Abstract
Histones comprise the major protein component of chromatin, the scaffold in which the eukaryotic genome is packaged, and are subject to many types of post-translational modifications (PTMs), especially on their flexible tails. These modifications may constitute a ‘histone code’ and could be used to manage epigenetic information that helps extend the genetic message beyond DNA sequences. This proposed code, read in part by histone PTM–binding ‘effector’ modules and their associated complexes, is predicted to define unique functional states of chromatin and/or regulate various chromatin-templated processes. A wealth of structural and functional data show how chromatin effector modules target their cognate covalent histone modifications. Here we summarize key features in molecular recognition of histone PTMs by a diverse family of ‘reader pockets’, highlighting specific readout mechanisms for individual marks, common themes and insights into the downstream functional consequences of the interactions. Changes in these interactions may have far-reaching implications for human biology and disease, notably cancer.
The vast majority of DNA in eukaryotic organisms is intimately wrapped around core histone proteins, forming chromatin, the physiological context in which the genomes of these organisms must function. Control of the dynamics of chromatin structure has been implicated widely in regulating both access to and interpretation of the associated DNA template1. For example, numerous studies suggest that gene expression patterns can be positively or negatively regulated by complexes of proteins that fine-tune the structural properties of chromatin, often through covalent PTMs of histone proteins or other chromatin-remodeling complexes2,3. As chromatin architecture may be transmissible to daughter cells along a particular developmental lineage, histones and their PTMs are likely candidates for epigenetic information carriers that propagate phenotypic determinants not encoded in the DNA sequence. More than 70 different sites for histone PTMs and eight types of histone PTMs have been reported, largely from the extensive application of mass spectrometry– and antibody-based detection techniques, as well as from metabolic-labeling studies1–3. Remarkably, almost two-thirds of potentially modifiable residues on histones have been characterized as PTM sites, and as more sensitive detection methods become available, this number is likely to increase. Despite the large number of histone PTMs, only a subset of amino acid residues in histones are known to be covalently modified, which include lysine (K), arginine (R), serine (S), threonine (T), tyrosine (Y), histidine (H) and glutamic acid (E)1. The majority of the PTMs are additions of relatively small yet chemically and structurally distinct moieties, such as acetyl, methyl and phosphate groups (Fig. 1a); these have been identified on sites ranging from the flexible tails to the internal globular domains of histones. In many cases, genome-wide studies using a combination of histone PTM–specific chromatin immunoprecipitation with either DNA microarray analysis (ChIP-chip) or sequencing analysis (ChIP-Seq) have yielded unprecedented insights into global correlations of specific histone PTMs with functional outcomes4–6. Such approaches clearly show that histone PTMs are sequestered to distinct regions of the genome. However, a gap currently exists in the mechanistic understanding of how the various histone PTMs that seemingly litter the chromatin landscape can be translated into specific biological outputs such as transcription, or used to extend the informational content of the genome from one generation to the next, as in epigenetic inheritance.
Figure 1.
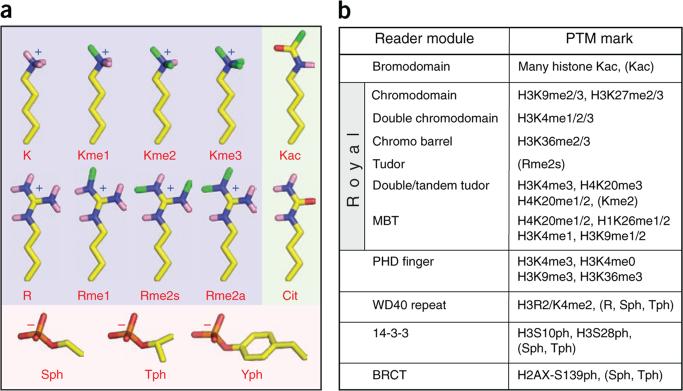
Histone post-translational modifications and their binding partners. (a) Stick models of different classes of post-translationally modified amino acid residuess, highlighting small chemical group side chains on histone tails. Yellow, carbon; blue, nitrogen; pink, polar hydrogen; red, oxygen; orange, phosphorus; green, methyl groups of post-translational modifications. Background is shaded by charge of side chains at physiological pH: light blue, positive; pink, negative; light green, uncharged. (b) Reading the histone (protein) code. Shown grouped by domain family are known chromatin-associated modules and the histone marks they have been reported to bind. Parentheses denote examples where structural information is known about related family members and their interactions with non-histone PTMs.
Although the detailed mechanisms by which cells may decipher a PTM-mediated histone code are currently the subject of much investigation, two overlapping models, the ‘direct’ and ‘effector-mediated’ models, have been proposed to make sense of the multitude of modification states and their connections to biology7,8. Examples of the direct model, in which histone PTMs directly affect chromatin compaction, include phosphorylation or acetylation on core histones that serves to attenuate the favorable coulombic interactions between basic histone proteins and the negative charge of the DNA9–13. The emerging effector-mediated paradigm posits that histone PTMs are ‘read’ by protein modules termed effectors, facilitating meaningful downstream events via the recruitment or stabilization of module-associated chromatin-templated machinery14,15. In the past decade, biochemical and biophysical assays have identified a wealth of conserved protein domains that specifically bind histone PTMs in a way that is dependent on both modification state and position within a histone sequence (Fig. 1b). By presenting unique combinations of PTMs for binding, modified histone tails may act as integrating platforms, permitting chromatin-associated complexes to receive information from upstream signaling cascades16.
In this review, we use selected examples of recently published structures to describe underlying themes governing the association of histone PTMs with their respective binding protein modules. We examine how residues lining PTM-interacting pockets work together to recognize specific chemical features of their cognate histone PTMs. Then we assess the structural similarities and differences among the diverse families of histone PTM reader pockets to derive several common principles underlying molecular recognition of various histone PTMs. We also examine how existing pockets could be engineered to alter the PTM specificity of a particular module. Finally, we discuss future directions for studies of chromatin reader biology, including how individual histone PTM-binding modules may work together to gain enhanced targeting specificity. Additional challenges include identification of binding partners for methylated arginine in histones, and links between histone PTM–binding modules and human disease.
Readout of acetyllysine marks by bromodomains
Acetylation was first identified on histones in 1964 as a potential regulator of RNA synthesis17. Back-to-back discoveries in 1996 showed that the steady-state balance of histone acetylation was catalyzed by enzymes with opposing activities (Gcn5p acetyltransferase and Rpd3p deacetylases) already known to be transcriptional regulators, thus firmly linking histone acetylation to transcription18,19. Certain lysine acetylation events are known to directly alter the physical properties of individual nucleosomes13 or disrupt their higher-order association10. Alternatively, lysine acetylation marks may be interpreted indirectly via the intermediacy of bromodomain effectors20. Bromodomains are protein modules found in chromatin-associated proteins, especially nuclear histone acetyltransferases (HATs) such as Gcn5p, and components of certain remodeling complexes, in which they have key roles in transcriptional activation and chromatin remodeling21.
Gcn5p bromodomain targets acetyllysine in H4 Lys16 context
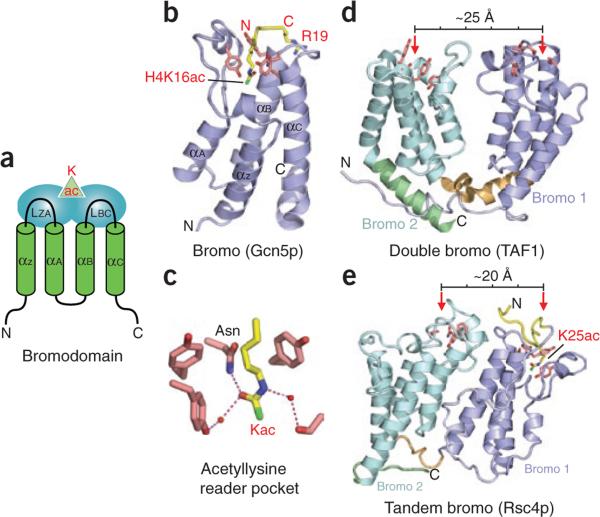
The prototypical bromodomain from the transcriptional coactivator ‘p300/CBP-associated factor’ (PCAF, the homolog of Saccharomyces cerevisiae Gcn5p) was the first histone-binding module to be structurally characterized. The NMR solution structure showed a left-handed antiparallel four-helix bundle with a hydrophobic binding pocket located at one end (Fig. 2a)20. An NMR solution structure of acetylhistamine (an analog of acetyllysine) bound to the PCAF bromodomain defined the intermolecular contacts in the binding pocket and established the importance of hydrophobic interactions to complex formation20. Additional details of acetyllysine recognition by bromodomains emerged in the high-resolution crystal structure of the Gcn5p bromodomain bound to a histone H4 peptide containing acetylated K16 (H4K16ac; Fig. 2b)22. The acetyllysine inserts into a deep and narrow binding pocket, which can accommodate the long side chain bearing a planar acetamide group. The acetyl modification is specifically anchored in place through a hydrogen bond involving its acetyl arbonyl and the amide nitrogen of a conserved asparagine residue lining the pocket, with the roughly antiparallel dipole alignment of these two amide moieties providing concomitant partial charge complementation (Fig. 2c). The pocket is lined by conserved aromatic, hydrophobic and uncharged residues, which are aligned, rendering the pocket essentially hydrophobic and neutral while retaining intrinsic hydrogen-bonding capacity at its base. Additional contacts within the binding pocket include a network of water-mediated intermolecular hydrogen bonds between the bound acetyllysine and protein main chain carbonyl groups. The histone peptide segments flanking the acetyllysine are also involved in recognition by the bromodomain; these specificity-determining intermolecular contacts are mediated by side chain interactions along a shallow depression on the bromodomain, a surface composed largely of two loops that span the helices (Fig. 2a). Interestingly, the Gcn5p protein also has HAT activity, immediately suggesting a potential mechanism for the spreading of acetylation marks in chromatin22,23.
Figure 2.
Readout of acetyllysine marks by bromodomains. (a) Topology of bromodomain fold. Green cylinders, core α-helices (αZ, αA, αB and αC); blue ovals, loops (LZA and LBC) that participate in acetyllysine reader pocket formation and also bear recognition elements for the modified peptide sequence; green triangle, approximate location of acetyllysine binding pocket. (b) Recognition of H4K16ac mark by the Gcn5p bromodomain (PDB 1E6I). Note insertion of the acetylated lysine side chain into a deep pocket generated at one end of the α-helix bundle. (c) Details of the Gcn5p acetyllysine binding pocket. Red spheres, water molecules; dashed lines, hydrogen bonds. For clarity, main chain portions of the cage residues and acetyllysine are omitted from stick models. (d,e) Examples of dual bromodomains. The TAF1 double bromodomain (d; PDB 1EQF) has been reported to function cooperatively in targeting properly separated diaceylated H4 tails24. By contrast, the tandem bromodomain–containing Rsc4p (e; PDB 2R10) has been shown to recognize histone H3K14ac using its second bromodomain (pale cyan), whereas its first bromodomain (slate) is involved in recognition of an autoregulatory acetyllysine modification (K25ac) of the Rsc4p protein itself25. Slate and pale cyan, N- and C-terminal bromodomains, respectively; red arrows, acetyllysine reader pockets; measuring bar, approximate distance separating the two pockets. In b, d, e and subsequent figures, all reader modules are depicted as ribbons, with key cage side chains shown as pink sticks; histone peptides bearing acetyllysine or other types of PTMs are in yellow, and modified residues are colored as in Figure 1a; N and C termini of the effector proteins are marked in black and those of histone peptides in red.
