SUMMARY
Histone methylation occurs on both lysine and arginine residues, and its dynamic regulation plays a critical role in chromatin biology. Here we identify the UHRF1 PHD finger (PHDUHRF1), an important regulator of DNA CpG methylation, as a histone H3 unmodified arginine 2 (H3R2) recognition modality. This conclusion is based on binding studies and cocrystal structures of PHDUHRF1 bound to histone H3 peptides, where the guanidinium group of unmodified R2 forms an extensive intermolecular hydrogen bond network, with methylation of H3R2, but not H3K4 or H3K9, disrupting complex formation. We have identified direct target genes of UHRF1 from microarray and ChIP studies. Importantly, we show that UHRF1's ability to repress its direct target gene expression is dependent on PHDUHRF1 binding to unmodified H3R2, thereby demonstrating the functional importance of this recognition event and supporting the potential for crosstalk between histone arginine methylation and UHRF1 function.
INTRODUCTION
Chromatin covalent modifications, which include DNA methylation and histone posttranslational modifications, play an important role in epigenetic regulation. Histone N-terminal tails undergo extensive modifications including methylation on lysine (K) and arginine (R) residues. Methylation of different lysine residues of histone H3 and H4 is recognized by a variety of protein modalities, including the plant homeodomain (PHD), PWWP, and chromodomains (Taverna et al., 2007). Such recognition mechanisms confer elaborate regulatory functions in a plethora of chromatin template-based biological processes including gene regulation, DNA replication, and recombination. Recent studies further demonstrate that both methylated and unmethylated lysine residues are recognized by specific protein modalities important for regulation of gene expression (Lan et al., 2007; Ooi et al., 2007; Shi et al., 2006). In contrast, significantly less is known about how histone arginine residues are recognized, although arginine methylation plays equally important roles (Bedford and Clark, 2009).
Here we report the identification of the PHD finger domain in UHRF1 (PHDUHRF1) as a histone H3 tail-binding module recognizing unmodified arginine residue 2 of histone H3 (H3R2). UHRF1 (ubiquitin-like, with PHD and RING finger domains 1) (also called NP95 and ICBP90) is required for the maintenance of CpG DNA methylation (Bostick et al., 2007; Sharif et al., 2007) and is composed of multiple protein modalities (Figure 1A), including SRA, which binds hemimethylated CpG (Bostick et al., 2007; Sharif et al., 2007), a Tudor domain that binds trimethylated histone H3 lysine 9 (H3K9me3) (Walker et al., 2008), as well as a PHD domain, whose histone binding partners remain unclear (Karagianni et al., 2008; Papait et al., 2008). UHRF1 is mainly localized to pericentromeric heterochromatin (PCH) (Papait et al., 2007), but recent studies suggest that UHRF1 also localizes to specific euchromatic regions, possibly playing a role in transcriptional repression (Daskalos et al., 2011; Kim et al., 2009). UHRF1 is believed to regulate PCH function as well as transcription of certain tumor suppressor genes (Daskalos et al., 2011). However, mechanisms underlying recruitment of UHRF1 to either heterochromatic or euchromatic regions remained largely unknown.
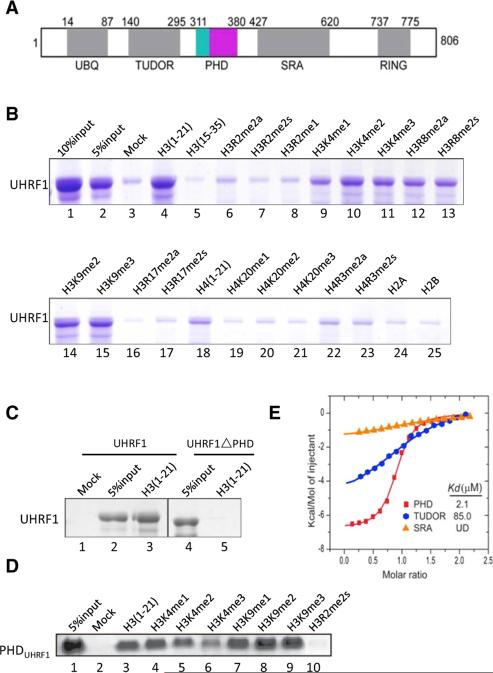
Figure 1. PHDUHRF1 Recognizes Unmodified Histone H3 Tail.
(A) Schematic representation of domain structure of human UHRF1. Numbers indicate amino acid positions at the boundaries of various domains.
(B–D) In vitro binding assays using various biotinylated histone peptides containing the indicated modifications. Either recombinant full-length UHRF1 or PHDUHRF1 was incubated with biotinylated histone peptides immobilized onto streptavidin Sepharose beads. Bound proteins were subjected to SDS-PAGE and stained by Coomassie blue.
(E) ITC plots for binding of histone H3(1-10) to Tudor, PHD, and SRA domains of URHF1 with dissociation constant (Kd) values indicated. UD, undetectable.
We show that in contrast to TudorUHRF1, which binds H3K9me3 (Walker et al., 2008), PHDUHRF1 specifically binds unmodified H3. Surprisingly, this binding is significantly reduced by H3R2 methylation but largely unaffected by H3K4 and H3K9 methylation, suggesting that PHDUHRF1 binds H3 via recognition of unmodified H3R2. This hypothesis is supported by the structure of PHDUHRF1 in complex with H3 peptides, which identified H3R2 as a major contact site for PHDUHRF1, together with the N-terminal amino group and side chain of the first alanine residue on H3, which likely helps anchor PHDUHRF1 and therefore contributes to the unmodified R2 recognition specificity. Isothermal titration calorimetry (ITC) provided binding affinities of PHDUHRF1 for either unmodified or modified H3 with methylation at R2, K4, and K9, reinforcing the notion that unmodified R2 is the major contact site for PHDUHRF1. Genome-wide expression microarray analysis coupled with chromatin immunoprecipitation (ChIP) identified a number of UHRF1 direct target genes whose expression is repressed by UHRF1. Importantly, point mutations that disrupt PHDUHRF1 binding to unmodified H3R2 also abrogated the ability of UHRF1 to repress target gene expression, while these mutations have no effect on UHRF1 PCH localization. Taken together, we have provided binding, structural, and functional data identifying PHDUHRF1 as an unmodified H3R2 binder. Our findings suggest that recognition of the unmodified H3R2 by PHDUHRF1 may represent an important mechanism for targeting UHRF1 to euchromatic regions and that histone H3R2 methylation may impact UHRF1 function by regulating its chromatin accessibility.
