Abstract
Argonaute proteins are conserved throughout all domains of life. Recently characterized prokaryotic Argonaute proteins (pAgos) participate in host defense by DNA interference, whereas eukaryotic Argonaute proteins (eAgos) control a wide range of processes by RNA interference. Here we review molecular mechanisms of guide and target binding by Argonaute proteins, and describe how the conformational changes induced by target binding lead to target cleavage. On the basis of structural comparisons and phylogenetic analyses of pAgos and eAgos, we reconstruct the evolutionary journey of the Argonaute proteins through the three domains of life and discuss how different structural features of pAgos and eAgos relate to their distinct physiological roles.
Argonaute (Ago) was first mentioned in a study describing a mutant in Arabidopsis thaliana1. Because the leaves of the mutant plant curled up like squid tentacles, the gene and corresponding protein were named after the octopus Argonauta argo. It later became clear that the Ago protein is the key player in eukaryotic RNA interference (RNAi) pathways (Box 1), in which Ago utilizes short 5′-phosphorylated RNA guides to target complementary RNA transcripts. The Ago proteins belong to the PIWI protein superfamily, defined by the presence of a PIWI (P element–induced wimpy testis) domain. In addition, all eAgos feature an N (N-terminal) domain, a PAZ (PIWI-Argonaute-Zwille) domain and a MID (middle) domain, along with two domain linkers, L1 and L2 (Fig. 1 and Box 2).
Box 1 RNA interference pathways.
Eukaryotic RNAi pathways (reviewed in refs. 61,73,88,89) include proteins with RNase III–like domains (Dicer and Drosha) that usually process dsRNA precursors into short dsRNA molecules (small interfering (si)RNAs). With phosphorylated 5′ ends and 2-nucleotide overhangs at the 3′ ends, siRNAs consist of a passenger strand and a guide strand, and the latter is selectively loaded into eAgos. After removal of the passenger strand, eAgo holds on to the guide strand, which enables eAgo to bind mRNA targets complementary to the guide. Binding of the guide strand to Ago results in helical preordering of the seed segments in the guide (nucleotides 2–7 or 2–8), which enhances the affnity for a matching target30. Target binding starts in this seed region (nucleation) and extends by zippering toward the 3′ end of the guide (propagation). This results in release of the 3′ end of the guide from Ago and induces conformational changes that result in target cleavage. The cleaved target strand is released, allowing Ago to bind and cleave additional targets. In the case of imperfect targets and/or catalytically inactive eAgos, binding of eAgo, alone or with associated proteins, results in repression of mRNA translation. Both processes eventually lead to silencing of gene expression.
Figure 1.
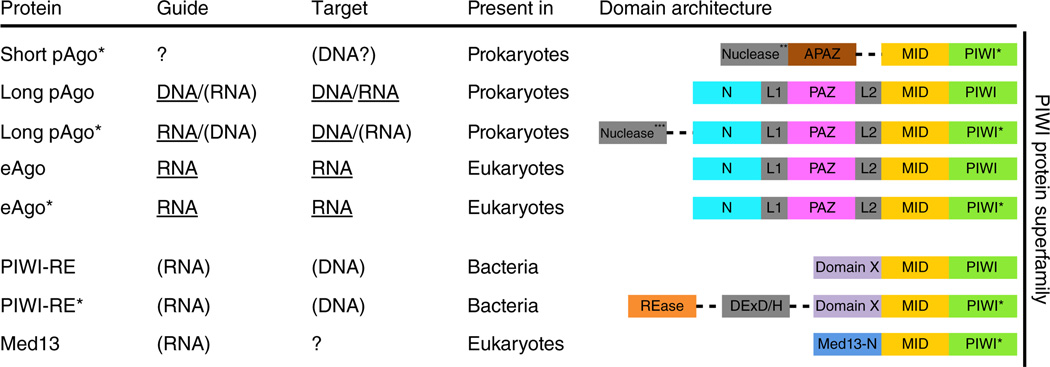
Domain architectures of the PIWI superfamily proteins. Dotted lines indicate separate genes located in the same (predicted) operon. *, Ago proteins with an incomplete DEDX catalytic tetrad in the PIWI domain. Guide and target usage is based on available biochemical data (underlined) or predicted (in parentheses). **, predicted nucleases from Sir2, Mrr or TIR protein families. ***, predicted nucleases from Sir2, Mrr, Cas4 or PLD protein families. REase, restriction endonuclease; DExD/H, superfamily II helicase (denoted after a signature amino acid motif).
Box 2 Ago domains have conserved structures and functions.
The core functions of each of the four structural domains of Ago proteins are well conserved in prokaryotes and eukaryotes.
The MID domain
This domain forms a basic nucleotide-binding pocket in which several conserved amino acids interact with the phosphate group at the 5′-end of the guide5,19–22. In addition to the specific binding of the sugar backbone of the 5′-end terminal nucleotide, at least some Agos recognize specific 5′-end bases11 using a structural feature termed the ‘nucleotide specifcity loop’21,77. The MID domain also stacks the guide in a helical conformation within its seed nucleotides (2–7 or 2–8), promoting target binding (reviewed in ref. 30).
The PIWI domain
This domain includes the RNase H–like active site of slicing Agos6,16,18,22. In the cleavage-compatible conformation, two divalent cations are bound by a DDX triad (where X is usually aspartic acid or histidine; in rare cases it is lysine89). The catalytic site is completed by a glutamate residue that resides on a mobile PIWI loop (the glutamate finger), forming the DEDX motif12,15.
The PAZ domain
This domain binds the 3′ end of the guide by interactions with the backbone of nucleotides 20 and 21 (refs. 7,12,26,27). This interaction is not essential for guide binding but protects the guide from degradation90.
The N domain
This domain is not involved in guide loading but plays a critical role in target cleavage35,66 and in the dissociation of cleaved strands35,91. During duplex RNA loading, the strand with the less stable 5′ end is retained as guide in Ago92,93. Removal of the other strand (passenger) can be slicer dependent (requires cleavage) or independent (requires mismatches or G•U wobble base pairs in the 5′ seed or in the middle of the 3′ region)94,95. In both pathways, the N domain functions as a wedge, disrupting guide-passenger base pairs at the 3′ end of the guide (active wedging) or by blocking guide-target base pairing downstream of nucleotide 16 of the guide as observed for TtAgo ternary complexes (passive wedging)11,91. The role of the N domain in target cleavage is indicated by work on human Ago 3 (hAGO3), which is unable to cleave targets in vitro, even though it has an intact catalytic site; hAGO3 can be activated when its N domain is swapped for that of hAGO2 (ref. 66). Similarly, target cleavage of activated hAGO1 is enhanced when its N domain is replaced by the counterpart of hAGO2 (ref. 35). How the N domain facilitates slicer activity is presently unclear
Many prokaryotic genomes also feature ago genes2–4. Long pAgos encompass the same domains as eAgos, whereas short pAgos consist of only the MID and PIWI domains (Fig. 1). Like eAgos, pAgos interact with 5′-phosphorylated oligonucleotides, but in contrast to eAgos, some pAgos have higher affinity for DNA guides than for RNA guides5–7. Both long and short variants of pAgos (Fig. 1) have been proposed to function in defense against mobile genetic elements4. Indeed, it was recently shown that both RNA-guided8 and DNA-guided9 pAgos interfere with foreign DNA in vivo.
A major challenge in the early days of RNAi research was to uncover structure-function relationships of Ago proteins. For practical reasons, initial efforts to obtain Ago structures focused on pAgos before their physiological role was known. Those studies provided valuable mechanistic insights into guide-target pairing and guide-mediated target cleavage7,10–12 (Box 1). More recently, structures of eAgos have also been solved13–15. Here we review the body of structural work on pAgos and eAgos, and compare the features that determine their differential functionalities, such as guide preference (DNA versus RNA), nucleolytic activity and docking sites for partner proteins. We also discuss phylogenetic analyses that provide insight into how Agos have changed during their evolutionary journey, from relatively simple host-defense proteins in prokaryotes, to key players in complex multiprotein regulatory pathways in eukaryotes.
Structures of Argonaute proteins. The first crystal structures determined were of the guide-free pAgos of Pyrococcus furiosus (PfAgo)16, Aquifex aeolicus (AaAgo)6,17 and short pAgo from Archaeoglobus fulgidus (AfAgo)18, which provided information about the overall structural organization of Agos. The long pAgos revealed a bilobal architecture, with the PAZ lobe (N, L1 and PAZ domains) connected by L2 to the PIWI lobe (MID and PIWI domains). The MID domain adopts a Rossmann-like fold with a characteristic nucleotide-binding pocket5,19–22. The PIWI domain adopts a typical RNase H fold6,16,18,22 with three catalytic aspartic acid residues, and the PAZ domain has an SH3-like barrel fold involved in nucleotide binding23–25.
