcat · [RF]/(K1/2 + [RF]).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Peter B. Moore at Yale University for a critical review of the manuscript and Professor Hani Zaher at Washington University in St. Louis for helpful discussions on kinetic experiments. We also thank the Biotechnology Center at University of Illinois at Urbana-Champaign for the mass spectrometric analysis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.053082.115.
REFERENCES
- Capecchi MR. 1967. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci 58: 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey CT, Tompkins R, Scolnick E, Caryk T, Nirenberg M. 1968. Sequential translation of trinucleotide codons for the initiation and termination of protein synthesis. Science 162: 135–138. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. 2010. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol 78: 796–808. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Matsumoto E, Aso H, Wada T, Kutsukake K, Sutou S, Abo T. 2011. Trans-translation-mediated tight regulation of the expression of the alternative ribosome-rescue factor ArfA in Escherichia coli. Genes Genet Syst 86: 151–163. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ito K, Kutsukake K, Abo T. 2012. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol Microbiol 86: 37–50. [DOI] [PubMed] [Google Scholar]
- Dincbas-Renqvist V, Engström A, Mora L, Heurgué-Hamard V, Buckingham R, Ehrenberg M. 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19: 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Wang J, Sternberg SH, MacDougall DD, Elvekrog MM, Pulukkunat DK, Englander MT, Gonzalez RL Jr. 2010. A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol 472: 221–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. 2000. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci 97: 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. 2012. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335: 1370–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sanchez F, Schaub RE, Janssen BD, Hayes CS. 2011. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol Microbiol 80: 1204–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgué-Hamard V, Champ S, Mora L, Merkulova-Rainon T, Kisselev LL, Buckingham RH. 2005. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J Biol Chem 280: 2439–2445. [DOI] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y. 1998. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci 95: 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y. 2000. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403: 680–684. [DOI] [PubMed] [Google Scholar]
- Ito K, Chadani Y, Nakamori K, Chiba S, Akiyama Y, Abo T. 2011. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One 6: e28413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. 2004. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol 338: 33–41. [DOI] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Ramakrishnan V. 2011. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci 108: 15798–15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Feaga HA. 2014. Resolving nonstop translation complexes is a matter of life or death. J Bacteriol 196: 2123–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. 2003. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421: 90–94. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF. 2010. Recognition of the amber UAG stop codon by release factor RF1. EMBO J 29: 2577–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, McDonald ME, Brunelle JL, Green R. 2014. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA 20: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlenkoetter S, Wintermeyer W, Rodnina MV. 2011. Different substrate-dependent transition states in the active site of the ribosome. Nature 476: 351–354. [DOI] [PubMed] [Google Scholar]
- Kurita D, Chadani Y, Muto A, Abo T, Himeno H. 2014. ArfA recognizes the lack of mRNA in the mRNA channel after RF2 binding for ribosome rescue. Nucleic Acids Res 42: 13339–13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857. [DOI] [PubMed] [Google Scholar]
- Modolell J, Vázquez. 1977. The inhibition of ribosomal translocation by viomycin. Eur J Biochem 81: 491–497. [DOI] [PubMed] [Google Scholar]
- Monteiro RA, Souza EM, Yates MG, Pedrosa FO, Chubatsu LS. 2000. Use of lactose to induce expression of soluble NifA protein domains of Herbaspirillum seropedicae in Escherichia coli. Can J Microbiol 46: 1087–1090. [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgué-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH. 2003. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol 47: 267–275. [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. 2012. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335: 1366–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. 2014. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res 42: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos AD, McDonald ME, Green R, Zaher HS. 2014. Distinct roles for release factor 1 and release factor 2 in translational quality control. J Biol Chem 289: 17589–17596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FVT IV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. 2005. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Rawat UBS, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. 2003. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90. [DOI] [PubMed] [Google Scholar]
- Santos N, Zhu J, Donohue JP, Korostelev AA, Noller HF. 2013. Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure 21: 1258–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub RE, Poole SJ, Garza-Sánchez F, Benbow S, Hayes CS. 2012. Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs. J Biol Chem 287: 29765–29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Mechulam Y, Ruff M, Mitschler A, Moras D, Blanquet S. 1996. Crystallization and preliminary x-ray analysis of Escherichia coli methionyl-tRNA(fMet) formyltransferase. Proteins 25: 139–141. [DOI] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M. 1968. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci 61: 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JJ, Green R. 2007. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell 28: 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y. 2012. ArfA recruits RF2 into stalled ribosomes. J Mol Biol 423: 624–631. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. 2000. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321. [DOI] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. 2010. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol 17: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. 2007. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell 27: 758–766. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. 2009. Mechanism of the translation termination reaction on the ribosome. Biochemistry 48: 11296–11303. [DOI] [PubMed] [Google Scholar]
- Walker SE, Fredrick K. 2008. Preparation and evaluation of acylated tRNAs. Methods 44: 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KS, Ito K, Noller HF, Nakamura Y. 2000. Functional sites of interaction between release factor RF1 and the ribosome. Nat Struct Biol 7: 866–870. [DOI] [PubMed] [Google Scholar]
- Wolfrum A, Brock S, Mac T, Grillenbeck N. 2003. Expression in E. coli and purification of Thermus thermophilus translation initiation factors IF1 and IF3. Protein Expr Purif 29: 15–23. [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizugichi Y, Nierhaus KH, Wittmann HG. 1978. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275: 460–461. [DOI] [PubMed] [Google Scholar]
- Youngman EM, He SL, Nikstad LJ, Green R. 2007. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell 28: 533–543. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell 10: 789–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.