RESULTS
PHDUHRF1 Recognizes Unmodified H3 Tail
As discussed above, UHRF1 is mainly localized to PCH, but it may also be present at euchromatic loci (Kim et al., 2009). Insight into how UHRF1 is recruited to these different regions of the genome is important for understanding mechanisms that underlie UHRF1-mediated biological processes. UHRF1 is composed of multiple protein modalities. In addition to the RING finger domain that mediates ubiquitylation (Citterio et al., 2004), it also contains Tudor, PHD, and SRA domains that mediate interactions of UHRF1 with histone and DNA, respectively (Bostick et al., 2007; Karagianni et al., 2008; Sharif et al., 2007; Walker et al., 2008) (Figure 1A). Previous studies suggested that PHDUHRF1 binds H3K9me3 (Karagianni et al., 2008). However, one recent study showed that H3K9me3 binding is also mediated by TudorUHRF1 (Walker et al., 2008). Thus, exactly what PHDUHRF1 binds remains unclear.
To address this issue, we first carried out in vitro binding assays using purified full-length UHRF1 and a collection of histone peptides with or without modifications. As shown in Figure 1B, UHRF1 specifically and robustly binds the unmodified, N-terminal histone H3 tail (aa 1–21) (lane 4), but not the more internal sequence of H3, either unmodified (aa 15–35, lane 5) or methylated on R17 (aa 15–35, lanes 16–17). UHRF1 also showed little or no binding to unmodified or methylated histone H4 (Figure 1B, lanes 18, 19–23), and to unmodified H2A or H2B (aa 1–21) (Figure 1B, lanes 24–25). Importantly, di- or trimethylation of H3K4 and H3K9, as well as H3R8 dimethylation, by and large did not significantly affect binding (Figure 1B, lanes 9–15). In contrast, methylation of H3R2 significantly reduced binding (Figure 1B, lanes 6–8), indicating that H3R2 may be a critical contact site for UHRF1. Deletion of the PHD domain from UHRF1 abrogated binding, suggesting that PHDUHRF1 is necessary for UHRF1 to bind H3 (Figure 1C, compare lane 5 with lane 3). Finally, the PHDUHRF1 domain alone was sufficient to bind unmodified H3, and binding was similarly impeded by methylation at H3R2 but largely unaffected by methylation at H3K9 (Figure 1D). ITC analysis determined the binding affinity (Kd) between PHDUHRF1 and unmodified H3 to be approximately 2.1 μM (Figure 1E). In contrast, TudorUHRF1, which mainly recognizes H3K9me3 (Walker et al., 2008), had significantly less affinity for the unmodified H3 (Kd = 85.0 μM). Furthermore, the SRAUHRF1 domain, which binds hemimethylated DNA, had no detectable binding to histone H3 (Figure 1E). Taken together, these findings suggest that among the various protein modalities present in UHRF1 (Figure 1A), PHDUHRF1 is responsible, and is both necessary and sufficient for binding unmodified H3, possibly via recognition of the unmodified R2 residue.
Crystal Structures of PHDUHRF1 in Free and H3(1-9) Bound States
The above biochemical and molecular investigations raised the exciting possibility that PHDUHRF1 may be an unmodified H3R2 “reader.” To understand the molecular mechanism of PHDUHRF1-mediated histone H3 recognition, we determined the crystal structures of PHDUHRF1 in the free state and in complex with histone H3(1-9) and H3(1-9)K4me3 peptides, with crystallographic statistics listed in Table 1.
Table 1.
Crystallographic Data Collection and Refinement Statistics of PHDUHRF1 in Free and Peptide-Bound States
| Crystal | UHRF1 PHD-H3(1-9) | UHRF1 PHD-H3(1-9)K4me3 | UHRF1 PHD Free State |
|---|---|---|---|
| Beamline | BNL X29 | APS 24ID-C | Home source |
| Wavelength | 1.2828 | 0.97918 | |
| Space group | P43212 | P43212 | I212121 |
| Unit cell | |||
| a, b, c (Å) | 42.62, 46.62, 183.48 | 42.62, 46.62, 183.48 | 53.74, 53.81, 128.48 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 20–1.80 (1.86–1.80)a | 20.0–1.95 (2.02–1.95)a | 20.0–2.65 (2.74–2.65)a |
| Rsym | 0.069 (0.54) | 0.12 (0.78) | 0.064 (0.84) |
| I/σ (I) | 38.3 (1.67) | 34.9 (3.23) | 41.9 (3.1) |
| Completeness (%) | 99.1 (92.1) | 98.7 (96.1) | 99.5 (99.1) |
| Redundancy | 13.1 (6.5) | 13.3 (13.7) | 7.0 (7.1) |
| Number of unique reflections | 17072 | 13122 | 5681 |
| Rwork/Rfree (%) | 21.5/24.3 | 21.3/24.7 | 24.8/29.4 |
| Number of nonhydrogen atoms | |||
| Protein | 1070 | 1035 | 948 |
| Peptide | 128 | 98 | - |
| Water | 122 | 47 | 7 |
| Zn | 8 | 8 | 8 |
| Average B factors (Å2) | |||
| Protein | 29.9 | 28.8 | 75.1 |
| Peptide | 38.6 | 35.6 | - |
| Zn | 34.3 | 25.3 | 76.9 |
| Water | 36.5 | 33.0 | 74.4 |
| Rmsd | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.012 |
| Bond angles (°) | 1.167 | 1.115 | 1.700 |
Highest-resolution shell (in Å) is shown in parentheses.
