Abstract
MicroRNAs (miRNAs) suppress targeting gene expression through blocking translation or triggering mRNA degradation and, in general, act in trans, through a partially complementary interaction with the 3′ untranslated region (3′ UTR) or coding regions of a target gene. Although it has been reported previously that some miRNAs suppress their target genes on the opposite strand with a fully complementary sequence (i.e., natural antisense miRNAs that act in cis), there is no report to systematically study such cis-antisense miRNAs in different animal species. Here we report that cis-antisense miRNAs do exist in different animal species: 48 in Caenorhabditis elegans, 17 in Drosophila, 36 in Mus musculus, and 52 in Homo sapiens using a systematical bioinformatics approach. We show that most of these cis-antisense miRNAs can efficiently reduce the expression levels of their target genes in human cells. We further investigate hsa-miR-3661, one of the predicted cis-antisense miRNAs, in detail and demonstrate that this miRNA directly targets the coding sequence of PPP2CA located on the opposite DNA strand and inhibits the PPP2CA expression. Taken together, these results indicate that cis-antisense miRNAs are conservative and functional in animal species including humans.
Keywords: miRNA, cis-antisense, gene expression, miR-3661, PPP2CA, human cells
INTRODUCTION
MicroRNAs (miRNAs) are a class of small RNA genes with 21–25 nt, which regulate their target genes in various species. Most animal miRNAs regulate the expression of other genes predominantly in trans through partially complementary interactions with 3′ untranslated regions (3′ UTRs) or transcript coding regions of the target genes. However, some animal miRNAs are discovered as transcribed antisense to exons of noncoding or protein-coding genes (Bartel 2004). Such antisense exonic miRNAs, together with the intronic miRNAs and the miRNAs in the sense strand of genes, are termed as intragenic miRNAs (Rodriguez et al. 2004; Hinske et al. 2010, 2014). The function of such antisense exonic miRNAs were investigated in mouse miR-126 and miR-136 that are transcribed antisense of a retrotransposon-like gene (RTL1) and a down-regulated RTL1 mRNA level through a fully complementary sequence match with the target gene (Seitz et al. 2003). These cis-antisense exonic miRNAs are similar to the natural antisense miRNAs (nat-miRNAs) reported later in the plant (Lu et al. 2008), but with one main difference: cis-antisense miRNAs are derived from precursors with a compact hairpin, whereas nat-miRNAs are derived from the precursors with large introns. To date, cis-antisense miRNAs were only reported in mouse; whether these cis-antisense miRNAs functionally exist in various animal species including humans has not been systematically studied.
To address this question, we performed a genome-wide analysis to identify miRNAs that are transcribed antisense to exons of known genes. As a result, we found that such cis-antisense miRNAs with a fully complementary antisense sequence against their target (protein-coding or noncoding) genes exist in different animal species: 48 in C. elegans, 17 in Drosophila, 36 in mouse, and 52 in the human genome (including miR-127/136 as reported [Seitz et al. 2003]). We then demonstrated that almost all of the cis-antisense miRNAs efficiently suppressed the expression levels of their target genes in human cells. We further studied in detail the effects of one cis-antisense miRNA, miR-3661, on its target gene, PPP2CA.
PP2Acα is the catalytic subunit of PP2A and encoded by the PPP2CA gene. PP2A is one of the major cellular serine–threonine phosphatases (Wera and Hemmings 1995). PP2A is a bona fide tumor suppressor consisting of multiple different subunits and targets a multitude of substrates (Janssens and Goris 2001; Sablina and Hahn 2008). PP2Acα is highly conserved from yeast to human. Loss-of-function mutations in PP2A subunits or up-regulation of PP2A-specific inhibitors that contribute to transformation were found in a variety of cancers (Eichhorn et al. 2009). One of the most important dephosphorylating substrates for PP2A is AKT (Andjelković et al. 1996; Meier et al. 1998; Chen et al. 1999; Sato et al. 2000; Ivaska et al. 2002; Resjö et al. 2002). PP2A negatively regulates the AKT pathway by dephosphorylating AKT itself and GSK-3β, an AKT-phosphorylated target (Shaw et al. 1997), which plays a critical role in cell proliferation, unlike the regulation of PP2A at the transcriptional level that has been reported in detail (Altiok et al. 1997; Chen et al. 2009; Liu et al. 2012). There are few reports on the regulation of PPP2CA at the post-transcriptional level, particularly via a miRNA-related regulation. Only recently, it was reported that miR-155 can negatively regulate PP2A (Lashine et al. 2015). Our results indicate that miR-3661 can regulate PPP2CA expression in human cells in a cis-antisense manner. These results demonstrate that the cis-antisense miRNAs conservatively and functionally exist in humans.