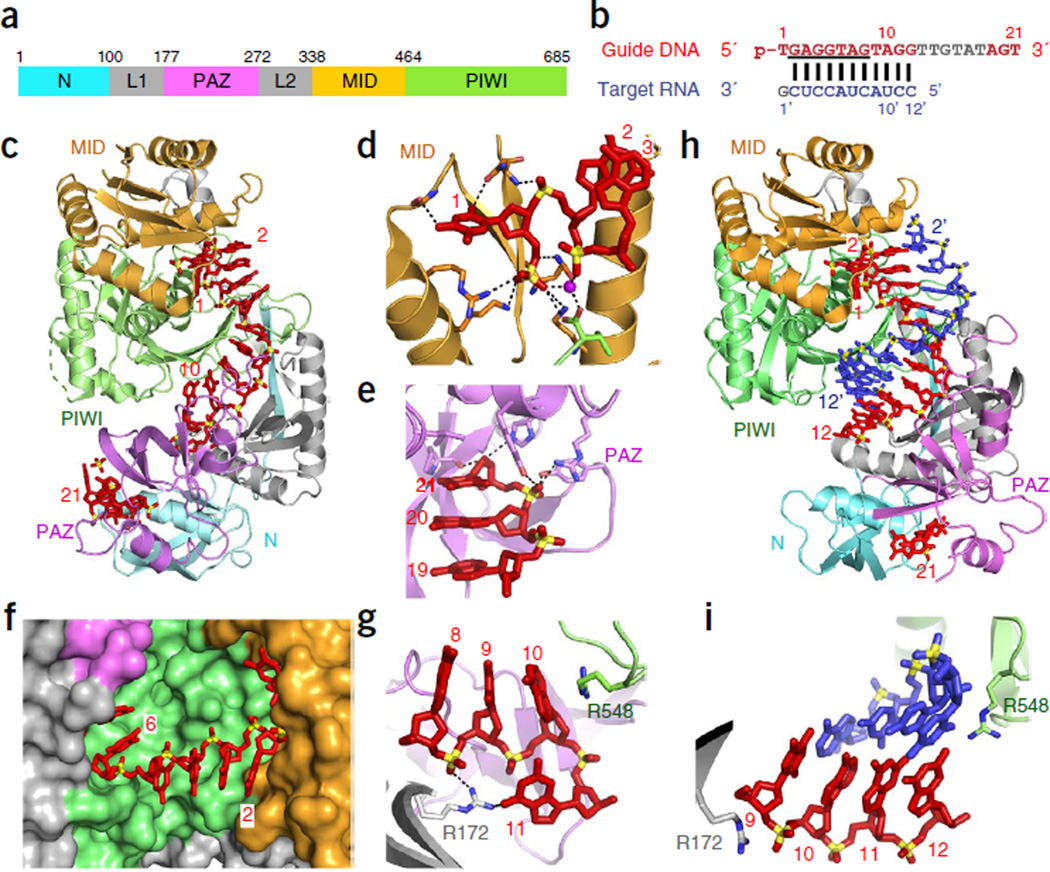
Binary structure of pAgo bound to guide strand. Initial attempts to produce complexes of long pAgos with 5′-phosphorylated guide RNAs were not successful. It was later found that several pAgos bind DNA guides with affinities two orders of magnitude higher than RNA guides5,6. Crystals of Thermus thermophilus pAgo (TtAgo; Fig. 2a) with a bound DNA guide (Fig. 2b) were eventually obtained at elevated temperatures (35–40 °C)7.
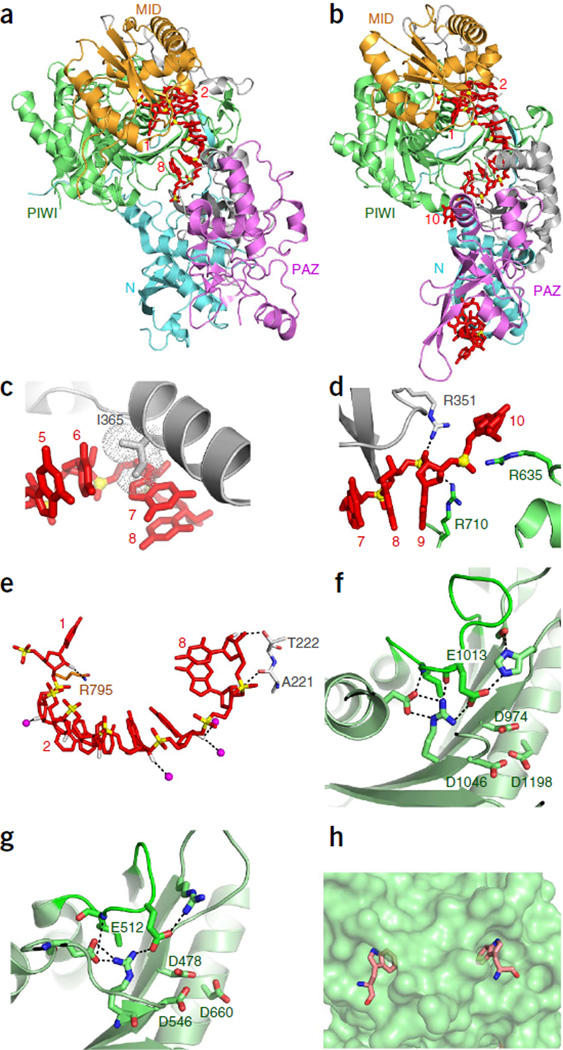
Figure 2.
TtAgo with 21-mer guide DNA (binary complex) and with complementary 12-mer target RNA (ternary complex) adopt cleavage-incompatible conformations. (a) Domain architecture of TtAgo. (b) Sequence of 5′-phosphorylated guide DNA (red, with disordered segment in gray and seed segment underlined) and complementary 12-mer target RNA (blue). (c–g) 3.0 Å structure of the binary complex of TtAgo bound to 5′-phosphorylated 21-mer guide DNA (PDB 3DLH). TtAgo in ribbon representation, domains colored as in a; guide DNA in red, in stick representation. (c) Overall view. (d) Insertion of the 5′-phosphate of the guide DNA into the MID pocket. (e) Insertion of the 2-nt 3′-end of the guide DNA into the PAZ pocket. (f) Outward directionality of bases 2–6 of the guide DNA in the binary complex of TtAgo with guide DNA, thereby aligning their Watson-Crick edges for pairing with target nucleic acids. (g) Bases 10 and 11 of the guide DNA are splayed apart as a result of insertion of an arginine side chain. (h,i) 2.6 Å structure of the ternary complex of TtAgo(D546N) bound to 5′-phosphorylated 21-mer guide DNA and complementary 12-mer target RNA (PDB 3HO1). Guide DNA (red) and target RNA (blue) are in stick representation. (h) Overall view. (i) Bases 10 and 11 of the guide DNA are stacked.
The 3.0 Å structure of TtAgo bound to a 5′-phosphorylated 21-mer guide DNA (Fig. 2c)7 showed that the guide strand contacts all domains of TtAgo, with the majority of the contacts involving interactions with the sugar-phosphate backbone of the guide DNA. The 5′-phosphorylated end was inserted into the nucleotide binding pocket in the MID domain (Fig. 2d), whereas the 3′ end of the guide was anchored in the PAZ domain (Fig. 2e), in agreement with previous structural reports on DNA complexes with the PIWI lobe5,19 or the PAZ domain26,27. Insertion of the 5′-phosphorylated end into TtAgo MID domain pocket strongly bends the guide strand, precluding base pairing of nucleotide 1 (Fig. 2d)5,19. Whereas residues lining the 5′ phosphate–binding pocket in the MID domain are critical for cleavage activity, substitution of the residues at the 3′ end–binding PAZ pocket showed little effect7. Guide nucleotides 2–10 in a helical arrangement (Fig. 2c), with bases 2–6 pointing outward and thus available for pairing with the target strand (Fig. 2f). These observations suggest that guide-target pairing could initiate (nucleate) in the ‘seed’ segment (nucleotides 2–8; Box 1), with the preformed helical conformation of the guide strand reducing the entropic penalty for duplex formation. Indeed, a guide DNA strand pairs with its target RNA with much higher affinity (~300-fold increase) when its seed fragment is associated with the A. fulgidus PIWI lobe, compared to protein-free pairing28. This higher affinity could enhance the fidelity of target recognition, as well as promote and stabilize the assembly of the active silencing complex. Notably, guide-target mismatches in the seed can have a pronounced impact on the affinity of guide-target recognition (reviewed in refs. 29,30). There are examples of exceptions in which the seed is not essential for target binding31, although the functional implications of these exceptions are not clear at present. In the binary TtAgo structure, the preordered guide helix is interrupted by two arginine residues that lock bases 10 and 11 into a unique orthogonal arrangement (Fig. 2g), whereas nucleotides 12–17 of the DNA guide are disordered and could not be traced.