Canonical PHD fingers are defined by a cross-bracketed two Zn finger domain architecture (Aasland et al., 1995). Sequence comparison with canonical PHD fingers reveals that PHDUHRF1 has an extra N-terminal motif containing three additional Cys residues when compared to the canonical sequence (see Figure S1A available online). Based on the 2.65Å crystal structure of PHDUHRF1 in the free state, the Pro311 to Val328 segment that is present N terminal to the canonical PHD finger adopts a knot-like fold designated the pre-PHD motif (cyan-colored fold in Figure S1B). Three Cys (315, 318, and 326) residues from the N-terminal Cys-rich pre-PHD motif and Cys329 from the N terminus of the canonical PHD finger coordinate a Zn ion (designated Zn1) to form the pre-PHD (Figure S1B), with a single helical turn connecting the pre-PHD and canonical PHD finger motifs, whose relative alignments are stabilized by hydrogen bonding interactions. Two symmetry-related monomers form a Zn-coordinated dimer for PHDUHRF1 in the free state, mediated by a single Zn ion (designated Zn4) (Figure S1C).
We have solved the 1.80Å structure of PHDUHRF1 cocrystallized with unmodified H3(1-9) peptide. Two noncrystallographic symmetry-related monomers form a Zn-coordinated dimer for PHDUHRF1 in the H3(1-9) bound state, mediated by a pair of Zn ions (designated Zn4) (Figure S1D). PHDUHRF1 contacts the first four residues of the bound H3(1-9) peptide, with residue Arg8 interacting with a symmetry-related molecule. The A1-R2-T3-K4 residues of the bound H3 peptide are docked in an antiparallel alignment with the β1 strand on the surface of the PHD finger through peptide-protein backbone interactions (Figure 2A). The side chain of Ala1 of the bound peptide is buried within a pocket formed by hydrophobic residues Leu344, Pro366, and Trp371 (Figure 2A). In addition, the amino terminus of the peptide interacts with the main-chain carbonyl oxygen of Glu368 (Figure 2A).
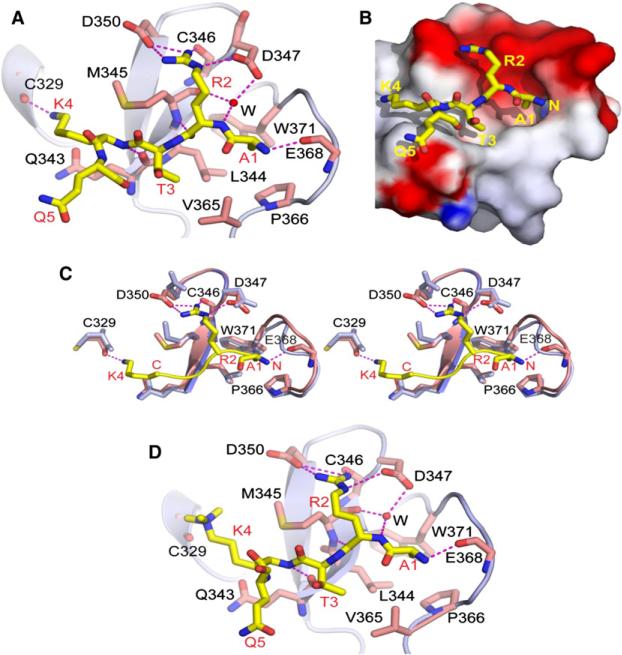
Figure 2. Crystal Structures of PHDUHRF1 Bound to H3(1-9) and H3(1-9)K4me3 Peptides.
(A) Shown is a ribbon (protein) and stick (peptide) representation of the 1.80Å crystal structure of PHDUHRF1 bound to H3(1-9) peptide. The PHDUHRF1 is colored in light blue, with the pre-PHD not shown in this view. The bound H3 peptide is colored in yellow, with interacting residues on PHDUHRF1 colored in magenta. Intermolecular interactions are depicted as magenta-colored dashed lines. A bridging water molecule involved in intermolecular recognition is shown as a red sphere.
(B) Shown is an electrostatic (protein) and stick (peptide) representation of the crystal structure of PHDUHRF1 bound to H3(1-9) peptide. The bound peptide is in yellow, and the side chain of R2 is positioned within the red-colored acidic surface patch of the protein.
(C) A stereo view of the superposition of the crystal structures of PHDUHRF1 in the free (light blue) and H3(1-9) peptide-bound (magenta) states. The H3(1-4) segment of the bound peptide is shown in yellow.
(D) A ribbon (protein) and stick (peptide) representation of the 1.95Å crystal structure of PHDUHRF1 bound to H3(1-9) K4me3 peptide.
See also Figures S1 and S6.
The Arg2 side chain is docked on the negatively charged surface groove in the PHD finger (Figure 2B), with the Asp347-Asp350 hairpin segment that connects the β1 and β2 strands of the PHD finger playing a pivotal role in unmodified Arg2 recognition, through hydrogen bond formation of its NεH atom with the side chain of Asp347 and both its NηH2 atoms hydrogen bond with the side chain of Asp350 (Figure 2A). The backbone carbonyl oxygen of Cys346 also forms a polar contact with the Nη2H proton of Arg2. In addition, an ordered water molecule mediates the network of hydrogen bond interactions such that it connects the backbone amide proton of Arg2 of the bound peptide with the main chain carbonyl oxygen of Met345 and the side chain of Asp347 of the protein (Figure 2A). Such an extensive array of hydrogen bonding interactions between Arg2 of the bound H3 peptide and the PHD finger provides an ample basis for the specificity of recognition of the unmodified H3R2 mark by PHDUHRF1. Upon peptide binding, the Glu368-Glu370 segment is reorganized to accommodate the Ala1 residue at the N terminus of the peptide, and the Asp350 side chain undergoes a conformational change (χ2 angle changes from −19° to 23°) to facilitate H3 Arg2 side chain recognition (stereo view in Figure 2C comparing free [blue] and bound [magenta] structures).