RESULTS
Fifty-two potential cis-antisense miRNAs and their antisense transcribed genes are identified in the human genome
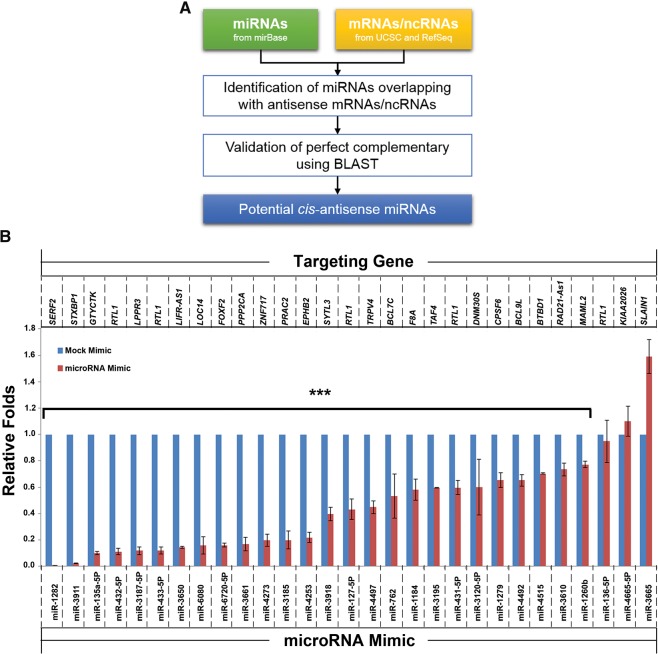
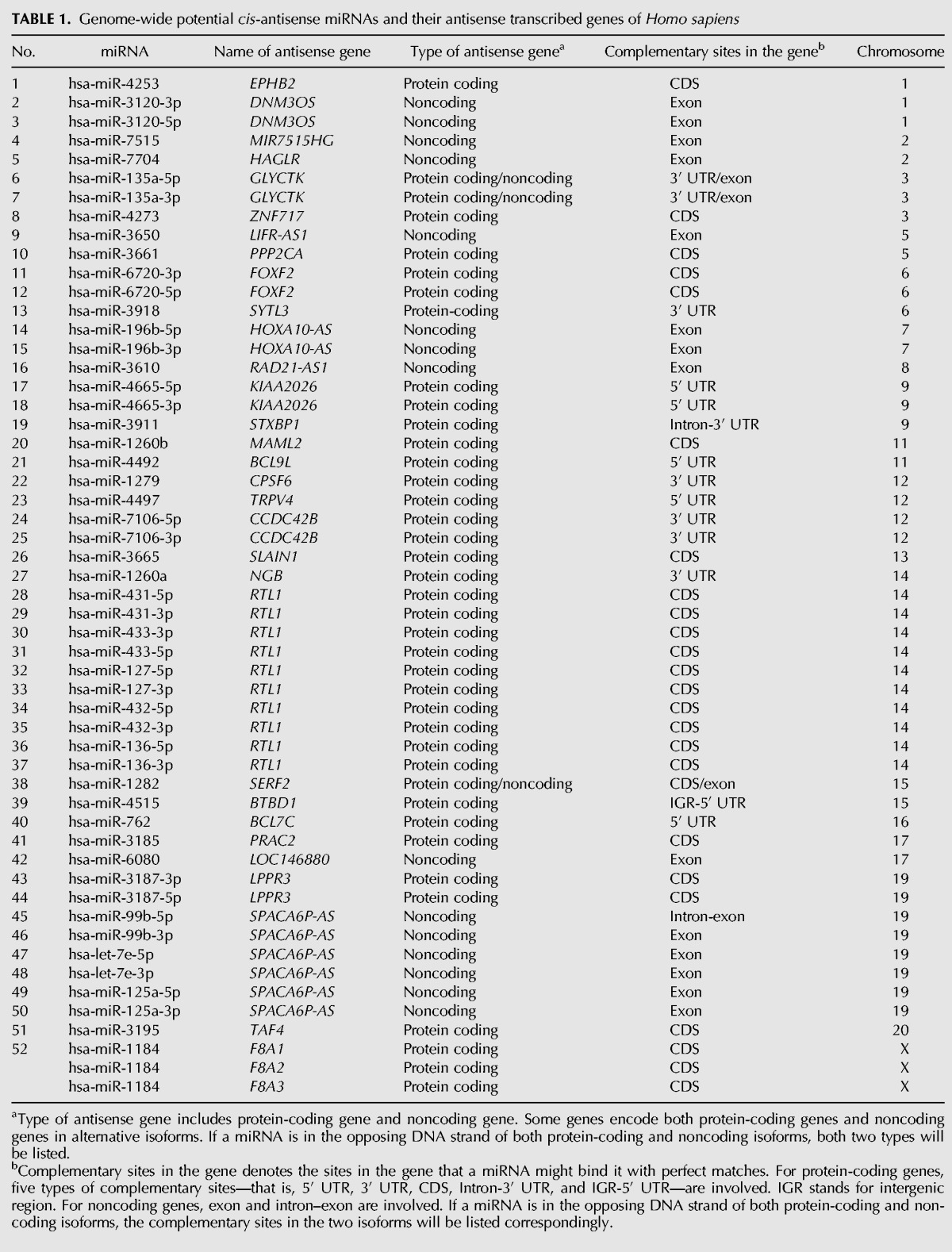
We performed a genome-wide analysis to identify miRNAs that are transcribed antisense to known genes (Fig. 1A). In the human (Homo sapiens) genome, 432 miRNAs were found in the antisense orientation of 313 protein-coding/noncoding genes. Among the 432 miRNAs, 380 miRNAs that are in the opposite DNA strands at the genomic locus of introns of 280 genes were excluded from this study. Finally 52 miRNAs (including miR-127/136 as reported [Seitz et al. 2003]) were identified as the candidates for suppressing the expression of their 33 antisense target genes in a cis-antisense manner with fully complementary sequences (Table 1). Among the 52 miRNAs, 25 miRNAs target coding sequence (CDS) (including miR-127/136 as reported [Seitz et al. 2003]) and 27 miRNAs target noncoding sequence including exons, noncoding genes 3′/5′ UTR of protein-coding genes (Table 1). To identify the cis-antisense miRNAs in other animal species, we applied the same analysis procedure to Mus musculus (M. musculus), Drosophila melanogaster (D. melanogaster), and Caenorhabditis elegans (C. elegans). As a result, we identified 235 miRNAs in M. musculus, 66 miRNAs in D. melanogaster, and 177 miRNAs in C. elegans genome, in the antisense orientation of protein-coding/noncoding genes, respectively. After excluding the miRNA transcribed antisense to the introns of genes, finally 36 (M. musculus), 17 (D. melanogaster), and, 48 (C. elegans) cis-antisense miRNAs were obtained (Supplemental Tables 1–3).