TAF1 double bromodomains target diacetylated H4 tails
Human TAF1 (formerly TAFII250) is the largest subunit of TFIID, a large multiprotein complex involved in initiating the assembly of the transcription machinery. Instead of recognizing a single acetyl PTM with a single bromodomain, TAF1 contains double bromodomains that bind multiply acetylated histone H4 peptides. The 2.1-Å crystal structure of these double bromodomains in the free state shows side-by-side arrangement of individual bromodomains in a four-helix bundle topology, with the two acetyllysine-binding pockets in the ‘V’-shaped scaffold separated by ~25 Å (indicated by red arrows, Fig. 2d)24. The N terminus of histone H4 has lysine residues at positions 5, 8, 12 and 16, and the 25-Å separation between binding pockets suggests bridging by about 7 residues. Of the marks examined, this double module binds most tightly to the dual acetyllysine mark H4K5acK12ac (Kd = 1.4 μM) and more weakly to single marks such as H4K16ac (Kd ≈ 39 μM). Further progress will require determination of the structures of these bromodomains in complex with a dually acetylated peptide to elucidate the molecular basis of dual recognition. Numerous examples of proteins containing multiple bromodomains exist (including the tandem modules outlined below), reinforcing the general theme wherein multiple modules provide PTM readers with increased specificity and binding affinity.
Rsc4p tandem bromodomains target histone and non-histone protein acetylation marks
A recently solved structure of the tandem bromodomains of Rsc4p from the yeast chromatin-remodeling complex RSC (Fig. 2e) suggests a different functional role from that of engagement with dual histone acetylation marks. Interestingly, the relative orientations of the two bromodomains differ in Rsc4p (Fig. 2e)25 and TAF1 (Fig. 2d)24. The Rsc4p tandem bromodomains appear to fold as a single structural unit, in contrast to those of TAF1, which appear to be relatively independent (bearing little interdomain contact surface). As a consequence of this more compact fold, the acetyllysine-binding pockets of individual bromodomains in Rsc4p are oriented on the same face and separated by ~20 Å (ref. 25). As Rsc4p was originally identified as an H3K14ac-binding module26, peptide soaking was used to study histone PTM binding. Unfortunately, of residues 6–18 in the H46–18K14ac peptide, only the side chain of K14ac could be traced in the 1.7-Å structure of the complex with the Rsc4p tandem bromodomains. In this complex, K14ac is inserted into the binding pocket of the second bromodomain (Fig. 2e, red arrow on left). For further examination of the histone tail interaction of H3K14ac peptides with Rsc4p, the H3 tail was fused to the Rsc4p N terminus and acetylation marks were incorporated by treatment with purified Gcn5p, a HAT known to acetylate H3K14 and several sites in H4 (ref. 23). Unexpectedly, K25 in Rsc4p was also acetylated, as observed in the 2.2-Å crystal structure of the complex. The structure shows this K25ac group inserted into the binding pocket of the first bromodomain (Fig. 2e, red arrow on right) and also shows the traceable backbone and side chains of flanking residues. The acetylated segment of Rsc4p matches the canonical Gcn5p-recognition motif (G-Kac-X-P). Interestingly, the Rsc4K25ac mark inhibits binding of the H3K14ac peptide to the binding pocket of the second bromodomain of Rsc4, as does incorporation of a phosphorylation mark at H3S10. This apparent mechanism for regulating the capacity to bind H3K14ac is supported by in vivo evidence: the Rsc4p K25A mutant strain shows modest growth defects in minimal media and at elevated temperatures, as well as exacerbated bromodomain-mutant phenotypes. Whether such negative feedback loops may also modulate other effector binding events remains an intriguing question.
Methylation marks and their recognition
In addition to the unmodified and acetylated states, histone lysine residues are also found in monomethylated (me1), dimethylated (me2) and trimethylated (me3) states in vivo, most often brought about by the balance of enzymatic activities of histone methyltransferases27 and opposing demethylases28,29. Genome-wide assessments show that these methylation states at particular lysine residues are enriched in certain regions of chromatin, implying that these marks have specialized and distinct biological functions30. Functionally, histone lysine methylation has been correlated with both gene activation and silencing, and the methylation states of distinct lysines along the H3 N-terminal tail provide a frequently cited example of this diversity. Within this defined region, H3K4me3 is highly correlated with active transcription start sites31, H3K9me3 is linked to constitutive heterochromatin, and H3K27me3 is linked to silent genes in euchromatic regions or facultative heterochromatin2,3. Interestingly, during stem-cell development, ‘bivalent domains’ are evident, in which H3K4me3 and H3K27me3 can be detected simultaneously on promoters of select genes32. How these coexisting dual marks are established, interpreted and resolved remains unclear; however, it is tempting to speculate that they are connected to epigenetic control of gene expression.
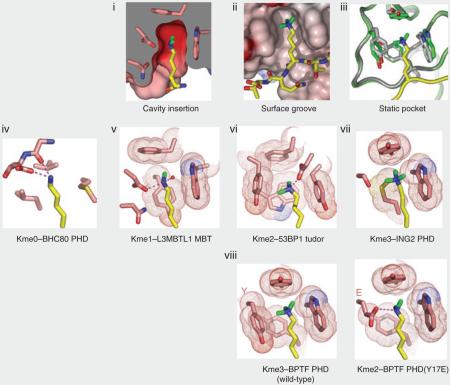
Whereas charge is ablated upon lysine acetylation, all methylated forms of lysine are anticipated to be cationic at physiological pH, and trimethyllysine always carries a positive charge. With the incremental addition of methyl groups, the hydrophobicity and the cation radius of the lysine methylammonium group increases, and its ability to donate hydrogen bonds concomitantly decreases. In the limiting case, trimethyllysine presents a Janus-faced moiety, an obligate cation enshrouded in a hydrophobic array of methyl and methylene units, requiring a partner with similar hydrophobic properties that is also able to accommodate the persistent positive charge. Thus, different methylation states yield considerable diversity in the physicochemical properties of lysine, enabling state-specific readout by different effector modules (Fig. 1a,b). The recognition of these lysine methylation states results from contacts made between the methylammonium moiety and aromatic residues in the binding partner that form a notional ‘cage’ about this functional group. Such aromatic cages engage the quaternary ammonium functional groups, with cation-π–type interactions dominating the energetics and hydrophobic desolvation effects having an appreciable but lesser role15,33,34; for recognition of lower methylation states (me2 and me1), hydrogen-bonding and steric exclusion are increasingly important35,36. Cation-π interactions are predominantly electrostatic in nature, occurring between a cation and an electrostatic potential field of a π-system34. Simplistically, the interaction can be thought of as a coulombic attraction between the quadrupole moment of the aromatic π-system and a cation34. As the quadrupole moment places partial negative charge above each face of the aromatic ring, favorable interactions with cations occur perpendicular to the aromatic plane within typical van der Waals contact distances34,37.
Two general classes of protein folds have evolutionarily converged to bind methyllysine with aromatic cages: members of the Royal superfamily of folds15,38 and PHD fingers39,40 share several molecular recognition features, detailed in separate sections below. Common features in the binding of histone methyllysine substrates are summarized in Boxes 1 and 2.
Royal superfamily readout of higher lysine methylation
The following sections present detailed case studies of methyllysine binding, organized by the class of effector and the methylation states preferentially bound. We first describe the Royal superfamily members that recognize higher lysine methylation states, Kme2 and Kme3.
Chromodomains target di- and trimethyllysine in H3 Lys9 and Lys27 contexts
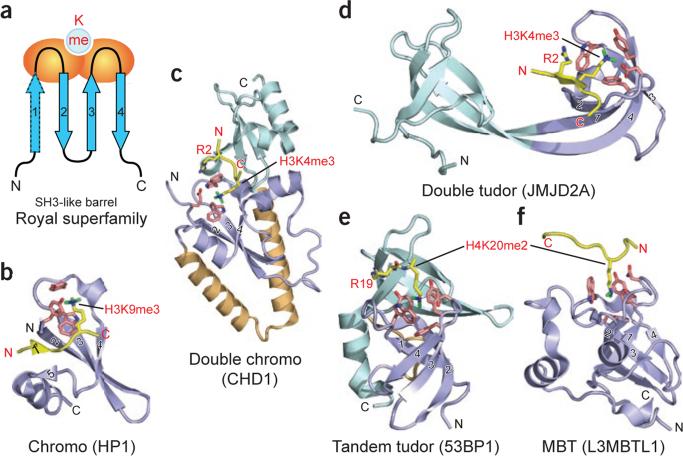
A short region of sequence similarity was identified in Drosophila melanogaster heterochromatin protein-1 (HP1) and Polycomb, regulators of chromatin structure that are involved in epigenetic repression, and termed the chromatin organization modifier domain, or chromodomain41. In vitro pull-down assays revealed an interaction between the chromodomain of mouse HP1 and H3K9me3 (refs. 42,43), supported by in situ immunofluorescence results showing that that H3K9me and HP1 colocalize to heterochromatic regions of D. melanogaster polytene chromosomes44. Molecular and mechanistic insights into chromodomain recognition of methyllysine marks has emerged from energetic, structural and mutagenic analyses of HP1 bound to H3K9me peptides45,46. The binding affinities are in the micromolar range for HP1 targeting H3K9me3 (Kd = 2.5 μM) and H3K9me2 (Kd = 7.0 μM), with no binding observed for the unmethylated counterpart. Both the crystal45 and NMR solution46 structures of the complexes show that the chromodomain adopts an incomplete β-barrel architecture with the methyllysine-binding pocket positioned at one end of the β-barrel (Fig. 3a). The tri- and dimethyllysines project into a surface declivity composed of three aromatic residues, two of which are orthogonally positioned with respect to a central side chain contributed by strand 3 (Fig. 3b) to define a cage-like enclosure. Therein, the methylammonium moieties are stabilized by cation-π interactions with the aromatic cage (see Box 1, panel iii)33,34. Mutation of any of these constituent residues greatly reduces binding affinity. Residues 5–10 of bound H1–15K9me3 adopt an extended conformation and interact by apparent induced-fit sandwiching between terminal β-strands of the HP1 chromodomain, thereby completing a five-stranded antiparallel β-sheet (see Box 2, panel i). Intermolecular contacts in the complex involve three side chains before and one side chain after the K9me mark. Mutation studies of both peptide and protein residues highlight the contributions of intermolecular contacts along the extended surface groove to sequence specificity of recognition. By contrast, the chromodomain of D. melanogaster Polycomb protein specifically targets H3K27me3 (refs. 47,48).
Figure 3.
Readout of methyllysine marks by Royal-superfamily modules. (a) Topology of the Royal-superfamily fold. Blue arrows, β-strands that form an incomplete β-barrel reminiscent of the SH3 domain fold; orange ovals, loops participating in methyllysine reader pocket formation; light blue circle, binding pocket at one end of the module. Classical chromo modules have only three core β-strands (labeled 2–4) and one orphaned extra strand (labeled 5). Upon complex formation, the histone peptide completes this five-stranded β-barrel fold by introducing an extra β-strand at position 1 prime, sandwiched between strands 2 and 5 (see b). This sandwiching binding mode occurs mainly with chromodomains. In other Royal superfamily members, the interactions are more varied; however, docking in the β-strand conformation to extend one edge of an existing β-sheet is not uncommon (see d). (b–f) Examples of known complex structures in the Royal superfamily, ranging from higher methylation state–specific readers (b–d) to lower methylation state–specific readers (e and f). Strands that form the SH3-like β-barrel are in slate, numbered as in a. Coordinates have PDB codes 1KNE (b), 2B2W (c), 2GFA (d), 2IG0 (e) and 2PQW (f).