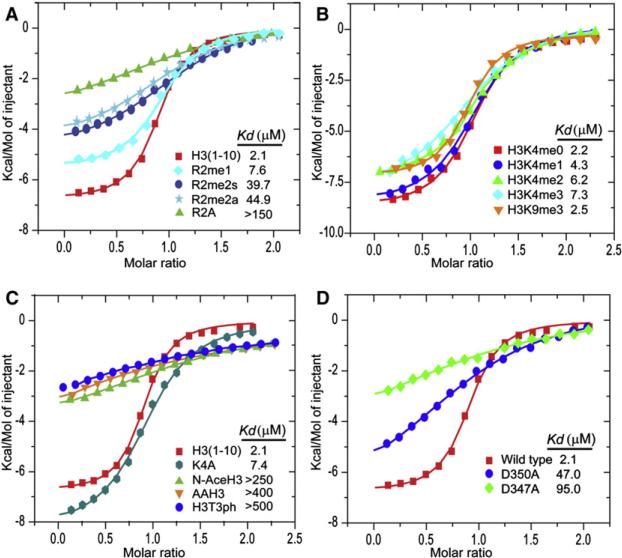
The observed extensive network of intermolecular contacts involving Arg2 establishes that the PHDUHRF1 recognizes unmodified H3R2 in the structure of its complex (Figure 2A). This structural observation is supported by significant reduction in binding affinity between PHDUHRF1 and H3 peptides (Kd = 2.1 μM) when Arg2 is either symmetrically (Kd = 39.7 μM) or asymmetrically dimethylated (Kd = 44.9 μM) or substituted by Ala (Kd > 150 μM), while a smaller drop in binding affinity is observed on monomethyation of Arg2 (Kd = 7.6 μM) (Figure 3A). Trimethylation at H3K9 by and large had no impact on binding (Kd = 2.5 μM), while trimethylation at H3K4 caused a modest reduction (Kd = 7.3 μM) (Figure 3B), consistent with the in vitro pull-down results with full-length UHRF1 (Figure 1B) and the 1.95Å crystal structure of PHDUHRF1 bound to H3(1-9)K4me3 peptide (Figure 2D). Further, addition of an Ala-Ala segment at the N terminus of the H3 peptide results in a large drop in binding affinity (Kd > 400 μM) (Figure 3C), as did acetylation of the N terminus (Kd > 250 μM) (Figure 3C), highlighting the important contribution of the N terminus of H3 to PHDUHRF1 H3 recognition (Figure 2A). These findings, together with binding studies on the R2A mutant (Kd > 150 μM) (Figure 3A), lend further support to the notion that binding of PHDUHRF1 to H3 is mediated mainly by unmodified R2, supplemented by recognition of the N terminus of H3. Indeed, Asp347 and Asp350, which form hydrogen bonds with the guanidinium group of H3R2 in the complex (Figure 2A), when mutated to Ala, also result in a significant reduction in the binding affinities to Kd = 47.0 μM for the D350A mutant and Kd = 95.0 μM for the D347A mutant (Figure 3D).
Figure 3. ITC Measurement of Interaction between PHDUHRF1 and Histone H3 Tail.
(A) Superposed exothermic ITC enthalpy plots for the binding of PHDUHRF1 to H3(1-10), H3(1-10)R2me1, H3(1-10)R2me2s, H3(1-10)R2me2a, and H3(1-10)R2A peptides. The insert lists the measured binding constants.
(B) Superposed exothermic ITC enthalpy plots for the binding of PHDUHRF1 to H3(1-15)K4me0, H3(1-15)K4me1, H3(1-15)K4me2, H3(1-15)K4me3, and H3(1-15)K9me3 peptides. The insert lists the measured binding constants.
(C) Superposed exothermic ITC enthalpy plots for the binding of PHDUHRF1 to H3(1-10), H3(1-10)K4A, H3AA(1-10), N-acetyl H3(1-15), and H3(1-15)T3ph peptides. The insert lists the measured binding constants.
(D) Comparison of exothermic enthalpy plots for wild-type with D350A and D347A mutants of PHDUHRF1 bound to H3(1-10) peptide.
Finally, PHDUHRF1 bound very weakly to histone H3 when T3 is phosphorylated (H3[1-15]T3ph) (Kd > 500 μM) (Figure 3C), indicative of complex destabilization from either steric and/or electrostatic repulsion following phosphorylation of T3 positioned adjacent to R2 in the H3 sequence.
PHDUHRF1 H3R2 Binding Is Critical for UHRF1-Regulated Gene Expression
The above biochemical and structural investigations revealed a protein modality that specifically recognizes unmodified R2 of H3, suggesting that this recognition may play an important role in directing UHRF1 to specific genomic locations. As discussed earlier, UHRF1 is found at PCH, but either deletion of PHDUHRF1 or mutations of the H3R2-binding amino acids (D347A and adjacent E348A) showed PCH localization comparable to that of wild-type UHRF1 (Figure 4A), suggesting that H3R2 binding is likely to be dispensable for UHRF1 PCH localization. This is perhaps not unexpected, because UHRF1 PCH localization is believed to involve SRAUHRF1, which binds hemi-methylated CpG (Bostick et al., 2007; Sharif et al., 2007), a hallmark modification of PCH (Richards and Elgin, 2002), although such a PCH-localizing role for SRAUHRF1 has been questioned by a more recent study (Rottach et al., 2010). Collectively, our findings suggest that while PHDUHRF1 appears dispensable for UHRF1 PCH localization, it could be critical for targeting UHRF1 to euchromatic locations.
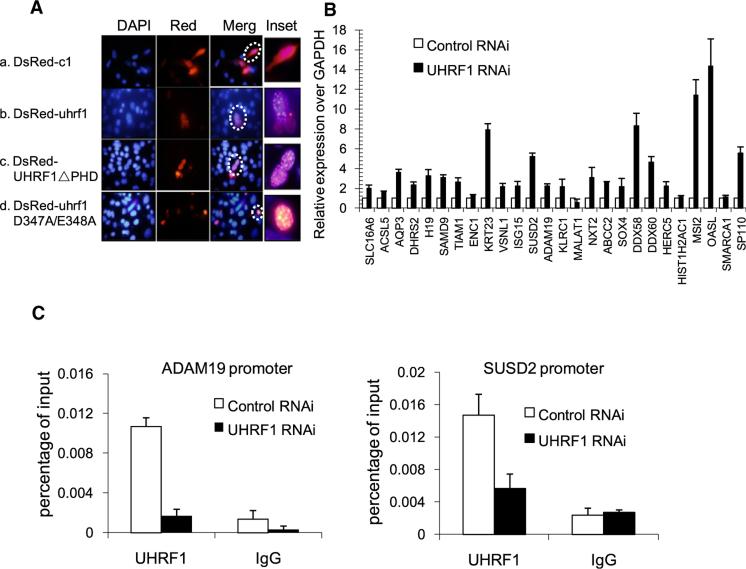
Figure 4. Identification of Target Genes Directly Regulated by UHRF1.