FIGURE 1.
Identification of cis-antisense miRNAs. (A) Flowchart of genome-wide identification of potential cis-antisense miRNAs. (B) Most of the identified cis-antisense miRNAs functionally suppress their targets in human cells. RNA levels of the cis-targeting genes (their names are listed at the top of the figure) were measured using 293FT cells transfected with the control RNA (mock mimic) and the indicated miRNAs mimic (their names are listed at the bottom of the figure) at 30 nM final concentration. Each assay was repeated in at least three independent experiments. The statistical analysis was performed by comparing the mRNA levels of target genes from the cells treated with miRNA mimic to that treated with control RNA (mock mimic). (***) P < 0.001.
TABLE 1.
Genome-wide potential cis-antisense miRNAs and their antisense transcribed genes of Homo sapiens
Most of the identified cis-antisense miRNAs functionally suppress their targets in humans
Next, we were interested in determining whether these human miRNAs can biologically suppress their antisense target genes. Among the 52 miRNAs, 30 miRNAs are from 15 precursors (5p and 3p strands of a pre-miRNA) targeting the same gene (Table 1), and 22 miRNA target genes individually. Therefore, we tested the 37 miRNAs for functional measurement. We obtained 29 commercially available miRNA mimics and the TaqMan assays of their target genes. We transfected these miRNA mimics to 293FT cells and measured their target expressions using a real-time PCR approach. The results showed that most miRNA mimics can efficiently suppress the expression of their targets (Fig. 1B); only three miRNA mimics could not suppress their antisense targets: miR-4665-5p targeting the 5′ UTR of KIAA2026, miR-136-5p targeting the CDS of RTL1, and miR-3665 targeting the CDS of SLAN1 (Table 1; Fig. 1B). These results indicate that most of the cis-antisense miRNAs are functional.
MiR-3661 specifically targets PPP2CA in human cells
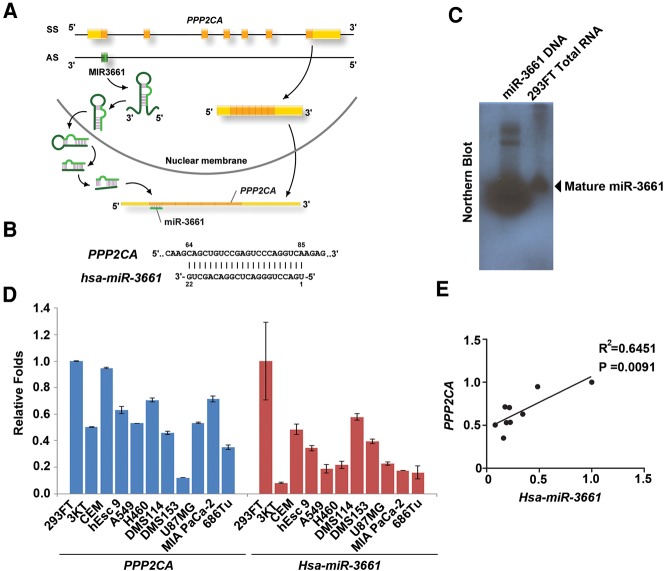
To further elucidate the functions of miRNAs acting in human cells, we chose miR-3661 because it targets PPP2CA that codes PP2Acα, an important catalytic subunit of PP2A. MiR-3661 was first identified in 2010 (Hansen et al. 2010) and then further confirmed by another group (Persson et al. 2011); however, its function remains unclear. We found that miR-3661 is located on the opposite strand of PPP2CA, the PP2Acα protein-coding region (Fig. 2A). A perfect complementary sequence exists between miR-3661 and the PPP2CA gene (Fig. 2B). The Northern blot data confirmed that miR-3661 is an expressed miRNA (Fig. 2C). To examine the relative expression pattern between miR-3661 and the PPP2CA gene, we compared the RNA levels of miR-3661 and PPP2CA in different human cell lines. The results showed a moderate positive correlation with high confidence between the expression of miR-3661 and the PPP2CA gene (Fig. 2D,E), suggesting that the miR-3661 and PPP2CA genes have a correlating expression profile.