Ternary structures with pAgo bound to guide and target strands. Crystal structures of TtAgo bound to a 5′-phosphorylated 21-mer DNA guide and complementary RNA targets of different lengths provided a major step in understanding Ago functionality. In order to prevent target cleavage, either mismatches were introduced in nucleotides 10 and 11 centered on the cleavage site10, or one of the three aspartic acid residues that line the cleavage pocket were substituted11. The ternary complex of TtAgo with a 12-mer target RNA (Fig. 2b) encompassed 11 Watson-Crick base pairs in an A conformation, spanning nucleotides 2–12 and including the seed segment and the cleavage site (Fig. 2h). In the guide strand, both the 5′ phosphate and the 3′ end remained anchored in their respective MID and PAZ pockets; in contrast, the orthogonal arrangement of bases 10 and 11 seen in the binary complex was disrupted in the ternary complex, where they appeared stacked and centered on the cleavage site (Fig. 2i). Pivot-like conformational transitions are observed for the N and PAZ domains from binary to ternary complex formation with the 12-mer RNA target10,11.
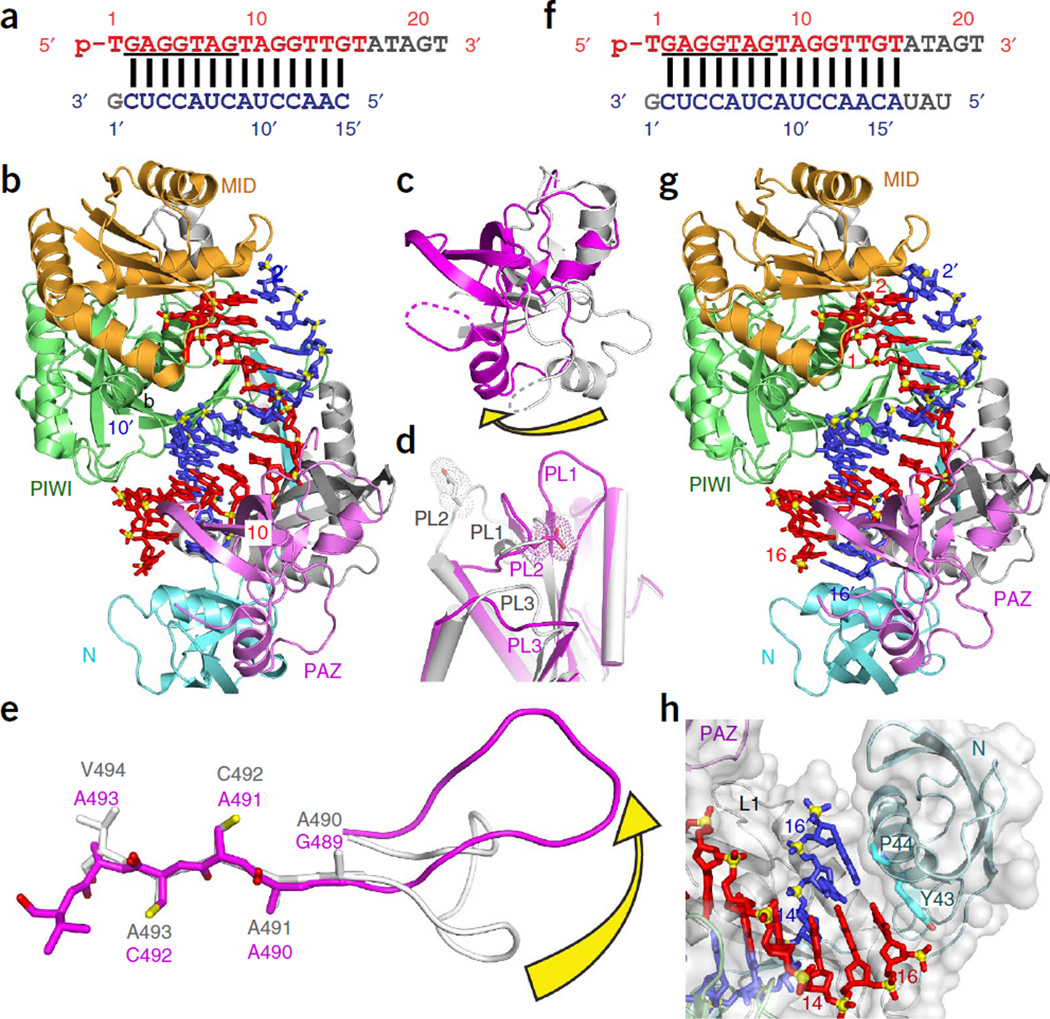
In structures of the ternary complexes of TtAgo with a 15-mer RNA target (3.0 Å resolution; Fig. 3a – e) or with a 19-mer RNA (2.8 Å resolution; Fig. 3f – h), the 5′-phosphate end of the guide remained anchored in the MID pocket, but the 3′ end of the guide was released from the PAZ pocket11. This release was required to overcome torsional constraints that accumulate during the propagation step (Box 1), as longer target segments enter the Ago interior to form an uninterrupted A-form duplex with the guide strand (14 base pairs (bp) with the 15-mer RNA target; 15 bp with the 19-mer RNA target). Release of the 3′ end of the guide is accompanied by rotation of the PAZ domain (Fig. 3c) and transitions within the nucleic acid–binding surface of the PIWI domain, namely movements in loops PL1, PL2 and PL3 (Fig. 3d), and a sliding and flipping of a β-strand (Fig. 3e). The ternary complex of TtAgo with the 19-mer target RNA shows that the N domain blocks guide-target pairing beyond position 16 (Fig. 3h)11. Altogether, the structures of TtAgo ternary complexes with RNA targets illustrate the conformational transitions during the progression from nucleation to propagation steps of guide–target duplex formation.
Figure 3.
TtAgo with 5′-phosphorylated 21-mer guide DNA and complementary 15-mer and 19-mer target RNAs (ternary complex) adopt cleavage-compatible conformations. (a) Sequences of 5′-phosphorylated guide DNA (red, with disordered segment in gray and seed segment underlined) and complementary 15-mer target RNA (blue). (b) 3.0 Å ternary complex of TtAgo(D546E) bound to guide DNA and 15-mer target RNA (PDB 3HJF). The guide DNA and target RNA are in a stick representation, with same colors as in a. (c–e) Conformational changes in TtAgo during its transition from ternary complex with 12-mer target RNA in a cleavage-incompatible conformation (silver; PDB 3HO1) to ternary complex with 15-mer target RNA in a cleavage-compatible conformation (magenta; PDB 3HJF). (c) Rotation of the PAZ domain. (d) Transitions in loops PL1, PL2 and PL3. (e) Rearrangement of the β-strand (Gly489 to Val494) of TtAgo by one residue and conformational transition in adjacent loop PL1. (f) Sequences of 5′-phosphorylated guide DNA (red, with disordered segment in gray and seed segment underlined) and complementary 19-mer target RNA (blue). (g) 2.8 Å ternary complex of TtAgo (D478A mutant) bound to guide DNA and 19-mer target RNA (PDB 3HK2). (h) The N domain blocks base pairing of the guide and the 19-mer target RNA beyond position 16 of the target strand.