(A) H3R2 binding is likely to be dispensable for UHRF1 PCH localization. dsRed-fused wild-type and mutant UHRF1 constructs were transfected into NIH 3T3 cells, and immunostaining was performed 2 days posttransfection. Representative cells were highlighted by dashed circles.
(B) Validation of microarray expression data with RT-qPCR. Twenty-six genes upregulated in our microarray were randomly selected for further validation. RNAs were prepared from plko.1-control shRNA and UHRF1-sh2-treated HCT116 cells and were reversed transcribed into cDNA for qRT-PCR. Error bars represent SEM calculated from three independent experiments.
(C) RNAi of UHRF1 results in decreased UHRF1 occupancy at its target genes (ADAM19 and SUSD2). Error bars represent SEM calculated from three independent experiments.
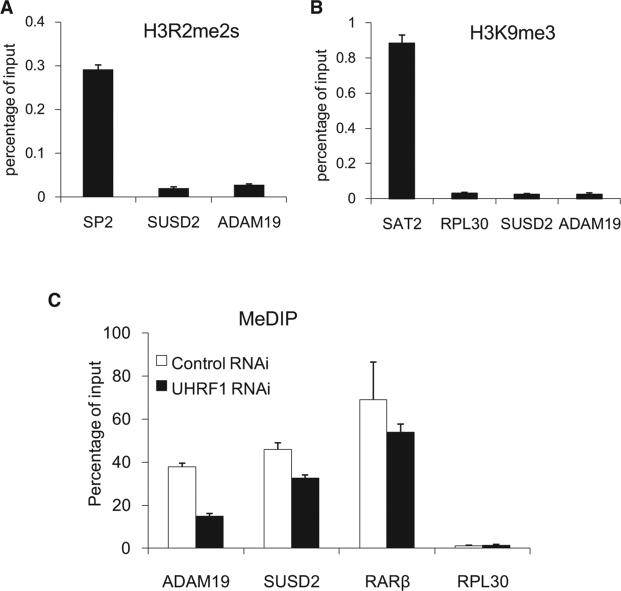
To determine whether H3R2 binding is important for UHRF1 regulation of euchromatic gene expression, we carried out microarray analysis to identify UHRF1-regulated genes using RNAs isolated from cells treated with either scrambled or two independent UHRF1 shRNAs (uhrf1-sh2 and uhrf1-sh5). Each uhrf1 shRNA caused both up- and downregulation of approximately 3000 genes (Figure S2). However, genes that show expression changes in response to both uhrf1 shRNAs were in smaller numbers (367 and 606 up- and downregulated genes, respectively) (Table S1). We considered only those genes that show differential expression in cells treated with both UHRF1 shRNAs as potential UHRF1-regulated genes. Given that UHRF1 is primarily a repressor of transcription (Daskalos et al., 2011; Kim et al., 2009), we focused our initial validation efforts on those genes that were upregulated in the absence of UHRF1. RT-qPCR showed that out of the 26 genes, 22 were upregulated in the uhrf1 RNAi cells, representing greater than 80% confirmation rate (Figure 4B). GO term analysis identified potential involvement of UHRF1-regulated genes in RNA processing and metabolism, as well as cell death (Figures S3A and S3B), while KEGG Pathway analysis suggested possible regulation of cancer pathways by UHRF1 (Figure S3C), which are consistent with the previous reports of possible roles for UHRF1 in apoptosis and tumorigen esis (Abbady et al., 2003; Hervouet et al., 2010; Tien et al., 2011). Importantly, we demonstrated by ChIP followed by quantitative PCR (ChIP-qPCR) that these two UHRF1-regulated genes are bound by UHRF1 near promoters (Figure 4C), as well as gene bodies (data not shown), indicating that their expression is directly regulated by UHRF1. Consistently, ChIP also showed that the promoter regions of these two genes lack H3R2 methylation (H3R2me2s) (Figure 5A). These two promoters also lack H3K9 trimethylation (Figure 5B), which may explain the dependency on the PHD domain for UHRF1 regulation of these genes (see below). We also investigated DNA methylation status of these promoters and determined whether UHRF1 is involved in the regulation by methylated DNA immunoprecipitation (MeDIP) assays (Weber et al., 2007). As shown in Figure 5C, the 5-methyl-C antibodies (Eurogentec) detected MeDIP signals at the promoters of ADAM19 and SUSD2 as well as that of RARβ, which is known to be methylated (Widschwendter et al., 2000) (also Figure S4). Consistently, the MeDIP signals are significantly higher than that of RPL30, which is an actively transcribed gene and therefore is expected to have no or low levels of DNA methylation. Interestingly, knockdown of UHRF1 reduced methylation signals at both promoters (Figure 5C), suggesting that UHRF1-mediated repression of these two genes may involve DNA methylation. While insight into whether and how these UHRF1-regulated genes may play a role in mediating UHRF1 biology requires additional studies, these results nevertheless suggest that both histone and DNA methylation may play a role in UHRF1-regulated gene repression and provide UHRF1-regulated target genes for the genetic complementation experiments described below.
Figure 5. UHRF1 Direct Target Genes, ADAM19 and SUSD2, Lack H3R2me2s and H3K9me3 but Have DNA Cytosine Methylation at Their Promoters.
(A) ADAM19 an SUSD2 promoters lack H3R2 symmetric dimethyation. H3R2me2s-specific polyclonal antibodies developed in the Guccione lab in Singapore were used for the ChIP experiments, and the Sp2 promoter was used as a positive control. Error bars represent SEM calculated from three independent experiments.