FIGURE 2.
MiR-3661 is located on the opposite strand of the PPP2CA coding region in the human genome. (A) The genomic view of the PPP2CA and miR-3661 genes: coding sequence (CDS), untranslated region (UTR), and miRNA regions are represented as orange, yellow, and green bars, respectively. (AS) antisense strand, (SS) sense strand. The model for cis-antisense miRNA biosynthesis and function: the miRNA processing initiated with the splicing of pri-miRNA transcripts to yield pre-miRNA hairpins that are transported to cytoplasm. After Dicer cleavage, the mature miRNAs bind to target RNAs and direct the translation inhibition or cleavage of the sense transcripts of their targets. (B) The perfect complementary between PPP2CA and miR-3661. The binding positions are shown above (PPP2CA) and below (miR-3661) the sequences. (C) The mature miR-3661 was identified in 293FT cells using a Northern blot. The left lane was the synthetic DNA sequence of the miR-3661, which was as the positive control, and the right lane was the total RNA of 293FT cells. (D) RNA levels of PPP2CA and miR-3661 were measured from various human cell lines as indicated. (E) Scatterplots illustrating the correlation of gene expressions between PPP2CA and Hsa-miR-3661 from various human cell lines as indicated.
Because PPP2CA has another isoform, PPP2CA-iso (Migueleti et al. 2012), and the two forms share the same miR-3661 target sequence, we then investigated whether PPP2CA-iso can also be efficiently targeted by miR-3661. For this purpose, we examined the effects of miR-3661 targeting PPP2CA and PPP2CA-iso in different cell lines: MRC5SV (transformed fibroblast), Beas2B (transformed lung epithelial cells), H1299 (lung tumor cells), and A549 (lung tumor cells). The results showed that similar to PPP2CA, PPP2CA-iso is also efficiently suppressed by miR-3661 (Fig. 3A), indicating that both PPP2CA and its isoform are good targets of miR-3661. In addition, the protein levels of PP2Acα were efficiently suppressed by miR-3661 in different cell lines as well (Fig. 3B). To further study the specific effect of miR-3661 on the suppression of PPP2CA, we compared the effects of miR-3661 and different miRNAs (including miR-3133, miR-4282, miR-3607, miR-4703, and miR-4277) that are predicted (using miRanda [Betel et al. 2008] and TargetScan software [Lewis et al. 2005]) to have a potential for targeting 3′ UTR of PPP2CA in 293FT cells. The results showed that only the miR-3661 mimic efficiently suppressed the PP2Acα protein level, and all other miRNA mimics, similar to the control RNA, had no effect (Fig. 3C). Combining the transfection of the miR-3661 mimic and the miR-3661 inhibitor blocked the suppression effect of the miR-3661 mimic alone on the PP2Acα protein level (Fig. 3D). Next, we designed and constructed a vector encoding a Flag-tagged PPP2CA mutated at the targeting sequence by miR-3661 (Fig. 3E, left panel). The data showed that the mutant Flag-PP2Acα protein level cannot be affected by the miR-3661 mimic although the wild-type Flag-PP2Acα can be suppressed successfully (Fig. 3E, right panel). These results clearly demonstrate that the effect of the miR-3661 on suppressing the PP2Acα protein level is specific.
FIGURE 3.
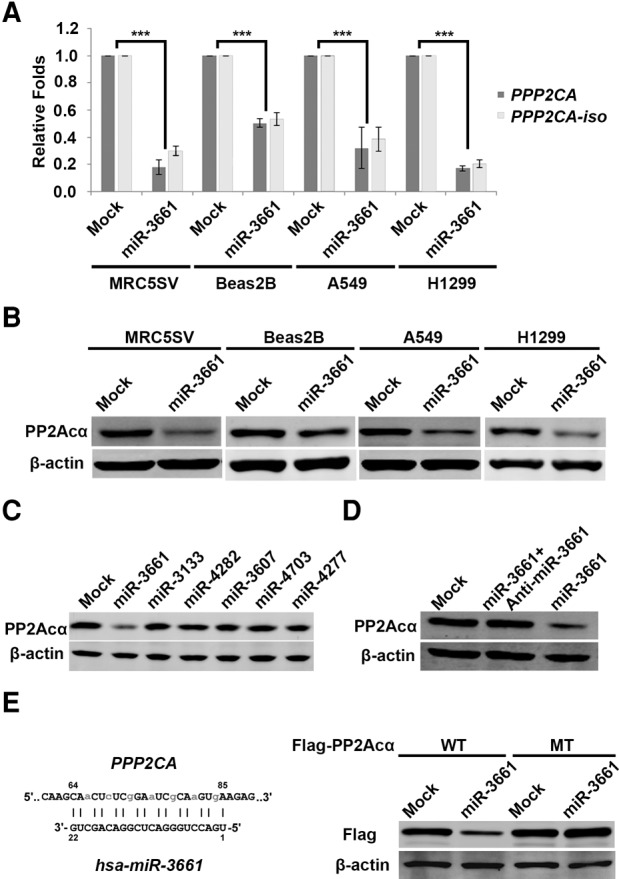
MiR-3661 specifically targets PPP2CA expression in human cells. (A) RNA levels of PPP2CA and PPP2CA-iso were measured from different human cell lines, MRC5SV, Beas2B, A549, and H1299, that were transfected with the control RNA (mock) or miR-3661 mimic at 30 nM final concentration. (B) The protein levels of PP2Acα were measured from MRC5SV, Beas2B, A549, and H1299 cells transfected with the control RNA (mock) or miR-3661 mimic at 30 nM final concentrations. β-actin was used as the internal loading control. (C) The protein levels of PP2Acα were measured from 293FT cells transfected with 30 nM of a control RNA or the predicted miRNAs as indicated. (D) The protein levels of PP2Acα were measured from 293FT cells transfected with the control RNA (mock), miR-3661 mimic alone or together with a miR-3661 inhibitor at 30 nM final concentration. (E) (Left panel) the top sequence is the mutant plasmid construction of PPP2CA, light gray: the mutated sites of the PPP2CA sequence targeted by Hsa-miR-3661 (bottom sequences). (Right panel) The protein levels of Flag-PP2Acα were measured from 293FT cells transfected with 30 nM of wild-type (WT) or mutant (MT) Flag-PP2Acα together with the mock or miR-3661.