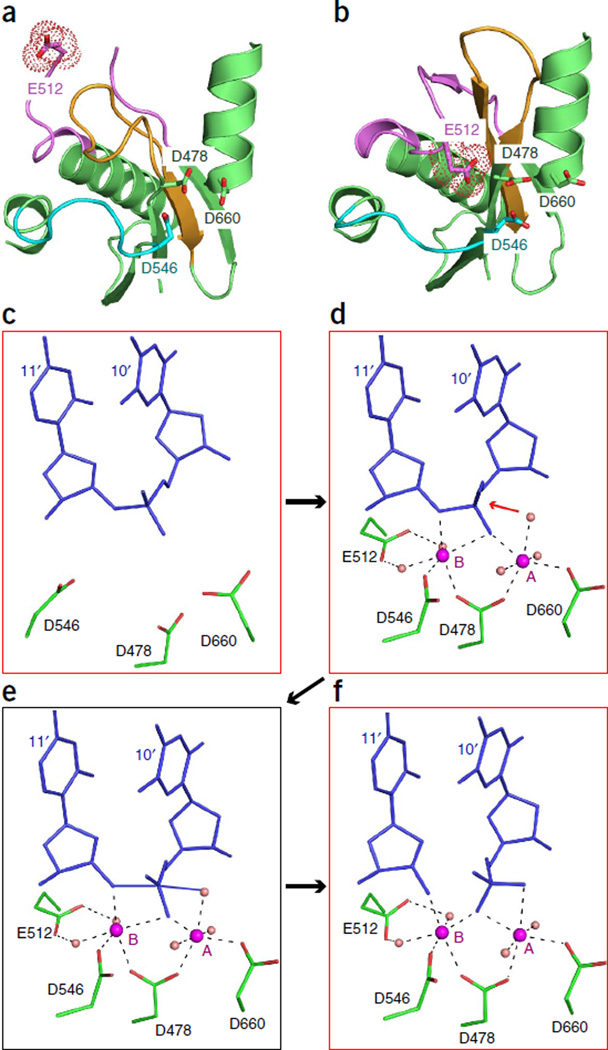
Structures of ternary complexes of DNA-guided TtAgo with target DNAs have been solved to a substantially higher resolution (2.2 Å)12 than those with RNA targets. A glutamic acid residue on loop PL2 (termed the ‘glutamate finger’15) is directed away from the catalytic pocket in the cleavage-incompatible conformation (i.e., in the guide-free protein, in the binary complex and in the ternary complex with 12-nucleotide targets; Fig. 4a). However, in the cleavage-compatible conformation (i.e., ternary complexes with targets at least 15-mer RNA or 16-mer DNA; Fig. 4b), this glutamic acid is inserted into the catalytic pocket and completes the catalytic tetrad12. The two-state model of Argonaute action has been confirmed by single-molecule fluorescence resonance energy transfer studies with Methanocaldococcus jannaschii pAgo (MjAgo)32. A pair of Mg2+ cations facilitate RNA hydrolysis in RNase H family nucleases33,34, but no metals were detectable in the cleavage-incompatible forms of TtAgo (Fig. 4c). In contrast, two Mg2+ cations were identified in the cleavage-compatible conformations in the complex with the 16-mer DNA target (Fig. 4d), where they bridged the catalytic aspartic acid residues and the cleavage site on the target strand, i.e., the backbone phosphate between nucleotides 10 and 11 of the target strand, where cleavage takes place. In addition, a water molecule was observed at a position allowing for an in-line attack on the cleavable phosphate group (Fig. 4d)12, and the catalytic glutamic acid is coordinated to the Mg2+ cation B through two bridging water molecules12; in contrast, in RNase H, the glutamic acid directly interacts with the divalent cation34. In TtAgo, insertion of the glutamic acid in the catalytic pocket results in cleavage of the target strand between the nucleotides that base pair with guide nucleotides 10 and 11 (ref. 12). These structural snapshots of ternary TtAgo complexes provide a model for Mg2+ cation– coordinated cleavage of the target strand (Fig. 4c – f).
Figure 4.
Structure-based insights into the cleavage mechanism of TtAgo. (a,b) Positioning of Glu512 (surface shown in a dotted representation) of TtAgo in the ternary complexes with 5′-phosphorylated 21-mer guide DNA and complementary 12-mer target DNA (a; Glu512 outside and directed away from the catalytic pocket, representative of a cleavage-incompatible conformation; PDB 4N47) and 19-mer target DNA (b; Glu512 inserted into the catalytic pocket, representative of a cleavage-compatible conformation; PDB 4NCB). (c–f), Proposed mechanism for Ago-mediated Mg2+-coordinated cleavage of target strand at the 10′–11′ step in the ternary complex of TtAgo with complementary DNA guide and DNA target strands. Crystal structure snapshots show cleavage-incompatible (c; PDB 4N47), cleavage-compatible (d; PDB 4NCB) and post-cleavage (f; PDB 4N76) states, along with with a proposed model of the transition state (e).
Binary structures of eAgos bound to guide strands. Sustained efforts have resulted in the successive crystallization and structural determination of budding yeast Kluyveromyces polysporus Ago (KpAgo, 3.2 Å; Fig. 5a)15, human AGO2 (hAGO2, 2.2 Å; Fig. 5b)13,14 and human AGO1 (hAGO1, 1.75–2.3 Å)35,36, all with fortuitously loaded heterogeneous RNA guides. hAGO1 and hAGO2 were also captured as binary complexes by replacing the co-purified RNA with a defined RNA guide: hAGO1 with let-7 at 2.1 Å, and hAGO2 with miR-20a at 2.2 Å13,35. Although the eAgo structures are currently restricted to binary complexes, biochemical studies have demonstrated the capacity of KpAgo to load RNA duplexes, which is followed by cleavage of the passenger strand, and eventually annealing and slicing of a complementary target strand15.
Figure 5.
Structures of binary complexes of KpAgo and hAGO2 bound to 5′-phosphorylated guide RNAs. (a) 3.2 Å structure of KpAgo (ribbon representation) with fortuitously loaded 5′-phosphorylated guide RNA (red, stick representation; PDB 4F1N). (b) 2.3 Å structure of hAGO2 (ribbon representation) with fortuitously loaded 5′-phosphorylated guide RNA (red, stick representation; PDB 4EI1). (c–e) Details of the 2.2 Å structure of hAGO2 bound to a defined, miR-20a 5′-phosphorylated guide RNA (PDB 4F3T). (c) Insertion of Ile365 (dotted circle), projecting from α-helix 7 of hAGO2, between bases 6 and 7 of the RNA guide strand. (d) Splaying apart of bases 9 and 10 of the guide RNA by insertion of Arg710 side chain. (e) Intermolecular contacts between 2′-OH groups of guide RNA and amino acid backbone and side chains of hAGO2; both direct and water-mediated (pink spheres) intermolecular hydrogen bonds are shown. (f,g) Intermolecular hydrogen bonding interactions stabilizing the conformation of the expanded and repositioned loop PL2 that inserts the glutamic acid finger into the catalytic pocket in the structure of the KpAgo binary complex with a fortuitously loaded 5′-phosphorylated guide RNA (f, PDB 4F1N) and in the structure of the TtAgo ternary complex with 5′-phosphorylated guide DNA and 19-mer target RNA (g, PDB 3HVR). (h) A pair of tryptophan-binding pockets on the surface of hAGO2 in its binary complex with a fortuitously loaded 5′-phosphorylated guide RNA (PDB 4EI3).
In these eAgo binary complex structures, both the bases and the phosphate backbone spanning the seed segment could readily be traced, even with bound heterogeneous RNA. Similar to DNA guides in TtAgo, the seed segments of eAgo-bound RNA guides adopt an A-like conformation, which in eAgos is facilitated by hydrogen bond interactions involving the 2′-OH and phosphate groups of the RNA guide to Ago. In all studied eAgo complexes, there is a kink between nucleotides 6 and 7 of the RNA guide, caused by the insertion of the side chain of an isoleucine residue (Fig. 5c). To allow guide pairing with RNA targets, this isoleucine side chain has to be displaced during ternary complex formation. Isoleucine or other hydrophobic residues are often found at this position, but they are not strictly conserved. The bases spanning the seed segment are stacked with an unusual tilting in the binary eAgo complexes13–15, requiring transition to a nontilted A-like helical state to facilitate pairing with the target strand in the ternary complex. Akin to the arginine-mediated perturbation of nucleotides 10 and 11 in TtAgo with a bound DNA guide7, base stacking at nucleotides 9 and 10 is perturbed in the complex of hAGO2 with RNA guide, with the kinked alignment stabilized by three arginine side chains (Fig. 5d)13. Yet another similarity with the binary TtAgo structures concerns the disordered middle part (nucleotides 11–19) of the guides in the eAgo binary complexes, whereas their 3′ ends are bound by the PAZ domain13,14.