(B) UHRF1 direct targets are associated with low levels of H3K9me3. The same primers used in Figure 4C were used for H3K9me3 ChIP. The SAT2 repetitive sequence representing heterochromatic regions and the housekeeping gene RPL30 were used as positive and negative controls, respectively. Error bars represent SEM calculated from three independent experiments.
(C) ADAM19 and SUSD2 promoters are cytosine methylated, and methylation appears to require UHRF1. The 5-methyl-C antibodies were used for MeDIP analysis. MeDIP results were expressed as values relative to their corresponding input. RARb and RPL30 were used as positive and negative controls, respectively. Error bars represent SD calculated from two independent experiments.
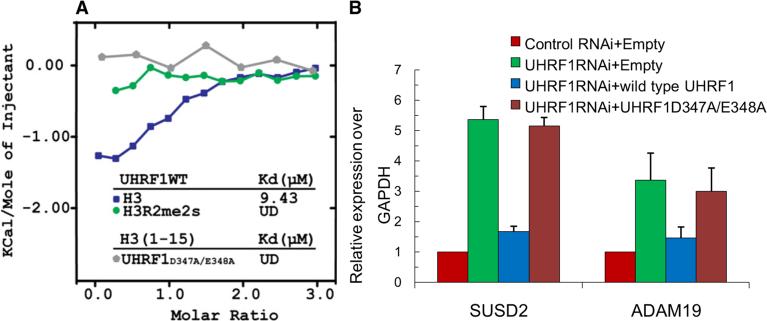
To determine the functional significance of PHDUHRF1 binding unmodified H3R2, we next carried out genetic complementation experiments using full-length, wild-type, and the R2-binding defective dual mutant (D347A and adjacent E348A), which showed undetectable H3 binding as determined by the ITC assay (Figure 6A). As shown in Figure 6B, wild-type, but not the binding-defective dual mutant of UHRF1 (D347A and adjacent E348A), restored repression of the two UHRF1-reguated genes, SUSD2 and ADAM19. Both the wild-type and the dual mutant proteins were comparably expressed, suggesting that the lack of rescue by the binding-defective mutant was not due to lack of expression (Figure S5). Taken together, these results suggest that the primary function of PHDUHRF1 is to localize UHRF1 to euchromatic targets by recognizing and binding the unmodified R2 residue.
Figure 6. UHRF1 Binding H3R2 Is Critical for Its Ability to Regulate Target Gene Expression.
(A) Superimposed ITC enthalpy plots for the binding of full-length UHRF1 (wild-type or mutant) and histone H3 peptides with the estimated binding affinity (Kd). UD, undetectable.
(B) Rescue experiments. RNAi-resistant wild-type UHRF1, but not the H3R2 binding-defective mutant of UHRF1 (D347A/E348A), restored repression of two UHRF1-reguated genes, SUSD2 and ADAM19, in UHRF1 RNAi cells. HCT116 stable cell lines were established that coexpress control or UHRF1 shRNA and/or indicated flag-tagged UHRF1 constructs. mRNA expression of SUSD2 and ADAM19 was measured by qPCR. GAPDH was used as an internal control. SEM was obtained from three independent experiments.
DISCUSSION
Comparison of N-Terminal H3 Peptide Recognition by PHDUHRF1 and WD40WDR5 Domains
In the present study, we have demonstrated that unmodified H3R2 recognition of PHDUHRF1, mediated predominantly by inter-side-chain hydrogen bonds, represents a mode of histone H3 tail recognition. Recognition of unmodified H3R2 by its binding pocket within PHDUHRF1 involves both an electrostatic and hydrogen bonding contribution between its guanidinium group and the side chain of acidic residues (Figure 2B), as well as a hydrophobic contribution between its aliphatic side chain and the side chain of Met345 (Figure 2A). Indeed, dimethylation of Arg2, or its replacement by Ala, results in a pronounced drop in binding affinity (Figure 3A).
Previous studies identified the WD40 domain of WDR5, a component of the SET1/MLL family of histone methyltransferases, as a reader of unmodified H3R2 (Couture et al., 2006; Han et al., 2006; Ruthenburg et al., 2006; Schuetz et al., 2006). Recognition of unmodified H3R2 by WD40WDR5 involves targeting of both the N terminus and insertion of the unmodified R2 side chain into the central cavity of the torroidal WD40 propeller fold, where it is oriented through direct and water-mediated hydrogen bonds and sandwiched between staggered Phe side chains (Figure S6B). However, there is an important distinction between the PHDUHRF1 (Figure S6A) and WD40WDR5 (Figure S6B) complexes, since unmodified H3R2 is targeted by PHDUHRF1 using a “surface groove” recognition mode, whose binding pocket is more accessible, while it is targeted by WD40WDR5 using a “cavity insertion” recognition mode, in which the mark is inserted and buried within a deep protein cleft, with greater potential for size-selective discrimination (Taverna et al., 2007).
It should be noted that TUDORJMJD2A (Huang et al., 2006; Lee et al., 2008) and ADDDNMT3A (Otani et al., 2009) that bind H3K4me3 and unmodified H3K4 marks, respectively, also display contacts with the unmodified H3R2 site. These TUDOR/ADD domains cannot be specified as readers of H3R2 marks since they do not form the extensive network of intermolecular hydrogen bonds to the guanidinium group of R2, as observed for the PHDUHRF1 (Figure 2A). Further, much smaller reductions in binding affinities were observed following either methylation of R2 or its replacement by Ala for ADDDNMT3A and TUDORJMJD2A compared to PHDUHRF1.
Unlike other complexes involved in unmodified K4 recognition (Lan et al., 2007; Ooi et al., 2007), where the ammonium group forms multiple hydrogen bonds with acidic side chains, the side chain of unmodified H3K4 forms a single hydrogen bond to the backbone carbonyl of Cys329 in the PHDUHRF1 complex (Figure 2A and Figure S6A). Thus, there is ample room to accommodate methylation modifications at H3K4, as validated by binding studies as a function of K4 methylation state (Figure 3B) and the crystal structure of H3(1-9)K4me3 peptide bound to PHDUHRF1 (Figure 2D).