Although the chromodomains of HP1 and Polycomb bind methylated lysines, this is not necessarily true for other chromodomain family members (see below), and it is likely that regions outside of the chromodomain are crucial in bringing about the final chromatin states. It is thought that the primary function of chromodomain binding to histone PTMs that are associated with repressive heterochromatin is to passively stabilize a dense conformation of the chromatin fiber by cross-linking nucleosomes49,50. This proposed bridging function remains unconfirmed, and the role of other interacting protein complexes and nucleic acids in HP1- and Polycomb-mediated repression is still uncertain.
Double chromodomains of CHD1 target methyllysine in H3 Lys4 context
Chromo helicase DNA-binding (CHD) proteins, composed of N-terminal double chromodomains, a central SWI2/SNF2 helicase and C-terminal DNA-binding domains, regulate ATP-dependent nucleosome assembly and mobilization at sites of transcriptional activity51. CHD1 has been implicated in transcription elongation–related functions, as it associates with transcriptionally-active ‘puff’ regions in D. melanogaster polytene chromosomes52. However, recent studies have shown that CHD1 also acts in nucleosome assembly in transcriptionally silent D. melanogaster embryos53, and thus the biological role of CHD1 may not be relegated exclusively to transcription-related processes. CHD1 was shown recently to be required for deposition of the H3 variant H3.3, in a DNA replication–independent fashion53. Although H3.3 shows enriched levels of histone marks commonly associated with H3K4me3 and transcription54,55, it remains to be determined whether the deposition activity is dependent on interaction with H3K4me3.
Chd1p double chromodomains were originally observed to bind H3K4me in experiments using budding yeast proteins56. Although the molecular details of the H3K4me2/3 interaction with the human ortholog of this protein (CHD1) have since been well characterized in structural and biophysical studies57, similar scrutiny applied to budding yeast protein has led to controversy58,59. The human CHD1 double chromodomains interact with H3K4 methylation sites associated with active chromatin, with measurable binding affinities for H3K4me3 (Kd = 5 μM) and H3K4me1 (17 μM) peptides, but no affinity for unmodified peptide57. The molecular mechanism underlying H3K4me recognition emerged from crystal structures of human CHD1 double chromodomains bound to H3K4me3 (Fig. 3c) and H3K4me1 peptides, which showed superimposable intermolecular contacts in the two complexes. Human CHD1 double chromodomains are bridged by a two-helix linker element that juxtaposes adjacent chromodomain folds to generate a continuous surface. The methylated lysine of a bound H3K4me peptide interacts with a pair of tryptophans through cation-π interactions in a structure analogous to a three-membered aromatic cage57. One of these tryptophans is sandwiched between the side chains of H3R2 and H3K4me3, and mutation of individual tryptophan residues results in a substantial reduction in binding affinity. The residue corresponding to the intervening tryptophan is replaced with a glutamate residue in the yeast protein, precluding a similar binding mode. In essence, the human CHD1 double chromodomains cooperate to create a novel recognition scaffold in which an acidic groove lies at the inter-chromodomain junction while, unique inserts block canonical binding to the chromodomain cage (Fig. 3c).
Double tudor domains of JMJD2A target trimethyllysine in H3 Lys4 context
Jumonji domain–containing protein-2A (JMJD2A, also known as JHDM3A) has JmjN and JmjC domains, which are required for its lysine demethylase activity, and two tudor domains, now classified as a double tudor domain60–62. JMJ2DA associates with histone deacetylases, but recent studies suggest that its function is also mediated by the demethylase activity imparted by Jmj domains. When a series of peptides representing various histone modification states were incubated with recombinant JMJD2A and subjected to mass spectrometric analysis, only H3K9me and H3K36me marks were found to be demethylated60,61. Importantly, depletion of JMJD2A by RNA interference (RNAi) was sufficient to cause a large increase in bulk levels of H3K9me3 and H3K36me3 in Caenorhabditis elegans61. Overexpression of JMJD2A results in partial mislocalization of HP1 (ref. 60), an H3K9me2/3-binding partner. How JMJD2A regulates gene transcription is poorly understood, and the fact that it demethylates both H3K9me3 and H3K36me3, albeit to different extents, will complicate the dissection of function. Indeed, the observation that a knockdown of JMJD2A results in both an increase in H3K9me and an increase in transcription of a target gene seems contradictory, given the correlation of H3K9me3 with gene silencing. The targeting of JMJD2A to regions enriched in H3K4me3 suggests a role in euchromatic gene regulation63.
The molecular basis for recognition has emerged from crystal structures of JMJD2A double tudor domains in the free state and bound to H3K4me3 (ref. 63). Strikingly, the double tudor domains adopt a saddle-shaped scaffold with interleaved, bilobal topology; each hybrid lobe adopts a canonical tudor domain fold spanned by two shared β-strands (Fig. 3d). Intriguingly, only the second of the two hybrid lobes is able to bind H3K4me3 peptide, owing to a more negative electrostatic potential and the apposition of aromatic caging residues to delimit a cleft within its scaffold (Kd = 10.4 μM). Two aromatic residues from the second tudor sequence motif and one aromatic residue from the first tudor sequence motif generate a composite aromatic cage located in the second hybrid lobe, which, together with an aspartate residue, form a binding pocket for insertion of the trimethyllysine residue63. Similar to other Royal family members that engage methyllysine, point mutations of the aromatic cage residues greatly reduce substrate-binding affinity. The N terminus of the H3K4me3 peptide (elaborated further in Box 2), along with residues H3R2 and H3T3, form intermolecular hydrogen bonds with residues found mainly in the first hybrid lobe of the interdigitated double tudor domains, which accounts for the observed binding specificity. Interestingly, array-based binding assays suggest that the JMJD2A double tudor domains also bind H4K20me3 (ref. 64). Direct comparison of the affinities and structural features of H4K20me3 complexes will require further study.
Royal superfamily readout of lower lysine methylation
Another subset of Royal superfamily members recognizes lower lysine methylation states, Kme1 and Kme2. These interactions are detailed below.
Tandem tudor domains of 53BP1 target mono- and dimethyllysine in H4 Lys20 context
Recent evidence suggests a role for histone lysine methylation in the recruitment of DNA damage repair proteins. Immunofluorescence imaging of cellular distribution showed that mammalian p53-binding protein (53BP1) and its fission yeast homolog Crb2p relocalize to double-strand breaks (DSBs) upon exposure to DNA-damaging agents65,66. Methylation-mediated recruitment of 53BP1 may extend to non-histone proteins as well, as a recent report shows that 53BP1 can also bind dimethylated K370 of the tumor suppressor p53 (p53K370me2)67. As the amino acid sequence surrounding p53K370 is divergent from that surrounding H4K20, structural comparison of these modes of interaction will be of great interest.
The 53BP1 protein contains tandem tudor domains that bind H4K20me2 (Kd = 19.7 μM) and H4K20me1 (Kd = 52.9 μM), but not unmodified or K20me3-containing H4 peptides36. Structures of the tandem tudor domain modules of 53BP1, both free and in complex with a short H4K20me2 peptide36, reveal the molecular basis for this sequence- and state-specific recognition of lower lysine methylation states of H4K20 (Fig. 3e). Unlike the double tudor domains of JMJD2A, the tandem tudor domains of 53BP1 form independently folded domains, of which mainly the first is involved in methyllysine recognition. During binding, the methylammonium group of K20me2 is positioned in a cage lined by four aromatic amino acids and an aspartate residue, while the guanidinium group of the adjacent R19 forms a cation-π stack over an adjacent tyrosine ring and is also contacted by phenylalanine contributed by the abutting second tudor domain. The specificity for lower lysine methylation is attributable to an intermolecular hydrogen bond between the dimethylammonium proton and the carboxylate group of the aspartate lining the binding pocket, coupled with steric exclusion of the corresponding trimethyllysine (see Box 1, panel vi). Furthermore, mutation of the aromatic residues or aspartate residue lining the binding pocket greatly reduces binding affinity. These residues are essential not only for interaction between H4K20me2 and 53BP1 in vitro but also for targeting of 53BP1 to DNA double-strand breaks in vivo36. This reveals that direct recognition of H4K20me2 by the tandem tudor domains of 53BP1 mediate processes leading to DNA double-strand break repair.
MBT repeats in L3MBTL1 target mono- and dimethyllysine independently of sequence context
Human lethal-(3) malignant brain tumor repeat–like protein-1 (L3MBTL1) belongs to a group of factors containing so called malignant brain tumor (MBT) repeats of ~70 amino acid residues, which function as transcriptional repressors and are often perturbed in hematopoietic malignancies68. L3MBTL1 is a homolog of the D. melanogaster l(3)mbt protein in the Polycomb group (PcG), whose members stably maintain gene repression over many cell cycles during development69. Human L3MBTL1 had been previously suggested to have an HDAC-independent role in repression of transcription70, and pull-down assays using tagged L3MBTL1 found that the associated proteins included, among others, the core histones and the linker histone H1 (ref. 71). To determine whether these interactions were contingent upon PTMs, a protein encompassing the three MBT repeats was mixed with either recombinant or native nucleosomes, and its association with chromatin was detected using sucrose-gradient sedimentation. As L3MBTL1 interacted with native rather than recombinant nucleosomes, affinity chromatography with histone peptides was used to pinpoint H4K20me1/2 and H1.bK26me1/2 as sites and states of modification. This approach is limited by the number of possible methylated substrates scanned, so it may not be comprehensive. The ability of L3MBTL1 to directly regulate chromatin compaction was assayed using electron microscopy; oligonucleosomes carrying the H1.bK26me2 mark attained a relatively compact configuration after incubation with L3MBTL1, presumably owing to multimerization of L3MBTL1 units. This compaction might render the DNA of L3MBTL1-bound nucleosomes inaccessible to the transcription machinery.
The three MBT repeats of L3MBTL1 have a tripartite propeller-like architecture in the crystal structure of the free state, with intercuspation between extended arms and cores of adjacent MBT modules72 (see Supplementary Fig. 1 online). In addition, each of the MBT domains of L3MBTL1 contains an aromatic cage motif composed of three aromatic residues and an aspartate residue. Quantitative binding measurements and crystallographic studies of the wild-type protein and pocket mutants firmly establish pocket 2 (associated with the second MBT repeat of L3MBTL1) as a methyllysine-binding module that selects mono- and dimethyllysine marks but discriminates against the trimethylated state35. Crystal structure–based comparison of mono- and dimethyllysine recognition by L3MBTL1 pocket 2 (Fig. 3f) establishes the principles underlying preference for lower lysine methylation states using a ‘cavity-insertion’ mode of recognition (see Box 1, panel i). The methylammonium proton(s) of mono- and dimethyllysine form a direct hydrogen bond to the carboxylate of the aspartate residue lining the aromatic cage. In addition, the dimensions of pocket 2, determined by the orientations of the side chains that project from gating and caging loops, restrict access of the larger trimethylammonium group. Mutations of caging loop residues adversely affect complex formation. Because L3MBTL1 promiscuously binds mono- and dimethyllysines in a wide variety of sequence contexts in vitro, defining the physiologically relevant targets of L3MBTL1 remains an outstanding problem35.