Differences between eAgo and pAgo complexes. Despite low sequence similarity between pAgos and eAgos (12% identity between various pAgos and hAGO2), their structural and functional features are remarkably similar (Box 2). Nevertheless, there are also notable structural differences that seem to correlate with distinct functionalities37. Whereas all characterized eAgos and some pAgos use RNA guides8, other pAgos use DNA guides5,6,9,32. The only chemical difference between RNA and DNA nucleotides is that at the 2′ position of the sugar ring, RNA has an OH group, whereas DNA has an H group. The eAgo PAZ domain does not bind the 2′-OH groups in the 3′ end of the RNA guide26,27, but some of the 2′-OH groups spanning the seed segment are specifically bound (either directly or via water-mediated hydrogen bonds) to the MID, L1 and PIWI domains (Fig. 5e)13–15,35,36. This indicates that the preference for an RNA guide is determined at the structural level, although those 2′-OH–binding residues are conserved only in a narrow group of fungal and metazoan eAgos. In addition, the 5′-phosphate binding pocket of the TtAgo MID domain is more hydrophobic than that of hAGO2, which might explain preference of TtAgo for DNA guides14. In the 5′ end–binding pocket of pAgos, the negative charge of two phosphates of the guide (nucleotides 1 and 3) and of the C-terminal carboxyl group of pAgo (which is inserted into the MID domain binding pocket) are neutralized by a bound divalent cation19 (reviewed in ref. 22). In contrast, fungal and metazoan eAgos use the ammonium group of a conserved lysine to neutralize this charge37.
In the KpAgo and hAGO2 binary complexes, the glutamate finger is inserted into the catalytic pocket, even in the absence of the target strand13–15. This is in contrast with pAgos, in which insertion of the glutamate finger to complete the catalytic tetrad follows extended guide-target base pairing, leading to the cleavage-compatible state. Notably, a hydrogen bond network in eAgo stabilizes the expanded and repositioned glutamate-containing loop in the activated state (Fig. 5f), with the same alignment in cleavage-compatible pAgo (Fig. 5g), thereby facilitating insertion of the glutamate finger into the binding pocket15. Further experimentation is required to define the constraints controlling cleavage activity of eAgo.
The catalytically active pAgos appear to function as stand-alone proteins, but eAgos interact with a range of proteins in a variety of RNAi pathways (see below). External insertion segments present in eAgos, but not in pAgos, likely provide binding surfaces for RNA-induced silencing complex (RISC) subunits15. KpAgo contains 19 insertion segments, of which 11 are conserved segments (cSs) found in all eAgos and 8 are variable segments (vSs) found only in a subset of eAgos15. At least some of the cSs are essential for silencing38 or appear to differentially affect the activity of eAgos. Although a gap between the two structural lobes is observed in TtAgo, cS1, cS3 and cS10 in KpAgo generate a subdomain that fills this gap15. The presence of this subdomain positions the N domain away from the nucleic acid–binding channel, which allows extensive guide-target pairing and accommodation15. In hAGO1, cS7 forms a surface that could sterically hinder the positioning of a fully paired guide-target RNA duplex in the catalytic site36. In the catalytically activated hAGO1(R805H) variant, activity is further increased upon swapping specific cS7 residues with those of hAGO2 (refs. 35,36). Structures of hAGO1 and hAGO2 have revealed that other cSs in the PIWI domain form two tryptophan-binding pockets, lined by aromatic and hydrophobic side chains (Fig. 5h), which are implicated in binding Gly-Trp (GW) repeats of TNRC6 family proteins (for example, GW182) that promote miRNA-mediated translation regulation (deadenylation) by hAGO1 (refs. 14,36,39). Indeed, GW-rich peptides from GW182 can target these pockets in eAgo40, with the distance between pockets matching the pairwise arrangement of tryptophan residues in GW proteins. Thus, eAgo-specific insertion segments play a role in the binding of interacting proteins and additionally can directly influence eAgo activity.
Evolution and function of Argonaute proteins
The evolutionary journey of the Agos has produced Ago protein families with distinct distribution patterns across the domains of life. Ago is encoded in ~65% of the sequenced eukaryotic genomes, dispersed over at least four of the five eukaryotic supergroups3,41. In contrast, a recent position-specific iterative basic local alignment search tool (PSI-BLAST) search of the RefSeq database (November 2013) using representative PIWI domain sequences as queries shows that Ago proteins are encoded in ~32% and ~9% of the available archaeal and bacterial genomes, respectively, and in 17 of 37 prokaryotic phyla. Similarly to most prokaryotic defense genes42, pAgo shows a patchy distribution, with at most 70% representation in any bacterial or archaeal phylum.
Both eAgos and pAgos belong to the PIWI-protein superfamily, which is defined by the presence of a PIWI domain and in some cases a PAZ domain2 (Fig. 1). The presence of the PIWI lobe in all Ago proteins detected so far implies that it is essential for Ago functionality4,41. We have thus used the sequences of only the MID and PIWI domains to build maximum-likelihood phylogenetic trees using the FastTree program43 (Fig. 6 and Supplementary Data 1–4). We discuss below how this phylogeny can be linked to the structural features that are either conserved or lost in the different families.
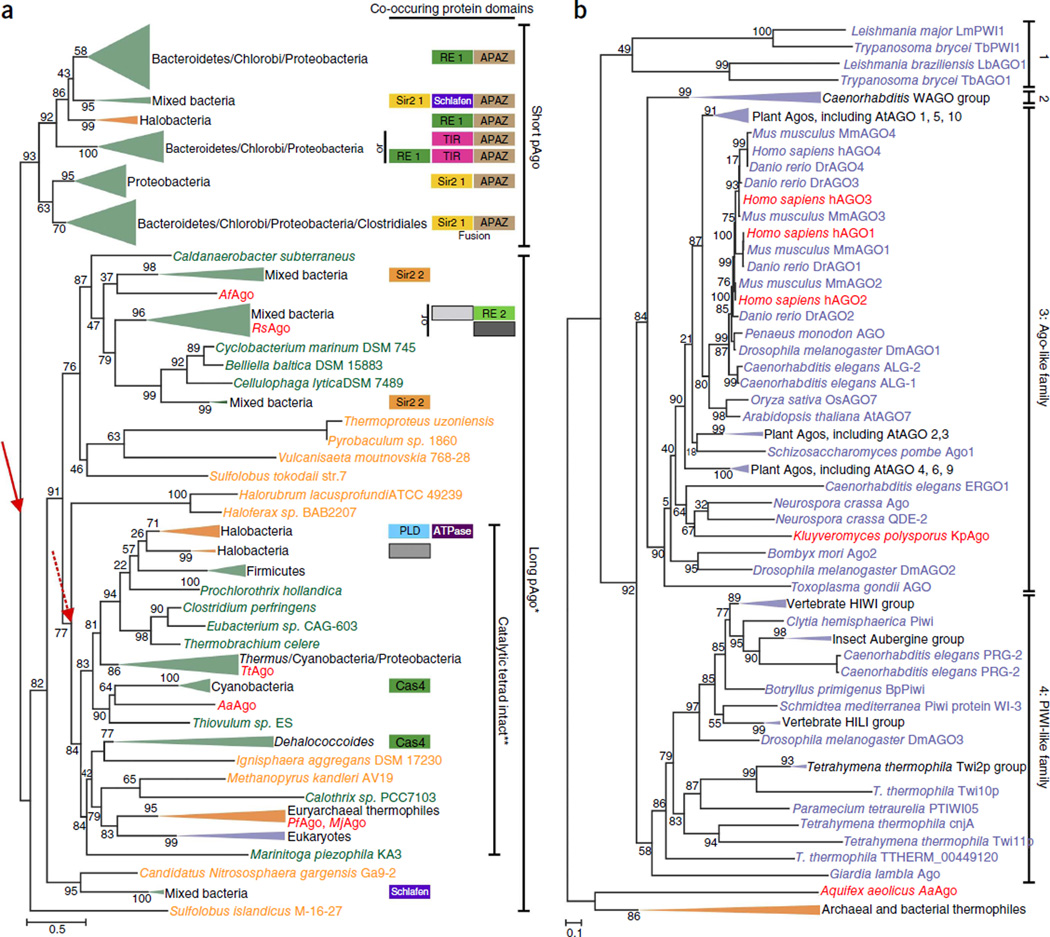
Figure 6.