PHDUHRF1 Is Critical for Targeting UHRF1 to Euchromatic Locations and for Regulation of Euchromatic Gene Expression
UHRF1 has been shown to be mainly involved in heterochromatin function (Karagianni et al., 2008; Papait et al., 2008; Papait et al., 2007) and in CpG methylation regulation during DNA replication (Bostick et al., 2007; Sharif et al., 2007). Although previous studies identified a handful of genes regulated by UHRF1, suggesting a possible role for UHRF1 in gene expression regulation (Kim et al., 2009), our study investigated such a role for UHRF1 at the genome-wide level. Our analysis identified thousands of potential UHRF1 target genes, supporting a general transcriptional role for UHRF1. Earlier studies implicated UHRF1 in cell proliferation and apoptosis regulation (Fang et al., 2009; Fujimori et al., 1998; Tien et al., 2011) and suggested that UHRF1 may function as a possible oncogene (Bronner et al., 2007). Our GO term analysis (Figure S3) identified possible tumor suppressor genes such as Axin2 and BMP4, among UHRF1-upregulated genes, as well as genes in apoptosis, raising the exciting possibility that these genes may mediate UHRF1's ability to regulate cell proliferation and cell death. Interestingly, a number of previously reported, UHRF1-regulated genes involved in cancer such as BRCA1 and p73 are not among the UHRF1-regulated genes identified in our analysis. This could be due to cell type or assay condition differences. Regardless, our studies support the notion that UHRF1 directly regulates the expression of a large number of genes, setting the stage for future studies directed toward providing further insights into how these genes may mediate UHRF1 biology.
It is important to note that our findings have provided molecular and structural insights into how UHRF1 is targeted to euchromatic regions to exert its transcriptional roles, namely by recognizing and binding the unmodified H3R2 residue. We believe that the primary function of PHDUHRF1 is to target UHRF1 to euchromatic regions lacking H3K9me3. For those genes that carry H3K9me3, we predict that TudorUHRF1, which binds H3K9me3, will predominate and relieve the dependency on PHDUHRF1. Consistent with this, we found that the presence of H3K9me3 counteracts the inhibitory effect of R2 dimethylation on UHRF1 binding to histone H3 in vitro (data not shown). In addition to the euchromatic targeting function of PHDUHRF1 reported here, previous studies also demonstrated the requirement of PHDUHRF1 in the regulation of PCH structure (Papait et al., 2008). Collectively, PHDUHRF1 appears to be important for targeting UHRF1 to euchromatic and for PCH function, although it is dispensable for PCH targeting.
Potential for Crosstalk between Histone Arginine Methylation and UHRF1 Function
In summary, we have provided binding, structural, and functional data highlighting the discovery of an effector module dedicated to the recognition of an unmodified arginine residue (R2) on histone H3. Our findings further suggest that any modifications of R2 may negatively impact on UHRF1 binding to H3, including methylation (Figures 2 and 3) and possibly deimination (Cuthbert et al., 2004), raising the possibility of crosstalk between UHRF1 chromatin binding and pathways that regulate H3R2 posttranslational modifications. Interestingly, R2 monomethylation exhibits a lesser impact on UHRF1 H3 binding (3.5-fold reduction, Figure 3A) compared to symmetrical and asymmetrical dimethylation (20-fold reduction in binding, Figure 3A), suggesting that subtle differences in R2 modifications may impart differential regulation on UHRF1 chromatin binding. The potential functional significance of mono- versus dimethylation of UHRF1 regulation remains unclear and awaits a better understanding of the methyltransferases involved in these modifications. Together with recent reports of readout of methylated arginine marks on histone (Yang et al., 2010) and protein (Liu et al., 2010a, 2010b) by TUDOR domains, these findings begin to uncover a potentially elaborate effector network for the recognition of differential methylation states on histone arginine residues.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
All constructs were generated using PCR-based cloning strategy, and all mutants were generated by the QuikChange Mutagenesis protocol (Strata-gene). DNA fragments of full-length UHRF1 (1–806), Tudor (139–298), and PHD (311–379) were subcloned into pGEX-6P-1 or pET-15b derivative encoding a 3C protease cleavage site. All constructs were expressed in Escherichia coli strain BL21(DE3) and purified using glutathione 4B column chromatography. The PHD finger (311–380), as well as TUDOR (140–295) and SRA (427–630), were also cloned into GST- and hexahistidine-sumo-tagged expression vectors, respectively. The expression conditions and sequential protein purification protocols are detailed in the Supplemental Experimental Procedures.
Peptide Pull-Down Assays
Biotinylated histone peptides (1 μg each) were used for the pull-down assays with either GST-fused PHD or his6-tagged full-length UHRF1 following the binding protocol described previously (Lan et al., 2007).
Antibodies
UHRF1 polyclonal antibodies were raised in house, while H3R2me2s antibodies were rabbit polyconal antibodies produced and quality controlled in the Gucioone lab in Singapore (eguccione@imcb.a-star.edu.sg).
ITC Measurements
Isothermal titration calorimetric experiments were performed using a VP-ITC calorimeter (MicroCal, LLC) at 25°C with MicroCal Origin software used for curve fitting, equilibrium dissociation constant, and molar ratio calculations. For detailed procedures, see the Supplemental Experimental Procedures.
Crystallization, X-Ray Data Collection, and Structure Determination
Crystallization trials of PHDUHRF1 bound to unmodified and methylated N-terminal H3 peptides were conducted at 18°C by the sitting drop vapor diffusion method using a MOSQUITO crystallization robot. The X-ray data set on crystals of PHDUHRF1 in free state, and bound to variants of H3 peptide, was collected at the beam lines mentioned in Table 1. The crystal structure of the complex of PHDUHRF1 bound to H3(1-9) was solved by single-wavelength anomalous dispersion (SAD) technique at Zn-peak wavelength. Subsequent structures of PHDUHRF1 and its complexes were solved by molecular replacement method using the structure of the PHDUHRF1 bound to H3(1-9) as a search model.