Readout of methyllysine marks by the PHD-finger family
According to genome-wide ChIP-chip analyses, H3K4me3 is associated with nucleosomes near the promoters and 5′ ends of highly transcribed genes, whereas H3K4me2 extends into the coding region of genes that are either active or poised for transcription4,5,32,73. For some time, it was unknown what the molecular connection was—if any—between H3K4me3/2 and transcriptional readout. In 2006, structural evidence was presented implicating plant homeodomain (PHD) fingers as tethering modules that bind H3K4me3, thereby recruiting or stabilizing downstream complexes39,40,74. Recently another connection between transcriptional activation and H3K4me2/3 was established by a study of the PHD finger in the TAF3 subunit of the basal transcription factor TFIID75. Importantly, supporting functional assays have yielded insight into the connection between H3K4me2 and positive and negative regulation of transcription (see below).
BPTF PHD finger targets di- and trimethyllysine in H3 Lys4 context
BPTF is the largest subunit of the nucleosomal remodeling factor (NURF) ATP-dependent chromatin-remodeling complex, which has been shown to stimulate transcription on nucleosomal templates in vitro76,77. Unbiased pull-down assays in nuclear extracts that contained short peptides representing differently modified histone tails demonstrated that the second PHD finger in BPTF interacts specifically with peptides modified with H3K4me3 and not with other PTMs78. In cell lines, global loss of H3K4me3 resulted in loss of chromatin association of BPTF, suggesting that H3K4me3 may tether BPTF to nucleosomes. In Xenopus laevis, loss of H3K4me3 results in developmental defects, and this phenotype could be recapitulated by loss of BPTF or by deletion or mutation of the H3K4me3-binding PHD finger in BPTF. Thus, H3K4me3 is probably coupled to BPTF (NURF)–dependent chromatin remodeling through a conserved PHD finger, in a pathway that probably controls transcription of key developmental genes (such as Hox genes).
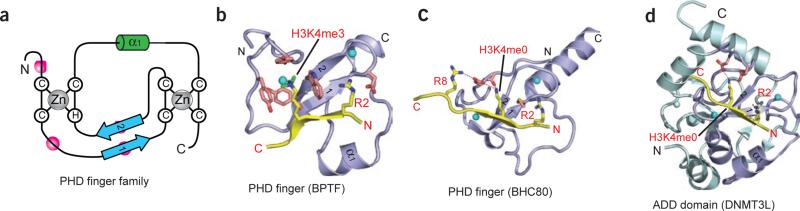
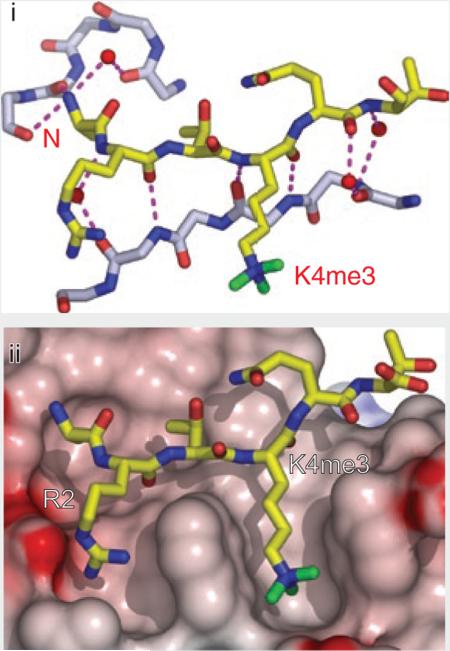
At its C terminus, human BPTF contains a bromodomain-proximal PHD finger, shown to be a reader of dimethyllysine and trimethyllysine marks in an H3K4 sequence context78. PHD fingers composed of a Cys4-His-Cys3–containing segment coordinated to two zinc ions (Fig. 4a) are modules found in a wide range of chromatin-associated proteins79. The BPTF PHD finger binds both H3K4me3- and H3K4me2-containing peptides (Kd = 2.7 and Kd = 5.0 μM, respectively), but it discriminates against their monomethylated and unmodified counterparts40. NMR and X-ray structures have been solved for the BPTF PHD finger in the free and H3K4me3 peptide–bound forms40 (Fig. 4b). The peptide docks onto the surface of the PHD finger in such a manner that it extends the antiparallel β-sheet of the PHD core by forming a third strand, with extensive contacts involving the N terminus of the peptide in the complex (see Box 2, panel i). Importantly, the R2 and K4me3 residues of the bound peptide are positioned in adjacent surface channels separated by a tryptophan indole group of the PHD finger (Fig. 4b and Box 2, panel ii). Mutations of these aromatic residues lining the cage greatly lower binding affinity, with the largest drop observed for a mutation of the tryptophan residue separating H3R2 and H3K4me3, in accordance with the developmental defects that arise from this mutation78. Similarly, mutations of residues lining the H3R2-binding channel also result in diminished binding affinity, reflecting disruption of electrostatic and hydrogen-bonding contacts. The trimethylammonium group of H3K4me3 is positioned within a cage of four aromatic amino acids and stabilized predominantly by cation-π interactions (Fig. 4b) associated with a ‘surface-groove’ mode of recognition (see Box 1, panel ii). There is minimal change in the spatial orientations of the aromatic cage residues upon complex formation; this static element may minimize conformational entropy loss of the module during binding to enhance the overall free-energy change (see Box 1, panel iii). The observed modest preference of the BPTF PHD finger for H3K4me3 over H3K4me2 can be reversed by replacing a specific tyrosine lining the aromatic cage by a glutamate (see Box 1, panel viii). Similar structural parameters governing PHD module recognition are found in chromatin regulators, including those dedicated to activating or repressive activities80 (see below).
Figure 4.
Readout of modified and unmodified histone lysine marks by PHD finger modules. (a) Topology of the PHD finger fold. Blue arrows, two small β-strands that bridge the interleaved zinc-finger motifs; labeled white circles, zinc-coordinated cysteine and histidine residues; green cylinder, short α-helix (α1) near the C terminus; pink circles, the caging residues for readout of methyllysine or unmodified lysine marks. (b) Specific recognition of the H3K4me3 mark by BPTF PHD finger (PDB 2F6J). The H3 peptide resides in a surface groove between α1 and β-strand 1 of BPTF PHD finger upon formation of the antiparallel β-sheet with the core β-strands. Note the full aromatic cage formed at the protein surface for trimethyl-specific readout. H3K4 site specificity is achieved by simultaneous recognition of H3K4me3 and H3R2, as well as by anchoring of the N terminus by the BPTF PHD finger. Similar strategies for readout of H3K4me3 marks have been observed for ING2 and Yng1p PHD fingers. Cyan spheres, zinc ions within the zinc-finger motifs. (c,d) Specific readout of unmodified H3K4 peptide by BHC80 PHD finger (c; PDB 2PUY) and DNMT3L ADD domain (a PHD finger–containing module; d; 2PVC). Both modules have a surface patch of acidic residues for unmodified H3K4 mark recognition. Similar β-sheet–formation and flanking residue–recognition strategies have been observed for site-specific readout of H3 peptides. In d, the PHD finger module (slate) is embedded within the cysteine-rich ADD domain, which contains an additional zinc-finger motif.
Yng1p PHD finger targets di- and trimethyllysine in H3 Lys4 context
Acetylation on histone H3 has also been connected to H3K4me3/2 and transcription through ChIP-chip assays4,81, but the connections between the pathways remained poorly understood. The nucleosomal acetyltransferase of histone H3 (NuA3) complex contains an H3-specific H3K14 HAT as well as a polypeptide, Yng1p, with a PHD finger that specifically interacts with H3K4me3, as verified by NMR structural studies of the complex74,82. Enhancement of H3K14 HAT activity on H3K4me3 peptides was dependent on an intact PHD finger, and in genome-wide ChIP-chip studies, Yng1p was largely colocalized with H3K4me3 and acetylation at 5′ regions of open reading frames (ORFs). Furthermore, mutation of the Yng1p PHD finger reduced transcription at a set of NuA3-targeted ORFs. Importantly, these studies imply a hierarchy of events based on the interaction between H3K4me3 and the PHD finger of Yng1p, in which association of the NuA3 complex promotes downstream acetylation and subsequent transcriptional events. Insight into additional downstream functions may come from recent studies wherein isotopic labeling was used to show that the human TFIID transcription initiation complex binds H3K4me3 through the PHD finger of the TAF3 subunit75 as previously predicted15. It was demonstrated that the TFIID interaction may be further enhanced when H3K9 and H3K14 are acetylated; this distribution of two acetylated lysine residues may serve as a substrate for the double bromodomains of the TAF1 subunit (described above, Fig. 2d). In this regard, NuA3 may function to deposit acetylation marks required for proper engagement of the TFIID basal transcription machinery.
ING2 PHD finger targets di- and trimethyllysine in H3 Lys4 context
In yeast, mutation of Set1p (the only known yeast H3K4 HMT) or point mutation of H3K4 to arginine produces defects in gene silencing, a result seemingly at odds with the strong correlation between H3K4me3 and transcription. Recent studies have partially resolved this paradox by showing that the PHD finger–containing human inhibitor of growth-2 (ING2) protein of the Sin3–histone deacetylase (HDAC) complex also interacts with H3K4me3 (refs. 39,83). After the PHD finger of ING2 was shown to be required for deacetylation of H3K14 in vitro, RNAi knockdown of the WD40-repeat protein WDR5 was used to reduce endogenous H3K4me3 levels, which resulted in loss of ING2 association with chromatin and the cyclin D1 promoter. In assays where doxorubicin treatment was used to induce ING2-dependent repression of cyclin D1, ING2 with mutated PHD fingers did not reduce transcript levels of cyclin D1, whereas reconstitution of the wild-type Sin3–HDAC complex greatly reduced transcription. Yeast also have orthologs of this HDAC complex (RPD3L), in which Pho23p has a PHD finger that binds H3K4me3 (ref. 83).
A crystallographic characterization of complex formation between ING2 H3K4me peptides and the PHD fingers of ING2 (ref. 39) independently reached the same conclusions about the basis of the recognition specificity reported above for the BPTF PHD finger–H3K4me3 complex40. The H3K4me-binding pocket is composed of two aromatic residues, with the partial aromatic cage completed by the side chains of serine and methionine residues. Mutational studies of binding site residues disrupted H3K4me3 binding in vitro and the ability of ING2 to induce apoptosis in vivo39,83.
Thus, although it may seem contradictory that complexes that either activate or repress transcription are recruited by the same mark, this may be a mechanism for the cell to rapidly shift between transcription states of cellular proliferation–associated genes upon DNA insult or other fluctuations in environmental or physiological conditions83,84. We favor the general view that multisubunit complexes have the potential to use multiple binding modules that may function to achieve appropriate targeting of the complex to chromatin or stabilization of its association74,80,84. Although many of these complexes read covalent histone modifications, unmodified histones and non-histone substrates are also physiologically relevant targets of such activities (see below).