Phylogenetic trees of Argonaute proteins. (a,b) Maximum-likelihood phylogenetic unrooted trees were built using the FastTree program43 using a multiple alignment of conserved blocks of MID and PIWI domains. The same program was also used to compute the bootstrap values (percentages) that are indicated for all internal branches. Green, Bacteria; orange, Archaea; purple, Eukaryota. Collapsed branches are shown as triangles of the corresponding color. Organisms of which Agos are discussed in this manuscript are colored red. (a) Phylogenetic analysis of pAgos and organization of the predicted operons. We clustered 487 pAgo proteins identified in Refseq by sequence similarity and selected a nonredundant representative set (261 pAgos and 8 selected eAgos). Red arrows indicate two alternative roots of the pAgo tree. *, long pAgo clade contains several short pAgos. **, not all eukaryotic eAgos have an intact catalytic tetrad. Domains associated with pAgos are shown as boxes on the right side of the tree. Homologous domains are shown by boxes of the same color or pattern. Sir2 1 and Sir2 2 are two distinct families of the predicted Sir2-like nuclease; RE1 and RE2 are two distinct families of restriction endonuclease superfamily. TIR, predicted nuclease of TIP family; Schlafen, predicted ATPase; APAZ, ‘analog of PAZ’ domain; Cas4, Cas4 subfamily of restriction endonuclease superfamily; PLD, predicted nuclease of phospholipase D superfamily. Gray boxes indicate distinct families of uncharacterized proteins. Short and long pAgos are not shown but present in all the operons. Slashes denote ‘and’. pAgo sequence alignment and uncollapsed phylogenetic tree are in Supplementary Data 1 and 2, respectively, and are described in Supplementary Note. (b) Phylogenetic analysis of a representative set of 177 eAgos. 1, Trypanosoma Ago family; 2, WAGO family. eAgo sequence alignment and uncollapsed phylogenetic tree are in Supplementary Data 3 and 4, respectively, and are described in Supplementary Note.
Evolution of prokaryotic Argonautes
The topology of the phylogenetic tree of pAgos and most of its sub-trees does not follow the prokaryote phylogeny derived by analysis of ribosomal RNA and other universal genes. This pattern suggests extensive horizontal gene transfer of pAgo-encoding genes, similar to the evolution of most prokaryotic defense genes42,44. The topology of the tree is congruent with the domain architectures of pAgo and the organization of the (predicted) operons containing pAgo genes (Fig. 6a, and Supplementary Data 1 and 2). As shown previously4, the tree can be confidently divided into two major branches: the short pAgo branch consists of short pAgos only, and the long pAgo branch contains all long pAgos and some short pAgos (for example, AfAgo). The latter variants are scattered over the long Ago branch, suggestive of multiple, independent truncation events4. Notably, long pAgos from several euryarchaeal species, mostly thermophiles, group with eAgos, supporting previous conclusions on the origin of eAgos from euryarchaeal pAgos3,4.
On the basis of the conservation of the four catalytic residues, only 28% of the long pAgos are predicted to be catalytically active; these predicted active pAgos form a monophyletic group (Fig. 6a), and the encoding genes often co-occur with predicted helicases. Predicted catalytically inactive long pAgo proteins often cluster in predicted operons with genes encoding putative nucleases (Box 3). Assuming that the ancestral pAgo was an active nuclease and that the primary split in the evolution of this family was the separation into short and long forms (Fig. 6a, solid red arrow), Agos were inactivated on multiple independent occasions, which resulted in loss of activity in all short pAgos and several groups of long Agos, including a subset of eAgos. Alternatively, as the root of maximum likelihood method–generated phylogenetic trees cannot be determined, the correct root position might be between the active and inactive forms (Fig. 6a, dotted red arrow); in this scenario, truncation of pAgo to yield the short forms would be a relatively late evolutionary event.
Box 3 Predicted nucleases and helicases associated with pAgos.
Genes encoding long pAgos with incomplete catalytic sites are often clustered in predicted operons with genes encoding putative nucleases of the Sir2 or Mrr families, predicted to be DNA-specifc nucleases, with different catalytic motifs96–98.
Genes encoding long pAgos with intact catalytic sites occasionally cluster with genes encoding Cas4-like or PLD domain nucleases. Cas4 is a clustered regularly interspaced short palindromic repeat (CRISPR)-associated nuclease/helicase, likely involved in the adaptation step of CRISPR-Cas host defense99,100, whereas phospholipase D (PLD) family nucleases are fused to a DNA/RNA helicase domain, a combination also found in bacterial restriction-modifcation systems101. Other predicted long pAgo operons encode Schlafen-like ATPases, which are putative DNA/RNA helicases102.
All genes encoding short pAgos are associated with a gene encoding the uncharacterized APAZ (analog of PAZ) domain (Fig. 1 and Fig. 6a). APAZ lacks detectable sequence similarity with the PAZ domain and has only been detected in the context of short pAgo genes, always fused to a (predicted) nuclease domain that may belong to the Sir2 or Mrr protein families (from different subfamilies than the ones associated with long pAgos) or to TIR domains4. The latter are predicted to possess nuclease activity4,103 and are involved in bacterial virulence104 or in eukaryotic antimicrobial and antiviral response, and in apoptosis105–107. In some prokaryotic genomes, the putative Sir2 nuclease is fused not only to the APAZ domain but also to pAgo itself (Fig. 6a). Less commonly, Sir2-APAZ domains contain an inserted Schlafen-like ATPase domain (Sir2-Schlafen-APAZ; Fig. 6a). Moreover, some short pAgo genes cluster with Mrr-TIR-APAZ gene fusions.
PIWI-RE proteins are fused to an uncharacterized N-terminal domain that does not appear to be related to either PAZ or APAZ41. In many genomes, genes encoding PIWI-RE are clustered with two genes, encoding a DinG-like helicase and a predicted restriction endonuclease41. Given that DinG family helicases specifcally act on R-loops108, the PIWI-RE proteins have been hypothesized to function as part of an RNA-guided restriction system41
Approximately 60% of the identified pAgos lack the PAZ lobe, and most of these short pAgos have incomplete catalytic tetrads. All genes in the short pAgo branch are associated with a gene encoding the uncharacterized APAZ (Analog of PAZ) domain (Fig. 1 and Fig. 6a)4. The APAZ domain does not have detectable sequence similarity with the PAZ domain and has not been detected in any context other than the short pAgo neighborhood. The N terminus of the APAZ domain is always fused to a (predicted) nuclease domain (Box 3)4.
A highly diverged family of short pAgo derivatives, designated PIWIRE for characteristic conserved arginine (R) and glutamic acid (E) residues41 (Fig. 1), appears in a few major bacterial lineages. Notably, the set of genomes encoding PIWI-RE or pAgo show almost no overlap41. Similar to short pAgos, most PIWI-RE proteins feature a seemingly inactive PIWI lobe. PIWI-RE proteins are fused to an uncharacterized N-terminal domain that does not appear to be related to PAZ or APAZ41. In many genomes, PIWI-RE-encoding genes cluster with DinG-like helicases and predicted nucleases (Box 3), and thus the PIWI-RE proteins have been hypothesized to be part of an RNA-guided restriction system41.
Function of prokaryotic Argonautes
The ability to cleave target nucleic acids in vitro has been investigated for four long pAgos from different branches in the Argonaute tree, namely TtAgo, AaAgo, MjAgo and Rhodobacter sphaeroides pAgo (RsAgo). TtAgo utilizes DNA guides to cleave single-stranded (ss)RNA, ssDNA and/or double-stranded (ds)DNA plasmid targets, the latter by independently nicking the two strands9. AaAgo utilizes ssDNA guides to cleave ssRNA strands, but its ability to cleave DNA has not been determined6,17. MjAgo utilizes ssDNA guides to cleave ssDNA strands but cannot cleave RNA targets32. No catalytic activity has been observed for RsAgo8, but it co-occurs with a predicted nuclease in R. sphaeroides.
Although the physiological functions of AaAgo and MjAgo have not yet been determined, both TtAgo and RsAgo play a demonstrated role in host defense8,9. TtAgo lowers plasmid transformation efficiency and intracellular plasmid concentrations in T. thermophilus9. Notably, RsAgo lowers intracellular plasmid concentrations in Escherichia coli but not in R. sphaeroides; however, it does interfere with plasmid-encoded RNA in R. sphaeroides8. As short DNA molecules complementary to the RNA guides associate with RsAgo in vivo, a yet-to-be-identified nuclease has been proposed to process DNA bound by RsAgo-RNA complexes8. TtAgo and RsAgo both acquire functional guides when expressed in E. coli8,9, which suggests that guide processing is performed either by pAgo itself or by common host factors. TtAgo utilizes 13–25-nt small interfering DNA (siDNA) guides and appears to depend on its own catalytic site for guide loading9, whereas catalytically inactive RsAgo acquires 15–19-nt RNA guides proposed to originate from degraded mRNAs8. Most guides acquired by TtAgo and RsAgo are complementary to foreign DNA, such as plasmids or insertion elements8,9.
The frequent association of homologous (predicted) nucleases with catalytically inactive long or short pAgos (Box 3) suggests a modular organization of pAgo-centered defense systems, with occasional recombination between loci encoding different variants of these systems (Fig. 6a). In some of these pathways, the long pAgo is predicted to possess both target recognition and nuclease activities. In other cases, catalytically inactive long or short pAgo might be responsible only for target recognition (using at least their MID and PIWI domains), whereas cleavage would be performed by other nucleases encoded in the same operons, which possibly physically interact with pAgo. The presence of additional non-nuclease genes near some genes encoding pAgos (Box 3) indicates the requirement for additional activities in those systems. Given that TtAgo requires unwinding of dsDNA targets for subsequent cleavage of each strand9, pAgo-associated helicases could play a role in enhancing the accessibility of dsDNA targets for pAgo-mediated cleavage.