Suppression of PP2Acα by miR-3661 increased its target phosphorylation in human cells
Next, we wanted to know whether pri-miR-3661 can be processed to a mature miRNA in cells and whether the mature miR-3661 can functionally target PP2Acα. For this purpose, we first constructed the vectors encoding pri-miR-3661 (miR-3661-1 and miR-3661-2) with different lengths at the 5′ and 3′ flanking sequences (Fig. 4A). After transfecting the plasmid into 293FT cells, both pri-miR-3661 plasmids (miR-3661-1 and miR-3661-2) resulted in a significant increase in the mature miR-3661 levels (Fig. 4B), supporting that pri-miR-3661 can be processed well to mature miR-3661 in the cells. At the same time, both pri-miR-3661 plasmid transfections efficiently decreased the levels of PPP2CA mRNA and protein, and increased the phosphorylation levels of AKT or GSK-3β (dephosphorylating substrates of PP2A) (Fig. 4C,D), supporting that pri-miR-3661 functionally targets PP2Acα. These results clearly indicate that the PP2A function is suppressed by miR-3661 to directly target PPP2CA.
FIGURE 4.
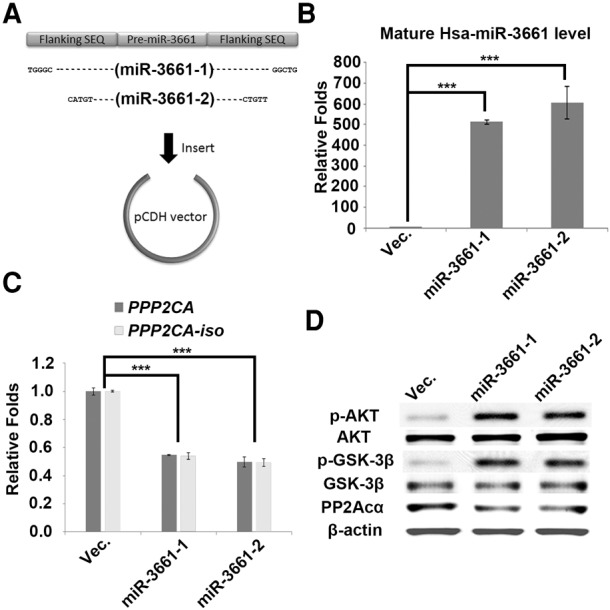
Suppression of PP2A by miR-3661 increases its target phosphorylation in human cells. (A) Schematic overview of pri-miR-3661 expression plasmids. (B) The RNA levels of Hsa-miR-3661 were measured from 293FT cells transfected with the control vector and two constructs of pri-miR-3661 expression plasmids. (C) The mRNA levels of PPP2CA and PPP2CA-iso were measured from 293FT cells transfected with the control vector and two constructs of pri-miR-3661 expression plasmids. (D) The protein levels of PP2Acα, phosphorylated AKT (p-AKT), AKT, phosphorylated GSK-3β (p-GSK-3β), and GSK-3β were measured from 293FT cells transfected with the control vector and two constructs of pri-miR-3661 expression plasmids.
DISCUSSION
Most miRNAs are found to suppress the expression of genes prevalently in trans. Some miRNAs are located within genomic regions of protein-coding genes or noncoding genes (Rodriguez et al. 2004; Hinske et al. 2010, 2014). These intragenic miRNAs were studied with their host genes and the results suggest that a complex transcription and regulation relationship exists between them (Monteys et al. 2010; Yan et al. 2011; Son et al. 2014). The subset of intragenic miRNAs, cis-antisense miRNAs that are transcribed antisense to exons of protein-coding or noncoding genes, is rarely studied. Although it has been reported that miR-127/136 in mouse cells is located on the opposite strand of RTL1 (Seitz et al. 2003), there is no report until now to systematically study whether the cis-antisense miRNAs functionally exist in different animal species. In this study, by systematically analyzing genomes from different animal species, we identified 52 cis-antisense miRNAs in the human (Homo sapiens), 48 in the C. elegans, 17 in the Drosophila, and 36 in the mouse (Mus musculus) genome. Then, we performed biological studies and showed that most cis-antisense miRNAs can suppress their targets well in human cells. We further investigated in detail the relationship between one of our predicted cis-antisense miRNAs, miR-3661, and its target PPP2CA in human cells. Our results showed that miR-3661 can efficiently suppress PPP2CA.