Readout of unmodified basic amino acids
Although complexes that interact with unmodified histone tails have been reported to be associated with hypoacetylation and silencing85, until recently the structural basis of this recognition had not been elucidated. Nevertheless, researchers made inroads by determining how unmodified residues are interpreted, structurally and functionally86–91. Collectively, these studies underscore an important concept: whereas some modifications are used to promote chromatin associations, in other instances the same mark can be used to weaken or block these interactions.
PHD finger of BHC80 targets lysine in H3 Lys4 context
Members of the corepressor complex include H3K4 lysine demethylase-1 (LSD1), HC80 and the corepressor CoREST; this complex is associated with transcriptional repression of neuron-specific genes92,93. In a striking observation, the PHD finger of BHC80 binds unmethylated H3K4 (Kd ≈ 30 μM) and discriminates strongly against its methylated counterparts87. The molecular basis for this unanticipated recognition specificity for unmethylated K4 (K4me0) in an H3 context was resolved by the 1.4-Å crystal structure of H31–10K4me0 bound to the BHC80 PHD finger87. Like its methylated counterparts, the peptide adopts an extended conformation and contributes another strand to the antiparallel β-sheet of the BHC80 PHD finger, with the N terminus of the H3 peptide recognized in a hydrogen bond cage formed by three backbone carbonyls. The free ζ-ammonium group of unmethylated H3K4 forms a pair of hydrogen bonds with two acceptors on the PHD finger: an aspartate side chain and the amide carbonyl of the residue preceding it (Fig. 4c and Box 1, panel iv). The recognition specificity for the unmodified lysine side chain is associated in part with presumed steric exclusion of methyl groups. The complex has a striking interdigitation of peptide and PHD finger side chains such that a methionine side chain of the PHD finger is interposed between the H3R2 and H3K4me0 side chains, and an aspartate side chain of the PHD finger is sandwiched between and hydrogen-bonded to the H3K4me0 and H3R8 side chains (Fig. 4c). The importance of these methionine and aspartate residues in the PHD finger to complex formation was validated by mutation studies, which resulted in complete loss of binding affinity.
At the functional level, derepression of LSD1 target genes accompanies BHC80 knockdown by RNAi87. Furthermore, ChIP experiments establish a reciprocal dependence of BHC80 and LSD1 on each other for association with chromatin. As BHC80 targeting to promoters is reduced in LSD1-knockdown cells, and the PHD finger binds the H3K4me0 product of the LSD1 reaction, the function of BHC80 is proposed to be downstream of LSD1 activity87. Interestingly, one component of the LSD1 complex, BRAF35, was shown to bind highly condensed mitotic chromatin; whether mitosis involves demethylation of H3K4 is not known94.
Cysteine-rich ADD domain of DNMT3L targets lysine in H3 Lys4 context
Cytosine methylation, performed by DNA methyltransferase (DNMT)-family enzymes containing C-terminal SAM-dependent methyltransferase and N-terminal cysteine-rich folds, is the only known epigenetic modification of DNA itself and is required for genomic imprinting (establishment of gene expression patterns through inherited DNA methylation patterns). DNA methylation is classified into two types: maintenance methylation (by DNMT1 homologs during replication)95,96 and de novo methylation (by DNMT3 homologs at cytosines of unmethylated CpGs during establishment of the germline-specific methylation imprinting)97. Germline-specific DNA methylation was shown to be defective in mice deficient in the DNMT3-like protein (DNMT3L), a DNMT3 family member that lacks intrinsic methyltransferase activity98.
Recently, it was shown that the N-terminal tail of histone H3 is engaged by the cysteine-rich zinc-binding domain of DNMT3L, but only when H3K4 is unmethylated. The 3.3-Å crystal structure of DNMT3L in the free state defined the folds and relative orientations of the impaired-methyltransferase and cysteine-rich domains86 (Fig. 4d). The cysteine-rich ATRX-DNMT-DNMT3L (ADD) domain is organized around three zinc ions and is structurally very similar to PHD finger domains in its core. In effect, the ADD domain is a PHD finger embedded within additional protein scaffolding99. Its highest binding affinity is for unmodified H3K4me0 (Kd = 2.1 μM), with the affinity dropping 20-fold for H3K4me1 (Kd = 36 μM) and with no measurable affinity for H3K4me2 and H3K4me3. H31–24 was soaked into the crystal of DNMT3L, and density was identified for a small portion of the bound peptide backbone. By analogy to the high-resolution structure of the N-terminal H3 peptide bound to BHC80 (ref. 87), in the low-resolution complex structure a hydrogen bond was proposed to exist between the side chain of K4 on the peptide and an aspartate residue on the cysteine-rich zinc-binding domain of DNMT3L86. Mutations that disrupt this interaction adversely impacted the binding affinity, and the observed discrimination against methylation at H3K4 was attributed to steric occlusion. The implications of these results are that DNA methylation status and heterochromatin formation may be negatively regulated by histone modifications.
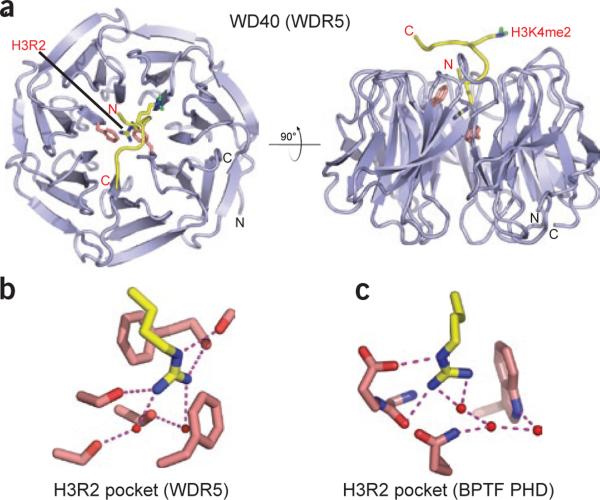
The WD40 protein WDR5 targets arginine in H3 Arg2 context
The WD40-repeat protein WDR5 is a common component of SET1-family histone methyltransferase complexes100. Initial biochemical studies suggested that WDR5 preferentially binds H3K4me2-containing peptides and nucleosomes101. More careful peptide binding analyses by a variety of methods provide a more complicated and controversial picture88–90, although a modest preference for H3K4me2 over other methyl forms is generally observed89,90. WDR5 has been shown to be essential for proper Hox gene activation and vertebrate development, and as described above, RNAi knockdown of WDR5 results in the global loss of H3K4me3 but not of H3K4me1/2 (ref. 101). The molecular basis for H3 tail recognition by WDR5 has emerged from crystal structures of WDR5 complexed to H3K4me2 peptides (best resolution up to 1.50 Å; Fig. 5a) by four research groups88–91. The complex is stabilized by a set of direct and water-mediated hydrogen bonds along the peptide-WDR5 interface, anchored at one end by the N-terminal alanine within a cavity formed by several aromatic residues. Strikingly, unmodified H3R2 inserts its side chain into the central channel of the toroidal WDR5 β-propeller fold, where it is sandwiched between staggered aromatic amino acids and oriented through direct and water-mediated hydrogen bonds (Fig. 5b). Comparing the intermolecular interactions of the H3R2 side chain in the H3K4me2–WDR5 complex (Fig. 5b) with those in the H3K4me3–BPTF PHD finger complex (Fig. 5c) reveals that the contacts made by WDR5 are more extensive and are sequestered from bulk solvent. As expected from the considerableH3R2 recognition interface, mono- and dimethylation of H3R2 abrogates binding affinity88, consistent with WDR5 being a reader of the unmodified arginine mark (for details, see below and a recent review15). Unexpectedly, in five of the six published H3K4me2 structures, the side chain of H3K4me2 is ordered and projects outward from the WDR5 core, with its dimethylammonium proton forming a water-mediated hydrogen bond to a glutamate residue. Two of the four research groups that produced these structures have also solved structures of WDR5 bound to H3K4me0, H3K4me1 and H3K4me3 peptides89,90. The bound methylated H3K4 peptides adopt essentially the same backbone conformation on WDR5 in all three methylation states, consistent with their comparable binding affinities in the low micromolar range88,89. One of the groups noted an apparent kinetic preference for the H3K4me2 state89, and another reported that WDR5 was better stabilized against thermal denaturation in the presence of H3K4me2 than in the presence of other methyl forms90.
Figure 5.
Readout of an unmodified arginine by the WD40 repeat of WDR5. (a) Top view (left) and side view (right) of WDR5 in complex with H3K4me2 peptide (PDB 2H6N). H3R2 and H3K4me2 are in stick representation. H3R2 is deeply buried in the central cavity of WDR5, whereas H3K4me2 is presented by WDR5 on the protein surface for further methylation. (b,c) Details of H3R2 readout by WDR5 in a cavity-insertion recognition mode (b) and by BPTF PHD finger in a surface-groove recognition mode (c).
The way in which WDR5 facilitates trimethylation of H3K4 remains poorly understood. In a recent study102, a Y191F mutation of WDR5 reduced binding of H3 by a factor of ~10, relative to wild-type binding, but maintained the stability of the MLL1 core complex; this resulted in greatly reduced H3K4 HMT activity102. Thus, the WDR5 interaction may be important in properly presenting the H3K4 residue to the active site of MLL1, an idea consistent with the solvent-exposed position of the H3K4 side chain in the WDR5 complexes. Furthermore, the difficulty of finding mutants that perturb H3 peptide binding without disrupting formation of the mixed lineage leukemia-1 (MLL1) core complex attests to an intimate packing association of the MLL C-terminal protein fragment—perhaps specifically its SET methyltransferase domain—with the peptide-proximal WDR5 surface102. Thus, if the SET domain were positioned atop WDR5, the WDR5 protein might act as a ‘presenter’ module to impart sequence and PTM state specificity to the catalytic methyltransferase domain of the complex. The notion of presenters as intermediaries is appealing because it accounts for sequential modification of lysines to increasingly methylated states. Furthermore, there are numerous other WD40-repeat proteins found in assorted chromatin-associated complexes103—for example, the WD40-repeat protein RbAp48 binds unmodified H3 in D. melanogaster NURF and presumably in other complexes78. It remains uncertain whether this developing theme of WD40-repeat presenter modules sensitive to particular PTM states is indeed a more general feature in chromatin biology.
Readout of phosphoserine marks
Many serine and threonine histone residues can be modified by phosphorylation, which, depending on the site, can be important in cell-cycle control, DNA damage repair and transcriptional regulation14. Phosphorylation of serine and threonine, as well as tyrosine, appends a bulky and negatively charged phosphate moiety to the hydroxyl group of the side chain, thereby substantially expanding the ion-pairing and hydrogen-bonding capacities of these residues. Effector-mediated recognition contingent upon protein phosphorylation is the paradigmatic example of PTM-regulated signal transduction14. Although many phosphate-binding modules have been described for non-histone proteins, information on phosphohistone effectors is limited to the two examples discussed below.