Evolution and function of eukaryotic Argonautes
We also reconstructed a phylogenetic tree using a representative set of eAgos with pAgo sequences as an outgroup (Fig. 6b, and Supplementary Data 3 and 4). In agreement with previous analyses45,46, eAgos can be divided into four major families: the Trypanosoma Ago family46, typified by Trypanosoma brucei; the WAGO family, typified by worm (Caenorhabditis elegans)-specific Agos; the Ago-like family, typified by Arabidopsis thaliana AGO1; and the PIWI family, typified by Drosophila melanogaster PIWI. The Ago-like and PIWI families are represented in several major groups of eukaryotes, indicating that at least one duplication of eAgo apparently antedated the last common ancestor of the extant eukaryotes. The other two families could have emerged as a result of additional, lineage-specific duplications. Another protein family belonging to the PIWI-protein superfamily was recently identified in eukaryotes41; these proteins have only the MID domain and an inactive PIWI domain, and are typified by Med13, a subunit of the transcription regulatory Mediator complex in mammals47.
The phylogenetic tree of eAgos generally follows the phylogeny of eukaryotes and, given the rarity of horizontal gene transfer in the evolution of eukaryotes, it appears that eAgos evolved solely by vertical inheritance. Thus, it has been inferred that a functional RNAi pathway, consisting of eAgo, Dicer and RNA-dependent RNA polymerase (RdRP), was present in the last eukaryotic common ancestor, where it most likely functioned in defense against viruses and transposons3. Dicer consists of RNase III, PAZ and DExD/H helicase domains, all with identifiable ancestors in prokaryotes, whereas RdRP apparently evolved from a group of so-far uncharacterized, predicted DNA-dependent RNA polymerases from bacteriophages3.
All eAgos function in larger protein networks that vary substantially between and within the different families, and eAgos have evolved into distinct players in these different networks. This diversification is the result of many sequence adaptations, which allow interactions with a multitude of proteins involved in either guide processing, guide loading, regulating eAgo activity or recruitment of additional proteins.
Trypanosoma Ago family. This eAgo family is mainly studied in T. brucei, in which long dsRNA guide precursors are expressed both from retrotransposons48 and chromosomal 147-bp tandem units49. These transcripts are processed either by a cytoplasmic Dicer (TbDCL1; ref. 50), which depends on TbRIF5 for activity51, or by a nuclear Dicer (TbDCL2; ref. 52). The exonuclease TbRIF4 is essential in converting the duplex siRNAs to ssRNA guides51. An N-terminal RGG domain allows TbAGO1-guide complexes to associate with polyribosomes, which results in efficient cleavage of retrotransposon transcripts53. Thus, like the prokaryotic RsAgo, Trypanosoma family Agos interfere with transposon activity.
WAGO family. The eAgos of this nematode-specific family generally act as so-called secondary Argonaute proteins, i.e., they are loaded with guide RNAs in response to the activity of the primary Ago protein54. In C. elegans, a primary Ago protein (for example, RDE-1 or PRG-1) is believed to recruit an RdRP to the targeted mRNA, which results in the synthesis of new guide RNAs, known as 22G RNAs (22-nt guides with 5′-ppp-G), that are used by WAGO proteins. As direct products from RdRP activity, 22G RNAs carry 5′-triphosphate (GTP) groups55,56, and it remains unclear how the WAGO proteins can accommodate this atypical guide RNA feature. The WAGO proteins execute a variety of silencing mechanisms, from target RNA destabilization54 to transcriptional silencing57. Absence of secondary Agos can be enough to desilence target expression54,57, which suggests that the action of the primary Ago is not sufficient for silencing. A notable case has been reported in which a WAGO protein seems to protect against silencing activities executed by other WAGO proteins58,59. Hence, apart from adapting to various mechanisms of guide RNA acquisition and target silencing, eAgos seem to play a role in counteracting or fine-tuning silencing.
Ago-like family. Guide RNAs are typically processed and loaded into Ago-like proteins by proteins such as Dicer (reviewed in refs. 60,61). In some cases (such as vertebrate AGO2 proteins), Ago-like proteins themselves perform secondary processing of preprocessed RNA hairpin structures, through their endonucleolytic activity62,63. Many Ago-like proteins use endogenous guide RNAs, known as microRNAs (miRNAs), to regulate gene expression, mainly by affecting mRNA translation elongation, acting as a road block for the ribosome, or by affecting polyadenylation of the mRNA by extensive interactions with 3′ untranslated region–processing machineries (reviewed in refs. 64,65). In these cases, the guide-target interactions are often characterized by limited, imperfect base-pairing that is incompatible with target RNA cleavage29. Thus, many eAgos act purely as sequence-specific RNA-binding proteins, whose sole function is to counteract the translation of specific mRNAs.
Once loaded with a guide, many Ago-like proteins function without involvement of other proteins. Based on the conservation of the four active site residues, ~90% of eAgos are predicted to be catalytically active. However, it should be noted that not all Agos with complete catalytic tetrads are catalytically active, as hAGO3 harbors all four residues but cannot cleave targets in vitro35,66. Catalytically inactive hAGO1 can be activated by minimal changes in the active site, with the activity further enhanced by mutations in either the N domain35,66 or cS7 (refs. 35,36). These findings are compatible with a scenario in which an ancestral eukaryote inherited an active long pAgo, whose catalytic function was subsequently lost in a subset of eAgos. In plants and in some animals, Ago-like proteins use target RNA cleavage as a gene-regulatory mechanism65 and have been shown to interfere with dsRNA viruses67–70; the latter role is reminiscent of the host-defense functions of pAgos. However, in contrast to pAgos, eAgo-like proteins depend on other proteins, such as Dicer, to process guides from the viral dsRNA genome. Even when confronted with similar guide-target RNA interactions, the kinetics of binding and releasing target RNA can vary widely between different eAgos71, which indicates that they have not only evolved to bind different protein-partners but also adapted biochemically to execute distinct functions.
PIWI family. The ancestral function of target cleavage is strongly conserved among the PIWI-like proteins. Many PIWI family members use their guide RNAs, known as piRNAs, to control the activity of transposable elements within the germ cells of animals72. In contrast to the Ago-like proteins, animal PIWI-like proteins are loaded through a pathway that includes ssRNA precursors (reviewed in ref 73). This process requires many different protein-protein and protein-RNA interactions, and takes place in extremely protein- and RNA-rich assemblies that flank nuclear pores. In some cases, this process involves a nuclease from the PLD family74 (Box 3); in others, a member of the PIWI-like family itself catalyzes precursor processing. These endonu-cleases generate 5′-phosphorylated RNA fragments that are bound by a PIWI-like protein. However, not all PIWI-like proteins employ such mechanisms: for example, the PIWI-like proteins in ciliates, which are involved in sequence-specific genome rearrangements, are loaded through Dicer-dependent pathways75,76. These variations illustrate the high flexibility in molecular mechanisms coupled to eAgos.
Some, but not all of the PIWI-like proteins display a strong preference for a uracil at the 5′ end of the loaded RNA, likely reflecting the presence of a nucleotide-specificity loop21,77, as described for some plant Ago proteins78. After loading of this piRNA intermediate, the 3′ end of the loaded RNA is likely trimmed by an exonuclease79 and then 2′-O-methylated80. Crystal structures of the PAZ domain of PIWI-like proteins have revealed the basis of preference for RNA guides with a 2′-O-methylation at their 3′ ends over those with unmodified 2′-OH groups81,82. The 2′-O-methyl modification has also been demonstrated in guide RNAs of some members of the Trypanosoma Ago and Ago-like families, including TbAGO1, DmAGO2 and all the Agos in plants83–85. A common property of these eAgos is that their guides extensively pair with their target RNAs, resulting in release of the 3′ end of the guide RNA from the PAZ domain and potentially rendering the guide RNA exposed to 3′ end–modifying activities. Indeed, in the absence of 2′-O-methylation, target recognition by these Ago-like proteins results in exonuclease trimming, adenylation and uridylation of the guide RNA86, which could all affect guide RNA stabilities87.