Among the 29 tested cis-antisense miRNAs, three (miR-4665-5p, miR-136-5p, and miR-3665) did not suppress their targets (Fig. 1B), which might be due to the following two reasons. One is that the three miRNAs may only inhibit the translation of the target genes, instead of trigging target degradation. The other is that the three miRNAs are all located near the 5′ end of the target genes and this region may form a complex structure that hinders the interaction of these miRNAs and their target RNAs. Target site accessibility has been found to be very important for miRNA recognition and targeting effectiveness (Kertesz et al. 2007). Of course, these possibilities need to be verified in future studies. Our results, the positive data from 26 among 29 tested miRNAs, indicate that most of the cis-antisense miRNAs are functional. We also investigated whether there were other miRNAs that had the potential to target 3′ UTR of PPP2CA. Using miRanda (Betel et al. 2008) and TargetScan software (Lewis et al. 2005), we predicted several miRNAs that target 3′ UTR of PPP2CA; however, none of them could efficiently suppress the PPP2CA expression as miR-3661 does (Fig. 3C). These results indicate that miR-3661 is the most efficient miRNA to target PPP2CA because of its perfect complementary to the target as compared with other 3′ UTR-targeting miRNAs.
Our results indicate that cis-antisense miRNAs function as a natural post-transcriptional regulator for the antisense transcribed genes. Because cis-antisense miRNAs and their antisense transcribed genes are on opposite strands, it is possible that they have their own transcriptional regulation. However, a correlational expression between miRNAs and their targets has been reported previously (Wang and Li 2009), although the underlying mechanism remains unclear. We also found a positive correlation between PPP2CA and miR-3661 (R2 = 0.6451; Fig. 2E). We thought that miR-3661 and PPP2CA might share the same enhancers or silencers or be under the same locus control region (LCR) because of the opposite strand location. Again, the prediction needs to be verified in future studies. In addition, our data do not exclude the other regulations of PPP2CA because of the importance of PP2A for normal physiologic cellular function. When PPP2CA was dramatically decreased by miR-3661, cells may have compensatively up-regulated PPP2CA via other pathways to maintain its normal function. A moderate positive correlation (R2 = 0.6451) between PPP2CA and miR-3661 also suggests the involvement of other regulation pathways. Thus, what we show in this study added miR-3661 as a new regulation factor to maintain the balance of PPP2CA, which contributes to the homeostasis of PP2A function.
In this study, we systematically investigated whether the antisense exonic miRNAs could regulate the target genes in the opposite strand in cis. The results showed that the cis-antisense miRNAs effectively suppressed the target genes transcribed in the opposite strand. However, cis-antisense miRNAs may also have other targets regulated in trans. For example, hsa-miR-135a-5p is validated to suppress the expression levels of antisense transcribed gene GLYCTK in our study (Fig. 1B). It was also reported to regulate the expression of JAK2, NR3C2, APC, HOXA10, and MYC in trans from miRTarBase, which contains the largest amount of validated miRNA–target interactions (Supplemental Fig. 1; Kozomara and Griffiths-Jones 2014). Whether cis-antisense miRNAs regulate cis- and trans-targets simultaneously or separately needs further verification.
Taken together, we report for the first time that cis-antisense miRNAs are functional in different animal species, particularly in humans. Such conserved cis-antisense regulation for some miRNAs may play an important role in adjusting their targeting gene expressions.
MATERIALS AND METHODS
Genome-wide identification of potential cis-antisense miRNAs and their antisense transcribed genes
The sequence and genomic annotations of human miRNAs were obtained from a miRBase database release 21 (Kozomara and Griffiths-Jones 2014). The sequences of protein-coding and noncoding genes were downloaded from the RefSeq database (Pruitt et al. 2014) and the genomic annotations from the UCSC genome browser (Karolchik et al. 2014). The genomic annotations of miRNAs and RefSeq genes were both based on the human genome assembly GRCh38. By comparing the genomic loci of miRNAs and exons of RefSeq genes, all the miRNAs that overlapped with the antisense exons were obtained for sequence analysis. The identified miRNAs were further validated by aligning each miRNA to the antisense gene sequences with the parameters “-word_size 11 -perc_identity 100 -strand minus -ungapped” (Altschul et al. 1990). The miRNAs that had a perfect complementary to the antisense mRNAs/ncRNAs were obtained as potential cis-antisense miRNAs. The same analysis procedure was performed to identify potential cis-antisense miRNAs in the other three species: M. musculus, D. melanogaster, and C. elegans. All the analyses were performed in the R programming language (version 3.0.2).