14-3-3 Proteins target phosphoserine in H3 Ser10 context
The 14-3-3 protein family has seven distinct isoforms (β, ε, γ, η, σ, τ and ζ), of which14-3-3γ is one of the most abundant. The mammalian 14-3-3 proteins (and Bmh1p and Bmh2p in budding yeast) constitute a family of ubiquitous and well-conserved phosphoserine-binding modules104,105 that regulate signal transduction, chromosome condensation and apoptotic cell death14,106. Serine phosphorylation has been observed on all core histone tails as well as on the linker histone H1. In addition to the considerable global increase in H3S10 phosphorylation (H3S10ph) observed during mitosis, transient H3 phosphorylation also has a role in immediate early gene induction mediated by mitogen- and stress-activated kinases 1 and 2 (MSK1/2)16,107–111. 14-3-3 proteins are known to hetero- and homodimerize, potentially allowing for interaction with at least two phosphoserine sites106. To our knowledge, however, no experimental evidence has yet been reported to support simultaneous engagement of two different histone tails on the same or distinct nucleosomes.
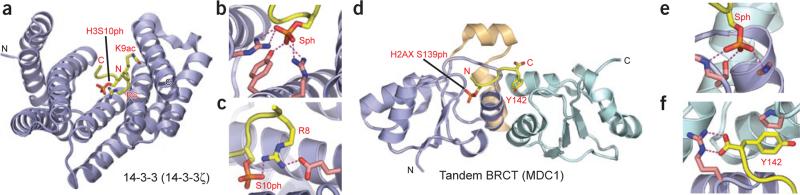
A combined structure-function study has identified 14-3-3 isoforms that target N-terminal histone H3 peptides in a phosphorylation-dependent manner112. H3 tails may be phosphorylated at S10 and S28, each of which is found in an Ala-Arg-Lys-Ser sequence motif. The 14-3-3 protein isoforms were identified by affinity purification as proteins in HeLa cell nuclear extracts that targeted phosphorylated H3 peptide tails, with complex formation demonstrated both in vitro and in vivo. The 14-3-3ξ isoform was found to have measurable affinities for binding to H3S10ph (Kd = 78 μM) and H3S28ph (Kd = 23 μM) peptides, and acetylation of K9 and K14 had a minimal effect on binding. Next, ChIP analysis was used to show in vivo that 14-3-3ξ–bound mononucleosomes containing phosphorylated, acetylated H3 peptides were inducibly recruited to c-fos– and c-jun–proximal nucleosomes after gene activation, concomitant with phosphoacetylation of H3 (ref. 112). Crystal structures have revealed molecular details of the binding pocket of 14-3-3 in its interaction with H31–20S10ph and H31–20S10phK9acK14ac peptides112 (Fig. 6a). The all α-helical 14-3-3 protein forms a dimer in the crystal, and the bound 7-A-R-Kac-Sph-T-G-G-Kac-14 peptide, in a partially extended conformation with a kink, spans the V-shaped binding channel. The complex is stabilized by intermolecular hydrogen bonds between the backbone of the bound peptide and the side chains of 14-3-3, with the phosphate forming multiple contacts and its charge being neutralized by the basic side chains of two arginine residues (Fig. 6b). The side chain of H3R8 forms an intramolecular hydrogen bond to the H3S10ph phosphate, as well as a hydrogen bond to a glutamate side chain of 14-3-3 (Fig. 6c). The backbone trajectory of the bound peptide changes direction at Gly-Gly, allowing it to exit from the binding-cleft constriction.
Figure 6.
Readout of phosphoserine marks by 14-3-3 and BRCT domains. (a) Overview of the complex structure of 14-3-3ζ bound to H3S10ph peptide (PDB 2C1J). Peptide is buried in the V-shaped 14-3-3 protein scaffold. The H3K9ac mark can be accommodated on the surface channel of 14-3-3. (b) Details of H3S10ph mark recognition around the phosphoserine-binding site within 14-3-3ζ. The phosphate group is anchored by multiple hydrogen-bonding and ion-pair networks. (c) Details of H3R8 recognition by 14-3-3ζ. Recognition of the guanidinium amino group of H3R8 contributes to sequence specificity of H3S10ph readout by 14-3-3ζ. The intramolecular contact to the phosphate in the bound peptide is stabilized by additional contacts involving each residue. Similar contacts are anticipated in phosphohistone complexes with other 14-3-3 isoforms. (d) Overview of structure of the MDC1 tandem BRCT domains in complex with a H2AXS139ph peptide (PDB 2AZM). Slate and pale cyan, N- and C-terminal BRCT domains, respectively; beige, helix-loop-helix linker. The peptide-binding site lies at the interface of the two domains. (e) Close-up view of the phosphoserine-binding site. The site is formed within the first BRCT domain and stabilized by several hydrogen bonds with pronounced coulombic character, involving side chains as well as main chains of several BRCT domain residues. (f) Close-up view of recognition of the C-terminal tyrosine (+3 position) at the interface of the two MDC1 BRCA domains. The free C-terminal carboxylate group is capped by an arginine side chain from the first BRCT domain. The tyrosine side chain is stacked against a proline ring from the second BRCT domain. This provides another example of terminus specificity as an apparent additional recognition determinant.
Tandem BRCT domains of MDC1 target phosophoserine in γH2AX
Cells respond to double-strand breaks in DNA, an extremely toxic lesion, by initiating a DNA damage response involving deployment of multiprotein complexes that signal for cell-cycle arrest and coordinate repair processes. DNA damage and repair ‘signatures’ in the form of covalent modifications of histones and related proteins are only beginning to be understood113. The human H2A variant H2AX has been shown to be phosphorylated by the ‘ataxia telangiectasia mutated’ kinase (ATM) at S139 (known as γH2AX) upon induction of double-strand breaks114,115. Mediator of DNA damage checkpoint protein -1 (MDC1, also known as NFBD1) was noted to colocalize with γH2AX and 53BP1 (see above) to specialized DNA damage-induced nuclear foci in a manner dependent on γH2AX116. Subsequent structural studies have illuminated a direct interaction between the phosphoepitope at the C terminus of γH2AX and the tandem BRCT repeats of MDC1 (refs. 117,118), as reviewed recently14.
A peptide spanning the γH2AX C-terminal tail (K-K-A-T-Q-A-Sph-Q-E-Y) binds the tandem BRCT domains of MDC1 with substantial affinity (Kd = 2.2 μM). Mutation studies validated S139 as the key phosphorylation site associated with this recognition and also highlighted the importance of the tyrosine residue at the +3 position relative to the phosphorylation site. The molecular basis for recognition of H2AXS139ph by the tandem BRCT domains of MDC1 was elucidated by the crystal structure of the complex118 (Fig. 6d). The compact α/β fold of each BRCT repeat is bridged by the helical linker segment. There is complementarity between the surfaces of the S139ph-modified γH2AX and the binding cleft that forms at the interface of the BRCT domains. The phosphate oxygens of the phosphoserine are hydrogen-bonded to main chain and side chain atoms of two structurally conserved residues on MDC1 (Fig. 6e). A key arginine in MDC1 forms a pair of hydrogen bonds to the C-terminal carboxylate of Y142 of γH2AX (Fig. 6f), and mutation of this arginine completely abolishes binding. Notably, the complex formed between MDC1 and the phosphoepitope of γH2AX118 shows similar molecular recognition features to those observed in the BRCA1–BACH1 complex119. When the interaction between MDC1 and γH2AX was disrupted, either by mutation of residues in MDC1 or by use of an unphosphorylatable H2AX S139A mutant or interaction-disrupting H2AX Y142A mutant, MDC1 was no longer recruited to damage foci. Immunofluorescence studies showed that MDC1 mislocalization also disrupted the recruitment of downstream DNA damage repair proteins such as 53BP1 and NBS1 (ref. 118), revealing the importance of this interaction.
Principles underlying phosphoserine recognition
In the structures of complexes of the H3S10ph peptide bound to 14-3-3 protein112 (Fig. 6b) and the H2AX S139ph peptide bound to the tandem BRCT domains of MDC1 (ref. 118 and Fig. 6e), the phosphate group attached to the serine is fully hydrogen-bonded to side chain and/or backbone atoms, at least one of which is positively charged to facilitate charge neutralization. The phosphoserine can also participate in a hydrogen-bonding network, as seen in the 14-3-3 complex, where a glutamate of the protein helps to orient an intramolecular contact within the peptide (Fig. 6c). An additional feature of the MDC1 complex hints at a more common theme: the register of the phosphoserine mark relative to the anchored C terminus of the bound γH2AX (Fig. 6f) seems to be important, analogous to N-terminal recognition for certain readers of unmodified and methylated lysine marks, discussed above. Interestingly, WD40-repeat proteins have also been structurally characterized as effectors that bind unmodified lysine or arginine and phosphoserine in signal transduction and protein-protein interaction interfaces120–122. In regard to this, the remaining challenge will be to identify more downstream effectors associated with chromatin that may interpret the information encoded by histone phosphorylation PTMs.
Phosphorylation-mediated cross-talk relationships
Other intriguing phosphate-related challenges remain for histone proteins. For example, phosphate-mediated ‘phospho-methyl switching’ has been proposed and shown experimentally to control the release of more stably bound chromatin effectors, such as HP1 bound to H3K9me during mitosis and probably additional effectors bound during other cell-cycle and developmentally regulated events123–125. Phospho-methyl switching has also been observed in non-histone proteins, for which it similarly toggles effector affinity126. Finally, we note that although some highly conserved phosphorylation sites in histone proteins are reasonably well studied (for example, H3S10ph or linker histone H1 phosphorylation during mitosis127), point mutations at these sites often yield little or no obvious phenotype in yeast128. It remains of interest to determine whether other phosphorylation sites in histone proteins carry redundant or backup functions, as well as to explore the potential functions of other, less well-studied phosphoacceptors (for example, tyrosine and histidine) in histone biology129. As new phosphorylation marks are identified, we predict that new cognate binding effectors will also be found that may conform to some of the principles of effector-mediated binding discussed in this review. In addition, important roles in the epigenetic landscape will probably also be discovered for phosphorylation-mediated direct effects and other cross-talk relationships.
Emerging themes and future challenges
The recent deluge of structural and functional data concerning the reading and writing of histone modifications has yielded tantalizing clues as to how the addition of small chemical entities to histones, and their removal, can translate into remarkable changes in chromatin structure and biology. In this section, we highlight emerging ideas that the field is poised to test using lessons from the observed interactions of histones with binding modules.
Linked effector modules and multivalent readout of histone marks
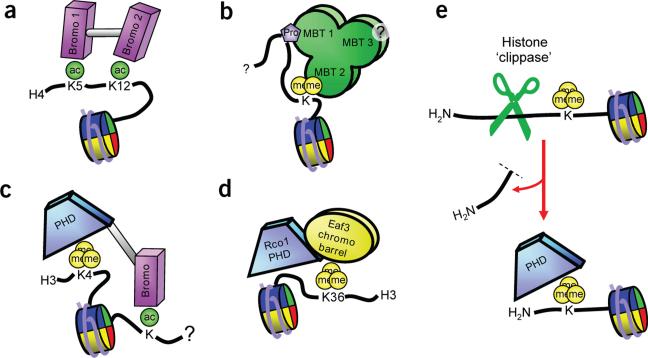
There are many examples of two or more putative histone-binding modules linked within the same protein or within the same multisubunit complex, allowing for combinatorial PTM readout at the histone or nucleosomal level84. For example, in the sections above we compare the architecture of dual domains such as the double chromodomains of CHD1 (Fig. 3c), double tudor domains of JMJD2A (Fig. 3d), tandem tudor domains of 53BP1 (Fig. 3e) and tandem BRCT domains of MDC1 (Fig. 6d). Interestingly, some of these dual domains bear a single binding site for their marks—a site that either is found only in one of the two domains (as in the JMJD2A double tudor domains) or is formed at the interface of the two domains (as in the CHD1 double chromodomains and the MDC1 tandem BRCT domains)—whereas other dual domains retain two separate binding sites. Importantly, the presence of two independent binding sites for modified histones establishes the possibility of simultaneous engagement of two different substrates. For instance, the TAF1 double bromodomain (Fig. 2d) may simultaneously engage dually acetylated histones such as those observed on the H4 tail (Fig. 7a). Another example is provided by the MBT repeats of L3MBTL1: two of the three repeats may be used simultaneously to engage two sites/marks in a histone tail (Fig. 7b and Supplementary Fig. 1). Pocket 2 of L3MBTL1 has been described as a mono- and dimethyllysine-binding module35,71, whereas pocket 1 contains a Pro-Ser binding motif35. Thus, these two binding determinants, separated by an appropriate distance in a histone peptide, could be simultaneously targeted, and this distance constraint may be satisfied by Pro30 and methylated K36 or K37 in histone H3.3. For both TAF1 and L3MBTL1, composite specificity may arise both from the molecular recognition properties of each binding pocket and from the spacing between the two pockets.