Additional details related to crystallization, data collection, and structure calculations are provided in the Supplemental Experimental Procedures. The crystallographic statistics for all structures presented above are listed in Table 1.
RNA Interference and Genetic Rescue
Lentiviral Plko.1 constructs expressing shRNAs (sequences listed in Table S2) were purchased from Open Biosystems. Viral particles were produced in 293T cells by following the recommended protocols (Addgene). Stable HCT116 cell lines were established with coexpressed Lentiviral plenti6.2, or plenti6.2-UHRF1 (wild-type or mutant) and plko.1 shRNAs (control or UHRF1 RNAi). Twenty-four hours after infection, puromycin and Blasticidin S hydrochloride were added at 1.6 μg/ml and 5 μg/ml, respectively, to select for pooled populations of stably infected cells. Cells were then harvested for RNA extraction after 6 days’ selection. Reverse transcription was performed with 1 μg RNA followed by real-time PCR. The primers for real-time PCR are listed in Table S3.
Gene Expression Microarray Analysis
Microarray analysis for global gene expression was performed using standard methods on the Affymetrix GeneChip System.
Methylated DNA Immunoprecipitation
After treatment with UHRF1shRNA and contol lentivirus for 14 days, genomic DNA was purified from HCT116 cells, sonicated, denatured, and incubated (8 μg) with a monoclonal antibody against 5-methyl-C (Eurogentec) (10 μl) at 4°C for 4 hr. The antibody-DNA complexes were captured by protein A/G beads, and the DNA enrichments in the MeDIP fraction were measured by real-time PCR.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yang Xiang, Degui Chen, and members of the Epigenetics Laboratory at Institutes of Biomedical Sciences, Fudan University, as well as Steve Jacobson and Greg Horowitz of UCLA Medical School and Mary Goll of the Memorial Sloan-Kettering Cancer Center for their interest and for helpful discussions. We thank Ernesto Guccione for the H3R2me2s antibodies and primers for Sp2 and Or Gozani for histone peptides. We thank the staff at beamlines 24ID-C and 24ID-E of the Advanced Photon Source at the Argonne National Laboratory, as well as the staff at BNL X29 of the Brookhaven National Laboratory for assistance with X-ray data collection. This work was supported by the Starr Foundation and a Leukemia-Lymphoma Program Project Grant (D.J.P) and by the “985” Program from the Chinese Ministry of Education and “973” State Key Development Program of Basic Research of China (2009CB825602, 2009CB825603) to the Epigenetics Laboratory at the Institute of Biomedical Sciences, Fudan University, Shanghai, China. Author contributions are as follows. E.R. was responsible for the ITC, crystallization, crystallographic data collection, and structure analysis, with assistance from H.L., under the supervision of D.J.P. The binding, genetic rescue, expression microarry analysis, cellular location of UHRF1 (wild-type and mutant), MeDIP, and ChIP experiments were undertaken by Z.W., H.M., and L.H. (under the supervision of F.L., Y.G.S., Y.X., and Y.S.). Y.L did the ITC experiment shown in Figure 6, and H.C. participated in the construction of various plasmids used in this study. R.G. and F.W. made technical and bioinformatics contributions, respectively. This manuscript was written by Y.S. and D.J.P. with input from the other authors. F.L. is an employee of Constellation Pharmaceuticals, Inc. Y.S. is a cofounder of Constellation Pharmaceuticals, Inc., and a member of its scientific advisory board.
Footnotes
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the following accession codes: PHDUHRF1 in the free state (3SOX), complex with H3(1-9) peptide (3SOU), and complex with bound H3(1-9)K4me3 peptide (3SOW). The UHRF1 microarray data have been deposited in the GEO database under the accession number GSE30478.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, three tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.molcel.2011.07.006.
REFERENCES
- Aasland R, Gibson TJ, Stewart AF. The Phd finger—implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Abbady AQ, Bronner C, Trotzier MA, Hopfner R, Bathami K, Muller CD, Jeanblanc M, Mousli M. ICBP90 expression is downregulated in apoptosis-induced Jurkat cells. Ann. N Y Acad. Sci. 2003;1010:300–303. doi: 10.1196/annals.1299.052. [DOI] [PubMed] [Google Scholar]
- Bedford M, Clark SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Daskalos A, Oleksiewicz U, Filia A, Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK, Liloglou T. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer. 2011;117:1027–1037. doi: 10.1002/cncr.25531. [DOI] [PubMed] [Google Scholar]
- Fang ZY, Xing FY, Bronner C, Teng ZP, Guo ZF. ICBP90 mediates the ERK1/2 signaling to regulate the proliferation of Jurkat T cells. Cell. Immunol. 2009;257:80–87. doi: 10.1016/j.cellimm.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- Han ZF, Guo L, Wang HY, Shen Y, Deng XW, Chai JJ. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS ONE. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010a;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. USA. 2010b;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Ikamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol. Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang WK, Graybosch DM, Li HT, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min JR. Structural basis for molecular recognition and presentation of histone H3 By WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi SI, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–925. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien AL, Senbanerjee S, Kulkarni A, Mudbhary R, Goudreau B, Ganesan S, Sadler KC, Ukomadu C. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem. J. 2011;435:175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AG, Jr., Xue S, Dong A, Li Y, Bountra C, Weigelt J, Arrowsmith CH, Edwards AM, Bochkarev A, Dhe-Paganon S. Cryptic tandem tudor domains in UHRF1 interact with H3K9ME and are important for pericentric heterochromatin replication. 2008 http://www.rcsb.org/pdb/3DB3.PDB.
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Berger J, Hermann M, Muller HM, Amberger A, Zeschnigk M, Widschwendter A, Abendstein B, Zeimet AG, Daxenbichler G, et al. Methylation and silencing of the retinoic acid receptor-beta2 gene in breast cancer. J. Natl. Cancer Inst. 2000;92:826–832. doi: 10.1093/jnci/92.10.826. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Lu Y, Espejo A, Wu JC, Xu W, Liang SD, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.