Cell lines and culture
Human cell lines—293FT (transformed embryo kidney cells, purchased from Invitrogen), MRC5SV (transformed human fibroblasts, as described previously [Liu et al. 2015]), U87MG (brain tumor cells, purchased from ATCCs), MIA PaCa-2 (pancreas carcinoma cells, purchased from ATCC), and TU686 (oral squamous cells, obtained from Dr. Georgia Chen's laboratory [Shin et al. 2013])—were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). CEM cells (leukemic cells, purchased from ATCC) were grown in MEM supplemented with 10% FBS. 3KT cells (hTERT-immortalized bronchial epithelial cells, obtained from Dr. Shay's laboratory [Ramirez et al. 2004]) were grown in keratinocyte-SFM medium (purchased from Invitrogen). Beas2B cells (transformed human brachial epithelial cells, purchased from ATCC) were grown in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham supplemented with 10% FBS. Small cell lung cancer cell lines, DMS114 and DMS153, were purchased from ATCC and grown in Waymouth's MB 752/1 medium supplemented with 10% FBS. Non-small cell lung cancer (NSCLC) cell lines, A549, H1299, and H460, were purchased from ATCC and grown in RPMI 1640 medium supplemented with 10% FBS. hESC-H9 cells (human embryo stem cells) were purchased from WISC Stem Cell Bank and grown on BD Matrigel hESC-qualified matrix-coated plates (BD Biosciences) in mTeSR 1 medium (purchased from Stem Cell Technologies).
Northern blot
The total RNA was extracted from 293FT cells. Twenty micrograms of total RNA was separated in prepared 12% Denaturing Urea Polyacrylamide Gel. After transferring to a nylon membrane (GE Healthcare), the membrane was hybridized with a special probe, 5-CAGCTGTCCGAGTCCCAGGTCA-Dig-3, to measure the mature miR-3661. At the same time, the synthesized DNA sequence of mature miR-3661 was used as a positive control. The membrane was soaked in a solution containing the DIG antibody according to the manufacturer's instructions (DIG Wash and Block Buffer Set; Roche). The signals were obtained by exposing the membrane to X-rays at room temperature as described previously (Hu et al. 2014).
Plasmid construction
The plasmid of encoding Hsa-miR-3661-1 and -2 was generated by inserting a PCR product from the human genomic library as a template with the primers (Forward1: 5-TGGGAATTCTGGGCGCCGGTCTCGGAGACTC-3; Reverse1: 5-TGCGG ATCCCAGCCTCAACAGCCGCCAGAAG-3; Forward2: 5-TGGGAATTCCATGTTGCAC CCTCCTCAC-3; and Reverse2: 5-TGCGGATCCAACAGCCGCCAGAAGTACAC-3) into the plasmid pCDH-CMV-MCS-EF1-Puro (SBI). The plasmid encoding Flag-human PPP2CA was generated by inserting a PCR product from a human cDNA library as the template with the primers (Forward: 5-TGAGAATTCAATGGACGAGAAGGTGTTC-3 and Reverse: 5-GTG GGATCCTTACAGGAAGTAGTCTGG-3) into the plasmid p3XFLAG-CMV (Sigma-Aldrich). Three rounds of PCR-mediated mutagenesis were done on p3XFlag-PPP2CA to generate Hsa-miR-3661 resistant PPP2CA expression plasmid with the primers (Forward: 5-CAaCTcTCgGAaTCgCAaGTgAAGAGCCTCTGCG AGAAG-3 and Reverse: 5-TCACTTGCGATTCCGAGAGTTGCTTGCACTCGTTCAGC-3), which is completely resistant to Hsa-miR-3661.
RNA isolation, RT-PCR, and real-time PCR
Total RNA was extracted from human cells using miRNeasy mini kits (QIAGEN). cDNA was synthesized using 500 ng of total RNA from each sample using the SuperScript VILO cDNA Synthesis Kit (Life Technologies), followed by triplicate qPCR reactions using TaqMan assays with the TaqMan Fast PCR Universal master mix on an Applied Biosystems 7500 Fast real-time PCR system. For miRNA, a TaqMan miRNA reverse transcription kit was used to prepare the products for the TaqMan miRNA assays as described previously (Liu et al. 2015). RNA levels of PPP2ACA and its isoform were measured using primers designed to detect either the standard (Forward1: 5-GAT CTT CTG TCT ACA TGG TGG TCT C-3 and Reverse1: 5-ACA CAT TGG ACC CTC ATG GGG AA-3) or the novel isoform (Forward1 and Reverse2: 5-CCA GTT ATA TCC CTC ATG GGG AAC-3). GAPDH was used as an internal control (forward primer: 5-TGC ACC ACC AAC TGC TTA GC-3; reverse primer: 5-GGC ATG GAC TGT GGT CAT GAG-3).
Western blot
Western blot was performed using standard techniques. The antibodies against human PP2Acα and β-actin were purchased from Santa Cruz Biotechnology Inc. The antibodies against human phosphorylated AKT (Thr308), AKT, phosphorylated GSK-3β (Ser9), and GSK-3β were purchased from Cell Signaling Technology Inc.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Georgia Chen for the cell lines and Ms. Doreen Theune for editing this manuscript. X.Y. acknowledges support from the China National High Technology Research and Development Program (2014AA020604), and Y.W. acknowledges support from the U.S. National Institutes of Health (CA186129, CA185882, and P30CA138292).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052894.115.