Figure 7.
Combinations of PTM-binding sites generate different specifities. (a) The double bromodomain motif in hTAF1 simultaneously binds proximal acetylated lysines on H3 or H4. (b) First and second modules of L3MBTL1 may simultaneously bind proline and dimethyllysine marks on histone tails. (c) The PHD finger of BPTF and the proximal bromodomain bind H3K4me3 and an acetyllysine simultaneously.
(d) The PHD finger of Rco1p increases the binding of the Eaf3p chromobarrel to H3K36me3 nucleosomes. (e) Schematic of histone-directed protease activity uncovering a cryptic PTM-binding site.
Also of interest are linked unique domains that can potentially bind multiple marks residing on the same or different histone tails. One example is the BPTF PHD finger–linked bromodomain, in which the linker adopts an α-helical conformation that separates the PHD-finger domain (which targets H3K4me3) and the bromodomain (which targets H4Kac) by a fixed distance and defines the relative orientations of their binding pockets40 (see Supplementary Fig. 1 for structure). The adjacent PHD finger and bromodomain of BPTF could be used to simultaneously engage both H3K4me3 and an acetylated lysine residue (Fig. 7c). In another intriguing case, the Rpd3S complex has two subunits that are required for in vitro interaction with H3K36me and hyperacetylated nucleosomes: the chromodomain of Eaf3p is a modest H3K36me3-binding effector130,131, and the PHD finger of Rco1p makes an additional contribution to nucleosomal binding that remains unclear132 (Fig. 7d). Moreover, many other complexes involved in chromatin regulation contain multiple prospective reader modules as subunits, setting the stage for readout of histone marks on nucleosomes by combinatorially assembled complexes84. Figure 7 schematically summarizes the ways in which multiple modules may collaborate to increase specificity for multiple marks in chromatin substrates. Individual recognition elements need not be restricted solely to the combinatorial recognition of histone marks, but could in addition make contributions through shape complementarity associated with molecular recognition between complementary surfaces (see Boxes 2 and 3). If, as has been proposed, a free N terminus is required for PHD fingers to engage H3K4me3 (see Box 2)15, a histone-specific endoprotease activity133 could uncover cryptic binding sites (Fig. 7e). In addition, sequence-specific and nonspecific DNA contacts may have a significant role as recruitment and avidity elements in multivalent interactions.
Given that these potential binding elements are arrayed in complexes capable of synchronous binding to a number of chromatin moieties, what are the anticipated functional consequences of multivalent substrate binding by chromatin-associated complexes? Multivalency may afford enhanced affinity, composite specificity and more dynamic control of residence times of chromatin-associated macromolecular assemblages15,84. With respect to the genome-wide localization of various histone PTMs, it is evident that discrete patterns of marks generally correlate with particular functional consequences5,73. Novel mass spectrometry approaches have recently detailed a wealth of previously undescribed coexisting histone marks that may serve as platforms for such engagement54,134. We have put forward the idea that these distinct patterns, coupled with other chromatin elements, serve to recruit and stabilize different macromolecular complexes to loci15,84. A single modification may often not suffice to transduce a particular effector-mediated downstream consequence: as noted above, modules that bind H3K4me3 are found in both complexes that activate transcription and complexes that repress it, begging the question of how the targeting specificity of a complex is achieved81,84. The biophysics of multivalency may resolve this conundrum—for example, an activating complex might bind H3K4me3 and additional activating PTMs, whereas a silencing complex might also bind H3K4me3 but simultaneously engage silencing PTMs.
Readers of mono- and dimethylarginine marks
Although interest in arginine methylation in histone proteins has arguably lagged behind the current interest in histone lysine methylation, this situation is not likely to last for long. The coactivators CARM1 and PRMT1 are arginine-directed methyltransferases targeting H3R17 and H4R3, respectively, and are well known to stimulate nuclear receptor–mediated transcription from chromatin templates both in vivo and in vitro135–137. Moreover, these activation reactions are reversible by demethylase and demethylimidase activities138,139, consistent with the well-established theme that it is the steady-state balance of methylation at these sites that is crucial for gene regulation. The arginine side chain can be monomethylated (Rme1), symmetrically dimethylated (Rme2s) or asymmetrically dimethylated (Rme2a) at the guanidinium η positions (Fig. 1a); however, knowledge about readers of methylarginine marks is limited, as no structure of a reader bound to this mark is yet available. We summarize below the available data on the survival of motor neuron (SMN) tudor domain and its putative recognition of the symmetrical dimethylarginine mark; we envision similar binding modes for other putative histone methylarginine effectors. The SMN protein is key in the assembly of spliceosomal small nuclear ribonucleoprotein (snRNP) complexes. NMR-based studies have established that the central tudor domain of SMN forms a barrel-like fold composed of sharply bent antiparallel β-sheets and that its outer surface has an overall negative charge140. The tudor domain of SMN binds the arginine/glycine-rich C-terminal tails of spliceosomal Sm core proteins, and this binding is facilitated after symmetrical dimethylation of arginine side chains141. NMR chemical shift and saturation transfer experiments indicate that the symmetrical dimethylarginine side chain is positioned near a cluster of conserved aromatic residues that form aromatic cages in structurally related methyllysine-binding effectors141. Spinal muscular atrophy results from specific mutations in the SMN gene, one of which is near this putative methylarginine-binding cage. The above data, though informative, have yet to provide molecular insights into the principles underlying methylarginine recognition by the SMN tudor domain.
Although knowledge of methylarginine effector modules is limited, certain arginines are known to have key roles in modulating the binding of other modules. Indeed, recent reports suggest that arginine methylation at H3R2 antagonizes H3K4 methylation142,143. Presumably, the mechanism for this disruption is traceable to the H3R2 modification–sensitive binding of WDR5 methyltransferase complexes, as an unmodified H3R2 is essential for productive WRD5 binding88, and WDR5 binding in turn is required for global H3K4 methylation101. Thus, as with other adjacent or nearby histone marks such as phosphomethyl switching of HP1 (refs. 123–125) and potentially other similar switches57,88,144, mutually exclusive cross-talk relationships seem to govern the final outcome of numerous effector-binding relationships (see review in this issue by Dent et al.145). We look forward not only to the identification of as yet unknown histone modification-binding modules, but to increased understanding of the molecular principles by which they operate and the functional consequences of these binding events.
Beyond chromatin biology
We predict that additional PTMs and PTM patterns will be identified with some regularity as new approaches and methods are applied to physiological states that are currently less well studied. Witness the relatively new idea that bivalent domains containing H3K4me and H3K27me marks (which turn transcription on and off, respectively) characterize certain developmentally poised genes in early embryonic stem cells32. Indeed, the known chromatin-based epigenetic landscape is rapidly changing shape and taking on new contours, and the ‘modification-effector road’ promises to be an exciting route with more unexpected turns.
In closing, we ask whether ‘pocket reading’ of chromatin modifications matters beyond its purely intellectual interest, and to what extent epigenetics underlies biological processes that affect human biology and disease. Traditionally, cancer research has focused on genetic changes such as amplifications, deletions and point mutations affecting molecules involved in growth control and cellular homeostasis. It has become increasingly clear that epigenetic changes, such as DNA and histone methylation, contribute to the progression of human disease (see ref. 146 and chapters 23 and 24 in ref. 1). We favor the general view that changes in the regulatory signals provided by chromatin modifications—changes that can result from alteration of either the enzyme systems that write this language or the protein modules designed to read it—lie at the heart of many pathological states and have far-reaching implications for human biology. For example, perturbations of the MLL1 protein, a methyltransferase associated with writing of H3K4 methylation marks at Hox gene clusters, are causative of a number of leukemias. However, MLL1 itself contains multiple PHD fingers in a region located well outside of its known catalytic SET domain, and the functions of these fingers are currently unknown. It will be important for future research to explore whether human disease can arise from changes in histone PTM–reading modules that act as either individual or combinatorial units of recognition.
Supplementary Material
BOX 1 Lessons learned from state-specific readout of methyllysine marks.
Structural studies of methyllysine effectors in cognate peptide–bound or apo forms suggest general mechanisms that are used by binding pockets to achieve specificity toward distinct methylation states. Here we illustrate these common features and their roles in engagement.
Surface-groove versus cavity-insertion recognition modes
The known structures of methyllysine effector complexes can be subdivided into those that use a cavity-insertion recognition mode (i), as observed in complexes of the lower methylation state–binding 53BP1 tandem tudor domains36 and L3MBTL1 (ref. 35), and those that use a surface-groove recognition mode (ii), as observed in complexes of the higher methylation state–binding HP1 chromodomain45,46,48, CHD1 double chromodomains57, JMJD2A double tudor domains63 and PHD finger39,40,74. In the cavity-insertion recognition mode, the methylammonium group of methyllysine is inserted into, and buried within, a deep protein cleft (i), potentially endowing the pocket with size-sensitive selection filter capabilities. In the surface-groove recognition mode, the binding pockets are both wider and more accessible (ii), such that the methyllysine side chain lies along a protein surface groove and the effector complexes have appreciable but less stringent preferences for certain lysine methylation states.
Static recognition pockets
Whereas the peptides can undergo β-strand backbone conformational changes induced during binding, the effector proteins often show little or no appreciable structural perturbation. The compact nature and apparent monolithic rigidity of effector folds precludes substantial movement as a consequence of ligand engagement. Furthermore, for most structurally characterized effectors, there seems to be little or no movement of side chains involved in recognition. In particular, aromatic cages in the bound form are very static with respect to their unliganded counterparts (iii). Preorganized cages have a clear energetic benefit over flexible binding sites in that the conformational entropy loss upon binding is minimized on the protein side. Notable exceptions to this rule include side chain movements observed upon complexation in WDR588,89 and 53BP1 (ref. 36).
Aromatic cage pockets
A common and striking feature of methyllysine reader domains is the positioning of the methylammonium moiety within an aromatic cage consisting of two to four aromatic residues, often supplemented by one or more acidic side chains. In contrast to the PHD fingers that recognize the unmodified lysine (which lack any semblance of aromatic caging residues; iv), the BPTF PHD finger has four aromatic residues (viii) and the ING2 finger has two aromatic residues and a hydrophobic methionine side chain (vii). The methylammonium group, which carries a diffuse positive charge, stacks over the partial negative charge permeating the face of the aromatic rings, with the electrostatic stabilization mediated by cation-π interactions33,34 (v–vii) and to a lesser extent by hydrophobic contacts.