Concluding remarks
Comparison of available pAgo and eAgo structures reveals that the domain architecture and the functions of individual domains are conserved throughout the three domains of life. The MID and PIWI domains are responsible for binding and helical preordering of the RNA or DNA guide. Short pAgos, with only these two domains, most likely function as guide-mediated target binders and depend on associated nucleases (and possibly helicases) for target cleavage and/or unwinding. Long pAgos and eAgos feature additional PAZ domain, which binds the 3′ end of the guide, and the N domain, which plays a role in unwinding of the guide–passenger duplex and interferes with guide-target base pairing toward the 3′ end of the guide.
The evolutionary journey of the Agos started in prokaryotes, through a fusion of a PIWI-like RNase H domain with a MID-like nucleic acid–binding domain, yielding the first guide-dependent short pAgo (Fig. 6). RNase H is a nearly ubiquitous nuclease that cleaves the RNA strand of a DNA-RNA duplex during replication in all domains of life. After the RNase H–MID fusion to generate a short Ago protein, there were additional associations with distinct interaction or enzymatic domains, often as N terminally–fused extensions, such as N-PAZ in long pAgos, nuclease-APAZ in short pAgos, or the unique N-terminal domain in PIWI-RE (Fig. 1). In different pAgo clades, these associations engendered multiple, independent variations, which resulted in active and inactive variants with different guide and target specificities. So far, two mechanistic pAgo functions have been characterized experimentally: DNA-guided DNA interference by TtAgo and MjAgo and RNA-guided DNA interference by RsAgo, an inactive pAgo variant associated with an uncharacterized nuclease. The TtAgo protein binds both DNA-RNA and DNA-DNA guide-target duplexes in an A-form helix, which is unusual for DNA duplexes. Notably, RNase H cleaves DNA-RNA helices, which also adopt the A conformation, suggesting that TtAgo retained the ancestral preference for an A-form helix in the course of evolution. The guide and target specificity of Argonaute variants cannot be currently predicted from their amino acid sequence. Most of the prokaryotic MID-PIWI– containing systems likely function in defense against invading DNA, whereby target cleavage is performed either by their PIWI domain or by co-occurring nucleases (Box 3). Given the variation of genes that cluster with pAgo, the functions of pAgos and partner proteins might extend beyond host-defense to various regulatory pathways.
A major step in Argonaute evolution appears to have been the transition from stand-alone proteins to multiprotein regulatory systems. Phylogenetic analyses indicate that the last eukaryotic common ancestor possessed not only an RNA-guided RNA-interfering Ago but also all other components essential for RNAi3. In the course of evolution, eAgos maintained the four domains and their respective functions (although some lost catalytic activity) but additionally acquired insertion segments that allowed optimization of specific protein-protein interactions, while maintaining the basic molecular mechanism of action. Thus, various eAgos evolved to interact with pathway-specific proteins, resulting in a variety of RNAi pathways involved in a wide range of cellular processes. The functions of many insertion segments are not yet known, and both structural and biochemical research is required to reveal their roles. Elucidation of these missing links will contribute to our growing understanding of the evolution, mechanism and physiology of Agos, and of the diverse defense and regulatory systems of prokaryotes and eukaryotes in which these proteins play crucial roles.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by grants from the Netherlands Organization of Scientific Research (NWO) to J.v.d.O. (NWO-TOP, 845.10.003) and the US National Institutes of Health to D.J.P. (TR01 GM104962). K.M. and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine). K.N. is supported by Precursory Research for Embryonic Science and Technology (PRESTO) from the Japan Science and Technology (JST) Agency.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bohmert K, et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 3.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, Wolf YI, van der Oost J, Koonin EV. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma JB, et al. Structural basis for 5′-end-specifc recognition of guide RNA by the A-fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specifc DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YL, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swarts DC, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YL, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YL, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng G, et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkayam E, et al. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 17.Rashid UJ, et al. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007;282:13824–13832. doi: 10.1074/jbc.M608619200. [DOI] [PubMed] [Google Scholar]
- 18.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boland A, Tritschler F, Heimstadt S, Izaurralde E, Weichenrieder O. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010;11:522–527. doi: 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 22.Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 24.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 25.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 26.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 28.Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunne T, Swarts DC, Brouns SJ. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014;22:74–83. doi: 10.1016/j.tim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Lal A, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zander A, Holzmeister P, Klose D, Tinnefeld P, Grohmann D. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014;11:45–56. doi: 10.4161/rna.27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Reports. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakanishi K, et al. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Reports. 2013;3:1893–1900. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn CD, Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013;38:263–271. doi: 10.1016/j.tibs.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. USA. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huntzinger E, et al. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaff J, et al. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. USA. 2013;110:E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burroughs AM, Iyer LM, Aravind L. Two novel PIWI families: roles in inter-genomic conflicts in bacteria and Mediator-dependent modulation of transcription in eukaryotes. Biol. Direct. 2013;8:13. doi: 10.1186/1745-6150-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia Silva MR, et al. Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene. 2010;466:26–35. doi: 10.1016/j.gene.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 49.Tschudi C, Shi H, Franklin JB, Ullu E. Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei. BMC Genomics. 2012;13:427. doi: 10.1186/1471-2164-13-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnes RL, Shi H, Kolev NG, Tschudi C, Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathog. 2012;8:e1002678. doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patrick KL, et al. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi H, Chamond N, Djikeng A, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: role of the n-terminal RGG domain and the polyribosome association of argonaute. J. Biol. Chem. 2009;284:36511–36520. doi: 10.1074/jbc.M109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yigit E, et al. Analysis of the C elegans. Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 55.Pak J, Fire A. Distinct populations of primary, secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 56.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 57.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seth M, et al. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Ender C, Meister G. Argonaute proteins at a glance. J. Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 61.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 62.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 65.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hauptmann J, et al. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 2013;20:814–817. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 67.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 68.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ketting RF. The many faces of RNAi. Dev. Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Nishimasu H, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 75.Mochizuki K, Gorovsky MAA. Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandoval PY, Swart EC, Arambasic M, Nowacki M. Functional diversification of Dicer-like proteins and small RNAs required for genome sculpting. Dev. Cell. 2014;28:174–188. doi: 10.1016/j.devcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 77.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012;31:3588–3595. doi: 10.1038/emboj.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 80.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 81.Simon B, et al. Recognition of 2′-O-methylated 3′-end of piRNA by the PAZ domain of a Piwi protein. Structure. 2011;19:172–180. doi: 10.1016/j.str.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 82.Tian Y, Simanshu DK, Ma JB, Patel DJ. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc. Natl. Acad. Sci. USA. 2011;108:903–910. doi: 10.1073/pnas.1017762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi H, et al. Role of the Trypanosoma brucei HEN1 family methyltransferase in small interfering RNA modification. Eukaryot. Cell. 2014;13:77–86. doi: 10.1128/EC.00233-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 86.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 88.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 89.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 90.Hur JK, Zinchenko MK, Djuranovic S, Green R. Regulation of Argonaute slicer activity by guide RNA 3′ end interactions with the N-terminal lobe. J. Biol. Chem. 2013;288:7829–7840. doi: 10.1074/jbc.M112.441030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 92.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 93.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 94.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 95.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kinch LN, Ginalski K, Rychlewski L, Grishin NV. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005;33:3598–3605. doi: 10.1093/nar/gki676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knizewski L, Kinch LN, Grishin NV, Rychlewski L, Ginalski K. Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches. BMC Struct. Biol. 2007;7:40. doi: 10.1186/1472-6807-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J, Kasciukovic T, White MF. The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012;7:e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lemak S, et al. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 2013;135:17476–17487. doi: 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grazulis S, et al. Structure of the metal-independent restriction enzyme BfI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl. Acad. Sci. USA. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004;16:1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 103.Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rana RR, Zhang M, Spear AM, Atkins HS, Byrne B. Bacterial TIR-containing proteins and host innate immune system evasion. Med. Microbiol. Immunol. (Berl.) 2013;202:1–10. doi: 10.1007/s00430-012-0253-2. [DOI] [PubMed] [Google Scholar]
- 105.Brikos C, O’Neill LA. Signalling of toll-like receptors. Handb. Exp. Pharmacol. 2008;183:21–50. doi: 10.1007/978-3-540-72167-3_2. [DOI] [PubMed] [Google Scholar]
- 106.Palsson-McDermott EM, O’Neill LA. Building an immune system from nine domains. Biochem. Soc. Trans. 2007;35:1437–1444. doi: 10.1042/BST0351437. [DOI] [PubMed] [Google Scholar]
- 107.Burch-Smith TM, Dinesh-Kumar SP. The functions of plant TIR domains. Sci. STKE. 2007;2007:e46. doi: 10.1126/stke.4012007pe46. [DOI] [PubMed] [Google Scholar]
- 108.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.