REFERENCES
- Altiok S, Xu M, Spiegelman BM. 1997. PPARγ induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev 11: 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andjelković M, Jakubowicz T, Cron P, Ming XF, Han JW, Hemmings BA. 1996. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci 93: 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Fucini RV, Olson AL, Hemmings BA, Pessin JE. 1999. Osmotic shock inhibits insulin signaling by maintaining Akt/protein kinase B in an inactive dephosphorylated state. Mol Cell Biol 19: 4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-G, Han W-J, Deng M, Qin J, Yuan D, Liu J-P, Xiao L, Gong L, Liang S, Zhang J, et al. 2009. Transcriptional regulation of PP2A-Aα is mediated by multiple factors including AP-2α, CREB, ETS-1, and SP-1. PLoS One 4: e7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn PJA, Creyghton MP, Bernards R. 2009. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795: 1–15. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Bramsen JB, Kjems J. 2010. Re-inspection of small RNA sequence datasets reveals several novel human miRNA genes. PLoS One 5: e10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. 2010. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics 11: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinske LC, França GS, Torres HA, Ohara DT, Lopes-Ramos CM, Heyn J, Reis LF, Ohno-Machado L, Kreth S, Galante PA. 2014. miRIAD—integrating microRNA inter- and intragenic data. Database (Oxford) 2014: pii: bau099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Ying X, Wang J, Piriyapongsa J, Jordan IK, Sheng J, Yu F, Zhao P, Li Y, Wang H, et al. 2014. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. Cancer Res 74: 2283–2294. [DOI] [PubMed] [Google Scholar]
- Ivaska J, Nissinen L, Immonen N, Eriksson JE, Kähäri V-M, Heino J. 2002. Integrin α2β1 promotes activation of protein phosphatase 2A and dephosphorylation of Akt and glycogen synthase kinase 3β. Mol Cell Biol 22: 1352–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353(Pt 3): 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42(Database issue): D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue): D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashine YA, Salah S, Aboelenein HR, Abdelaziz AI. 2015. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus 24: 240–247. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Liu J, Ji W, Sun S, Zhang L, Chen HG, Mao Y, Liu L, Zhang X, Gong L, Deng M, et al. 2012. The PP2A-Aβ gene is regulated by multiple transcriptional factors including Ets-1, SP1/SP3, and RXRα/β. Curr Mol Med 12: 982–994. [DOI] [PubMed] [Google Scholar]
- Liu B, Liu M, Wang J, Zhang X, Wang X, Wang P, Wang H, Li W, Wang Y. 2015. DICER-dependent biogenesis of let-7 miRNAs affects human cell response to DNA damage via targeting p21/p27. Nucleic Acids Res 43: 1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lu C, Jeong D-H, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. 2008. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci 105: 4951–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Thelen M, Hemmings BA. 1998. Inactivation and dephosphorylation of protein kinase Bα (PKBα) promoted by hyperosmotic stress. EMBO J 17: 7294–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueleti DL, Smetana JH, Nunes HF, Kobarg J, Zanchin NI. 2012. Identification and characterization of an alternatively spliced isoform of the human protein phosphatase 2Aα catalytic subunit. J Biol Chem 287: 4853–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. 2010. Structure and activity of putative intronic miRNA promoters. RNA 16: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, et al. 2011. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res 71: 78–86. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42(Database issue): D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, DiMaio JM, et al. 2004. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64: 9027–9034. [DOI] [PubMed] [Google Scholar]
- Resjö S, Göransson O, Härndahl L, Zolnierowicz S, Manganiello V, Degerman E. 2002. Protein phosphatase 2A is the main phosphatase involved in the regulation of protein kinase B in rat adipocytes. Cell Signal 14: 231–238. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina AA, Hahn WC. 2008. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev 27: 137–146. [DOI] [PubMed] [Google Scholar]
- Sato S, Fujita N, Tsuruo T. 2000. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci 97: 10832–10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin S, Dalbert S, Paulsen M, Bachellerie J, Ferguson-Smith A, Cavaillé J. 2003. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet 34: 261–262. [DOI] [PubMed] [Google Scholar]
- Shaw M, Cohen P, Alessi DR. 1997. Further evidence that the inhibition of glycogen synthase kinase-3β by IGF-1 is mediated by PDK1/PKB-induced phosphorylation of Ser-9 and not by dephosphorylation of Tyr-216. FEBS Lett 416: 307–311. [DOI] [PubMed] [Google Scholar]
- Shin DM, Zhang H, Saba NF, Chen AY, Nannapaneni S, Amin AR, Müller S, Lewis M, Sica G, Kono S, et al. 2013. Chemoprevention of head and neck cancer by simultaneous blocking of epidermal growth factor receptor and cyclooxygenase-2 signaling pathways: preclinical and clinical studies. Clin Cancer Res 19: 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son YH, Ka S, Kim AY, Kim JB. 2014. Regulation of adipocyte differentiation via microRNAs. Endocrinol Metab (Seoul) 29: 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-P, Li K-B. 2009. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics 10: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wera S, Hemmings B. 1995. Serine/threonine protein phosphatases. Biochem J 311(Pt 1): 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Hao H, Elton TS, Liu Z, Ou H. 2011. Intronic microRNA suppresses endothelial nitric oxide synthase expression and endothelial cell proliferation via inhibition of STAT3 signaling. Mol Cell Biochem 357: 9–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.