Structural basis for lower lysine methylation state–specific readout
The tandem tudor domains of 53BP1 (ref. 36) and the second MBT pocket of L3MBTL1 (ref. 35) both bind mono- and dimethyllysine within an aromatic cage, using the cavity-insertion recognition mode (i) to discriminate against the trimethyllysine counterpart (compare to surface-groove recognition, ii). The specificity for lower lysine methylation state readout originates in a direct hydrogen bond and electrostatic interaction between the methylammonium proton and the carboxylate of an aspartate residue lining the walls of the aromatic cage (v and vi). Furthermore, trimethyllysine binding would be disfavored by an anticipated steric repulsion between the additional methyl group and the aspartate side chain. In addition, the limited dimensions of the binding pockets in 53BP1 and L3MBTL1 complexes, both at the entrance and within the channel, seem to restrict the access of the larger trimethylammonium group.
Engineering methyllysine recognition specificity
The wild-type BPTF PHD finger binds methylated lysines in an H3K4 sequence context with affinities in the order Kme3 > Kme2 >> Kme1 > Kme0, as measured by NMR titration, surface plasmon resonance and fluorescence polarization studies40. It was anticipated that the availability of a properly positioned carboxylate side chain could allow fine-tuning of the state-specific readout of methylated lysines. Indeed, a single Tyr-to-Glu substitution in the aromatic cage motif of the BPTF PHD finger reverses the binding preference from H3K4me3 to H3K4me2 (ref. 35). The crystal structure of the Y17E mutant BPTF PHD finger (viii) establishes that the specificity change is associated with hydrogen-bonding between the methylammonium proton and the carboxylate group of the glutamate side chain in the mutant. In this instance, the stringency of discrimination by surface-groove recognition does not match that described above for cavity-insertion recognition because of the steric demands associated with the latter recognition mode.
BOX 2 Lessons learned from sequence-specific readout of methyllysine marks.
The sequence context in which a histone modification is presented to an effector module is crucial to the fidelity of molecular readout. Here we describe general constraints on modification-proximal residues of the substrate peptide for common recognition elements in binding modules.
Induced β-sheet intermolecular alignment
The methyllysine-containing peptides are unstructured in the free state but undergo induced-fit conformational transitions upon forming complexes with effector domains. In most cases, the bound peptide adopts an induced-fit β-strand conformation and pairs through antiparallel alignment along the exposed edge of an existing β-sheet scaffold of the effector domain (i)15,40. This pairing alignment in turn projects the side chains of methyllysine and its non-nearest neighbor residues at the −2 and +2 positions toward the effector domain, with binding affinity and specificity determined by steric compatibility and the network of intermolecular hydrogen-bonding and electrostatic interactions.
Register of marks relative to a free peptide termini
The register of the methyllysine mark relative to the terminus of the histone tail could also be a general contributing factor in effector recognition15. This is likely to be important only when the terminus is near the modification recognized, as occurs for the H3K4me mark. In such cases, the positively charged N terminus is anchored in its own binding pocket (i and ii), which in essence acts as a block against N-terminal extensions characteristic of longer substrates. Similarly, C-terminal recognition seems to be important for the H2AXS139ph modification (Fig. 6f). It remains unclear to what extent, if any, early reports of developmentally regulated histone proteolysis foreshadow physiologically relevant chromatin-binding events, one function of which could be to generate free ends, enabling the binding of modules that would otherwise be excluded by additional peptide extensions.
Specificity of BPTF and ING2 PHD fingers for trimethyllysine
The arrangement of tryptophan-separated binding channels (ii and Fig. 4b) readily explains the specificity of the BPTF40 and ING239 PHD fingers for H3K4me3, as this sequence has H3R2 and H3K4me3 separated by one residue (T3), whereas binding of H3K9me3 and H3K27me3 is disfavored because an arginine is adjacent to Kme3 in both these sequences. Deletion of T3 greatly reduces binding in the ING2-like Yng1p PHD finger74, suggesting that T3 functions as a spacer40.
BOX 3 Contribution of shape complementarity to molecular recognition.
Although this review focuses largely on histone PTMs (mainly hydrophilic in nature) and their binding modules, it does not escape our attention that non-PTM hydrophobic interactions may contribute to the engagement and function of non-histone proteins at chromatin, especially at the level of multivalent interactions.
ETO MYND domain targets proline-rich hydrophobic motifs in SMRT
NMR solution studies establish that myeloid-Nervy-DEAF-1 (MYND) domains, which contain a Cys6-His-Cys zinc-chelating motif, adopt an interleaved zinc-chelating topology reminiscent of PHD- and RING-finger folds147,148. Binding studies have implicated certain MYND family members in recognition of hydrophobic stretches in peptides. Indeed, the ETO MYND domain interacts with the NPPPLIS peptide of the silencing mediator of retinoic and thyroid hormone receptor (SMRT)148. The SMRT peptide binds in a kinked, semi-extended conformation to a hydrophobic declivity in the MYND domain by forming an antiparallel β-sheet—that is, the two–stranded β-sheet core of the MYND domain is extended to its three-stranded counterpart by alignment of the adopted β-strand contributed by the C-terminal portion of the SMRT peptide. The side chain of the leucine in the bound NPPPLIS sequence inserts into a hydrophobic pocket (i and ii), while the first proline ring of the sequence packs against a tryptophan ring of the MYND domain (i). The complex is further stabilized by intermolecular hydrogen bonds between side chains of glutamine and serine residues of the MYND domain and CO backbone groups of the peptide (i). Hydrophobic desolvation effects probably dominate the energetics of binding and, together with the numerous hydrogen-bonding contacts, may dominate the specificity as well. In addition, the conformational restriction of proline-rich peptide sequence may minimize the entropic binding penalty while at the same time positioning the peptide backbone so that it extends out at the C-terminal end for intermolecular hydrogen bond formation in an antiparallel β-sheet context. Certain mutations of MYND residues involved in SMRT peptide recognition reduce binding affinity and attenuate the impact of the leukemogenic AML1-ETO fusion on gene expression, differentiation and proliferation148.
ACKNOWLEDGMENTS
We apologize to all of the researchers whose important contributions could not be acknowledged because of space constraints. We thank members of the Patel and Allis laboratories as well as the anonymous reviewers for critically reading the manuscript, and A. VanDemark (University of Pittsburgh) and C.P. Hill (University of Utah) for providing coordinates for Rsc4p in Figure 2e. D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust and the Dewitt Wallace and Maloris Foundations, C.D.A. and S.D.T are supported by US National Institutes of Health grants GM53512 and GM63959 and by funds from The Rockefeller University, and A.J.R. is supported by a postdoctoral fellowship from the Irvington Foundation.
Footnotes
Note added in proof: A paper was just published149 reporting that JMJD6 is a histone arginine demethylase.
Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Allis CD, Jenuwein T, Reinberg D, Caparros ML, editors. Epigenetics. Cold Spring Harbor Laboratory Press; Woodbury, New York: 2006. [Google Scholar]
- 2.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shogren-Knaak M, et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 11.Ahn SH, et al. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 15.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 17.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 19.Brownell JE, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 20.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 22.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo MH, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 25.VanDemark AP, et al. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetyla tion. Mol. Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Swigut T, Wysocka J. H3K27 demethylase at last: what it means for memory and plasticity of gene expression in developmental processes. Cell. 2007;131:29–32. [Google Scholar]
- 30.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 31.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Yuung RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–78. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 33.Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc. Natl. Acad. Sci. USA. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma JC, Dougherty DA. Cation-π Interaction. Chem. Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 35.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats and an engineered PHD finger. Mol. Cell. doi: 10.1016/j.molcel.2007.10.023. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burley SK, Petsko GA. Weakly polar interactions in proteins. Adv. Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 38.Maurer-Stroh S, et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 39.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant home-odomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 43.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs SA, et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 47.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromo-domain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen AL, et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 50.Zhao T, Heyduk T, Allis CD, Eissenberg JC. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
- 51.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 52.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 53.Konev AY, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taverna SD, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. USA. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hake SB, et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 56.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodo-main links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 57.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 58.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okuda M, Horikoshi M, Nishimura Y. Structural polymorphism of chromodomains in Chd1. J. Mol. Biol. 2007;365:1047–1062. doi: 10.1016/j.jmb.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 60.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 61.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 62.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 63.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 66.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 67.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 68.Koga H, et al. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- 69.Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- 70.Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J. Biol. Chem. 2003;278:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- 71.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 72.Wang WK, et al. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 74.Taverna SD, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 76.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 77.Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- 78.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 79.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Doyon Y, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 81.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 82.Martin DG, et al. The Yng1p PHD finger is a methyl-histone binding module that recognizes lysine 4 methylated histone H3. Mol. Cell. Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruthenburg AJ, Li H, Taverna SD, Patel DJ, Allis CD. Multivalent readout of histone modifications by linked effector modules on a nucleosome scaffold. Nat. Rev. Mol. Cell Biol. in the press. [Google Scholar]
- 85.Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 86.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 89.Ruthenburg AJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schuetz A, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han Z, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 92.Hakimi MA, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 94.Marmorstein LY, et al. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 95.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 96.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 98.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 99.Argentaro A, et al. Structural consequences of disease-causing mutations in the ATRXDNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 102.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 103.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 104.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14–3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 105.Yaffe MB, et al. The structural basis for 14–3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 106.Dougherty MK, Morrison DK. Unlocking the code of 14–3-3. J. Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 107.Thomson S, et al. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Soloaga A, et al. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 110.Duncan EA, Anest V, Cogswell P, Baldwin AS. The kinases MSK1 and MSK2 are required for epidermal growth factor-induced, but not tumor necrosis factor-induced, histone H3 Ser10 phosphorylation. J. Biol. Chem. 2006;281:12521–12525. doi: 10.1074/jbc.M513333200. [DOI] [PubMed] [Google Scholar]
- 111.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 112.Macdonald N, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14–3-3. Mol. Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 113.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 114.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 115.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates his-tone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 116.Peng A, Chen PL. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 2003;278:8873–8876. doi: 10.1074/jbc.C300001200. [DOI] [PubMed] [Google Scholar]
- 117.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J. Biol. Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 118.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 119.Clapperton JA, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 120.Wu G, et al. Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 121.Xiong JP, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 123.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 124.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 125.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 126.Sampath SC, et al. Methylation of histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 127.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 128.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 129.Chen CC, Smith DL, Bruegger BB, Halpern RM, Smith RA. Occurrence and distribution of acid-labile histone phosphates in regenerating rat liver. Biochemistry. 1974;13:3785–3789. doi: 10.1021/bi00715a026. [DOI] [PubMed] [Google Scholar]
- 130.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 131.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 132.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 133.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 134.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 135.Strahl BD, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 136.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 137.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 138.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 139.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 140.Selenko P, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 141.Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 142.Kirmizis A, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guccione E, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 144.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 145.Latham JA, Dent SYR. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 146.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 147.Spadaccini R, Perrin H, Bottomley MJ, Ansieau S, Sattler M. Structure and functional analysis of the MYND domain. J. Mol. Biol. 2006;358:498–508. doi: 10.1016/j.jmb.2008.01.071. [DOI] [PubMed] [Google Scholar]
- 148.Liu Y, et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.